Introduction
The field of genetics is rapidly evolving, and as our knowledge of the application of genetic mutations to the field of heart disease expands, so does its role in the treatment and counseling of cardiac patients. This is particularly true in the case of cardiomyopathy, and in the tragic circumstance of sudden cardiac death.
In this paper, we report a novel and previously unreported rare variant involving the Desmoplakin (DSP) gene. This variant was isolated in a patient who presented with aborted sudden cardiac death and was subsequently found to have a nonischemic cardiomyopathy.
Case report
The patient is a 54-year-old man who was previously extremely healthy. The patient was an endurance athlete, running 10–12 miles a day with no functional limitations. Unfortunately, during the last week of July 2013, he suffered unheralded syncope and aborted sudden cardiac death while running. Fortunately he had spontaneous return of circulation, and subsequently made a full recovery. He noted in retrospect 2 prior instances where his heart raced suddenly while exercising, associated with lightheadedness, but these were self-limited and were not further investigated.
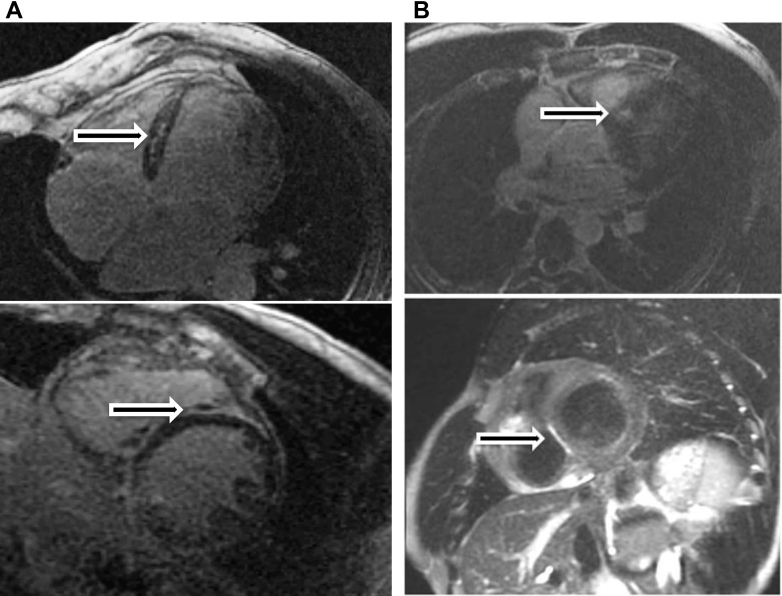
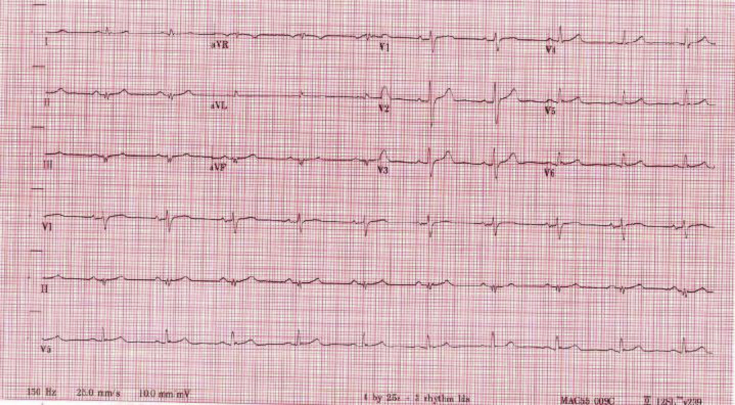
His left ventricular (LV) ejection fraction at that time was approximately 25%, and cardiac magnetic resonance imaging (MRI) demonstrated multiple regions of scar involving the left ventricle, normal right ventricular (RV) size and function, significant LV dilation, and delayed enhancement (Figure 1A). The diagnosis of a nonischemic cardiomyopathy was made after coronary angiography demonstrated no evidence of epicardial coronary artery disease. A family history revealed that his sedentary twin brother was also diagnosed with a nonischemic cardiomyopathy, which was incidentally discovered when sampling a colleague’s imaging machine. An electrocardiogram performed shortly after initial presentation demonstrated low-voltage QRS, left anterior fascicular block, and inferior Q waves (Figure 2). Based upon these events, the patient was then implanted with a dual-chamber implantable cardioverter-defibrillator for the secondary prevention of sudden cardiac death. Heart failure medications were initiated for this ACC/AHA stage B patient, who had NYHA functional class I symptoms, for underlying LV dysfunction. A graded exercise stress test was performed shortly after initial presentation, which demonstrated excellent functional capacity. The patient was able to exercise for 13 minutes and 48 seconds on a full Bruce protocol, achieving 16.7 metabolic equivalents.
Figure 1.
A: Cardiac magnetic resonance imaging (MRI) obtained of the proband patient demonstrates severe left ventricular (LV) global hypokinesis, LV dilation, and midmyocardial delayed enhancement in the basal to apical inferoseptal wall. B: Cardiac MRI obtained of the patient’s son demonstrates predominantly subepicardial delayed enhancement in the basal to midanteroseptal and inferoseptal wall.
Figure 2.
Electrocardiogram demonstrates low-voltage QRS, left anterior fascicular block, and inferior Q waves.
Given the likely familial nature of his cardiomyopathy, a comprehensive cardiomyopathy panel (GeneDx; Gaithersburg, MD) was obtained. Direct sequencing of the coding region of the DSP gene demonstrated the c.3735_3741dupAAATCGA rare variant. This variant causes a shift in reading frame starting at codon Aspartic Acid 1248, changing it to a lysine, and creating a premature stop codon at position 7 of the new reading frame (Asp1248LysfsX7). This may result in loss of protein from this allele through either truncated protein product or nonsense-mediated mRNA decay.1, 2 Other DSP gene variants have been previously implicated as one of the causative mutations in arrhythmogenic RV cardiomyopathy3; however, the c.3735_3741dupAAATCGA mutation has not previously been reported as pathogenic.
Upon identification of a disease-causing mutation, genetic testing was performed on all available relatives and offspring (Figure 3). The results identified the causative mutation to be present in 1 asymptomatic sibling, in the proband’s twin brother with known cardiomyopathy, and in 2 of the patient’s children. One of the patient’s gene mutation–positive children is a current member of the United States Army Special Forces. To assist with risk stratification, the patient’s son underwent a cardiac MRI (Figure 1B). This study showed preserved LV function but evidence of subepicardial delayed gadolinium enhancement.
Figure 3.
Family pedigree. The c.3735_3741dupAAATCGA mutation identified in the presented patient was also detected in 2 of the patient’s siblings and in 2 of the patient’s children. DSP = Desmoplakin; LGE = late gadolinium enhancement; MRI = magnetic resonance imaging.
The proband patient continues to receive close follow-up. To date, he has done well and remains active, with NYHA class I symptoms. Unfortunately, 3 years later, his LV dysfunction persists despite guideline-directed medical therapy, and his most recent LV ejection fraction was estimated at 30%–35% by echocardiogram.
Discussion
The management of patients with a genetic heart disease condition, and their family members, is complex. As our understanding of the genetic contributors to cardiac disease evolves, so does our ability to confirm disease in the proband and to advise family members in regard to their cardiac risk. This is particularly true in the rare but tragic circumstance involving sudden cardiac death.
In an effort to contribute to the evolving field of the genetics of sudden cardiac arrest, we report a novel rare variant in the DSP gene, which was discovered in a patient who was resuscitated following sudden cardiac arrest. The presence of nonischemic cardiomyopathy and similar myocardial scar pattern in family members sharing the same mutation within the DSP gene supports the possible disease-causative nature of this variant.
The DSP gene encodes the protein desmoplakin, which is a critical component of desmosomes, the specialized proteins that form intercellular junctions, thereby imparting mechanical strength from cell to neighboring cell. As a result, a complex latticework is formed by virtue of anchoring intermediate filaments to the cytoplasmic membranes of adjacent cells. Mutations in desmosomal genes may compromise either the intercellular desmosomal adhesion or intermediate filament function.4
Interrupted intercellular adhesion, as a consequence of DSP gene mutations, predisposes to multiple phenotypic expressions (ie, dermatologic, autoimmune, and cardiac disorders). Desmosomal disorders cause cardiac-specific disease as a consequence of myocyte detachment and death,2 and this is the process implicated in the pathogenesis of arrhythmogenic right ventricular cardiomyopathy (ARVC). ARVC-causative mutations have been reported in virtually all desmosomal proteins. These proteins include desmoplakin, plakoglobin, plakophilin-2, and desmoglein-2. The presence of mutations affecting desmosomal proteins in ARVC supports the central role for impaired cell adhesion in the pathogenesis of this disorder.4, 5
It is increasingly becoming recognized that arrhythmogenic cardiomyopathy presents in a spectrum of patterns of myocardial involvement. Classical ARVC presents with predominantly RV involvement in the form of fibro-fatty infiltration, ventricular dilation, ventricular tachyarrhythmia, and RV systolic dysfunction.6, 7 However, it is increasingly evident that in contrast to the classic right predominant variant, arrhythmogenic cardiomyopathy may present with varying degrees of biventricular or predominantly LV involvement (arrhythmogenic LV cardiomyopathy).6, 7 In recognition of the heterogeneous clinical expression, it has been suggested that the term “arrhythmogenic right ventricular cardiomyopathy” be simplified to “arrhythmogenic cardiomyopathy.” This helps to explain the phenotype of the patient presented in this report, who demonstrates a disproportionate degree of LV dilation and dysfunction, as compared to the normal RV.
We acknowledge that despite the evidence presented to support the pathogenic nature of the c.3735_3741dupAAATCGA variant, as identified within the proband and in other family members, proof of causality is not definitive. According to the Exome Aggregation Consortium data set, there are cases that involve phenotypically normal patients who possess protein-altering terminal DSP variants,8 which argues that terminal DSP variants may be tolerated. However, despite this evidence supporting the clinical quiescence of terminal DSP loss-of-function mutations, there are published cases reporting that terminal DSP loss-of-function variants can be pathogenic.9, 10, 11 For example, in a report authored by Uzumcu and colleagues,9 a nonsense mutation located within exon 23 of the DSP gene (R1267X) resulted in both the truncation of the DSP isoform (confirmed by Western blot testing) and an early-onset cardiomyopathy with features similar to Naxos disease. In addition, a genetic variant analysis performed on 40 patients who died owing to sudden unexplained nocturnal death syndrome identified 10 DSP gene missense mutations.10 Of the 10 mutations involving the DSP gene, 6 variants were terminal mutations (3 variants resided within exon 23 and 3 variants resided within exon 24). Furthermore, of the over 60,000 phenotypically normal patients represented within the Exome Aggregation Consortium data set, no patients were found to have the c.3735_3741dupAAATCGA variant. This variant, however, was isolated in a single patient with ARVC who was sequenced by the Oxford Medical Genetics Laboratory.12 These case reports and sequencing database results help provide evidence supporting the rarity and potential pathogenic nature of terminal DSP loss-of-function mutations, similar to the mutation described within the body of our report.
An additional aspect of this case that deserves attention is the incomplete penetrance and variable expressivity of the mutation discussed within this case report. Of the 5 family members identified as having the c.3735_3741dupAAATCGA mutation, 3 have objective evidence of LV dysfunction and/or late gadolinium enhancement while 2 members are thus far unaffected. However, variable penetrance and the expressive nature of this mutation are not surprising. Incomplete (age-dependent) penetrance and variable expressivity is a well-described aspect of arrhythmogenic cardiomyopathy. Penetrance, in this disorder, has been estimated to be as low as 20%–30%.13
Counseling the patient’s genotype-positive, phenotype-negative son, who is employed as a member of the United States Army Special Forces community, in regard to the safety of ongoing military service has proven to be a challenging aspect of this case. By traditional technologies such as echocardiography, Holter monitoring, treadmill stress testing, and electrocardiography, there is no abnormality seen in this patient. When we apply advanced imaging techniques, early signs of disease can be seen in the form of late gadolinium enhancement on MRI. Technology has pushed the boundary of when disease detection occurs, often before sequalae are experienced. The point at which drastic lifestyle changes should be made is unknown. Does a child with an identified genetic mutation for arrhythmogenic cardiomyopathy thus warrant activity restriction from childhood, and be restricted from playing vigorous sports? Does a professional athlete then stop all activity at the peak of an arduous career because of early preclinical detection? This case highlights some of the difficulties present in the field of cardiac genetics and the unique challenges faced in the military medical field.
There are limited data upon which to formulate management plans for asymptomatic carriers of arrhythmogenic cardiomyopathy–associated genetic variants beyond longitudinal observation.14 Animal studies suggest that endurance training may accelerate the development of RV dysfunction and arrhythmia15; however, the evidence is incomplete and the natural history of this disease in the left ventricle is not completely understood. Thus, although avoidance of highly strenuous activity in asymptomatic arrhythmogenic cardiomyopathy–implicated genetic variants is logical, the exact timing of such restrictions is unclear. Ultimately, after extensive counseling and shared decision making, we decided to continue unrestricted service with annual reassessment with Holter monitoring and cardiac MRI. Any progression of his imaging abnormalities or arrhythmias would trigger more severe physical activity restrictions.
Conclusion
In conclusion, we present a patient with clinical features of arrhythmogenic cardiomyopathy presenting with aborted sudden cardiac death. The patient’s cardiac testing demonstrated predominant LV dysfunction consistent with the phenotypic expression of arrhythmogenic cardiomyopathy. Subsequent genetic testing revealed a previously unreported rare genetic variant, resulting in a premature stop codon in exon 23 of the DSP gene. The identification of this unique variant, which is associated with familial lineage of arrhythmogenic cardiomyopathy, helps to expand our understanding of the clinical heterogeneity and genetic basis for arrhythmogenic cardiomyopathy. This case also demonstrates some of the difficulties in implementing the data provided by advanced diagnostics into clinical practice.
Key Teaching Points.
-
•
The Desmoplakin (DSP) gene encodes the protein desmoplakin, which is a critical component of desmosomes, the specialized proteins that form intercellular junctions.
-
•
Desmosomal disorders cause cardiac-specific disease as a consequence of myocyte detachment and death, and this is the process implicated in the pathogenesis of arrhythmogenic right ventricular cardiomyopathy.
-
•
In contrast to the classic right predominant variant, arrhythmogenic cardiomyopathy may present with varying degrees of biventricular or predominantly left ventricular involvement (arrhythmogenic left ventricular cardiomyopathy).
-
•
Incomplete (age-dependent) penetrance and variable expressivity is a well-described aspect of arrhythmogenic cardiomyopathy.
-
•
There are limited data upon which to formulate management plans for asymptomatic carriers of arrhythmogenic cardiomyopathy–associated genetic variants beyond longitudinal observation.
References
- 1.Hug N., Longman D., Cáceres J. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzarini E., Jongbloed J.D., Pilichou K., Thiene G., Basso C., Bikker H., Charbon B., Swertz M., van Tintelen J.P., van der Zwaag P.A. The ARVD/C Genetic Variants Database: 2014 update. Hum Mutat. 2015;36:403–410. doi: 10.1002/humu.22765. [DOI] [PubMed] [Google Scholar]
- 4.Norman M., Simpson M., Mogensen J., Shaw A., Hughes S., Syrris P., Sen-Chowdhry S., Rowland E., Crosby A., McKenna W.J. Novel mutation in Desmoplakin causes arrhythmogenic left ventricular cardiomyopathy. Circulation. 2005;112:636–642. doi: 10.1161/CIRCULATIONAHA.104.532234. [DOI] [PubMed] [Google Scholar]
- 5.Sen-Chowdhry S., Syrris P., McKenna W.J. Role of genetic analysis in the management of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J Am Coll Cardiol. 2007;50:1813–1821. doi: 10.1016/j.jacc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Corrado D., Basso C., Thiene G., McKenna W.J., Davies M.J., Fontaliran F., Nava A., Silvestri F., Blomstrom-Lundqvist C., Wlodarska E.K., Fontaine G., Camerini F. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J Am Coll Cardiol. 1997;30:1512–1520. doi: 10.1016/s0735-1097(97)00332-x. [DOI] [PubMed] [Google Scholar]
- 7.Sen-Chowdry S., Syrris P., Prasad S., Hughes S., Merrifield R., Ward D., Pennell D., McKenna W. Left-dominant arrhythmogenic cardiomyopathy. J Am Coll Cardiol. 2008;52:2175–2187. doi: 10.1016/j.jacc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 8.ExAC Browser (Beta) | Exome Aggregation Consortium. Available at: http://exac.broadinstitute.org. accessed March 15, 2017.
- 9.Uzumcu A., Norgett E.E., Dindar A. Loss of Desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J Med Genet. 2006;43:e5. doi: 10.1136/jmg.2005.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q., Chen Y., Peng L., Gao R., Liu N., Jiang P., Liu C., Tang S., Quan L., Makielski J., Cheng J. Identification of rare variants of DSP gene in sudden unexplained nocturnal death syndrome in the Southern Chinese Han population. Int J Legal Med. 2016;130:317–322. doi: 10.1007/s00414-015-1275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z., Bowles N.E., Scherer S.E. Desmosomal dysfunction due to mutations in Desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Res. 2006;99:646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 12.Walsh R., Thomson K.L., Ware J.S. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen-Chowdry S., Syrris P., McKenna W.J. Genetics of right ventricular cardiomyopathy. J Cardiovasc Electrophysiol. 2005;16:927–935. doi: 10.1111/j.1540-8167.2005.40842.x. [DOI] [PubMed] [Google Scholar]
- 14.Ackerman M., Priori S., Willems S. HRS/EHRA Expert Consensus Statement on the state of genetic testing for the channelopathies and cardiomyopathies. Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 15.Kirchhof P., Fabritz L., Zwiener M. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation. 2006;114:1799–1806. doi: 10.1161/CIRCULATIONAHA.106.624502. [DOI] [PubMed] [Google Scholar]