Abstract
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disease caused by expansion of a CAG trinucleotide repeat in HTT, resulting in an extended polyglutamine tract in huntingtin. We and others have previously determined that the HD-causing expansion occurs on multiple different haplotype backbones, reflecting more than one ancestral origin of the same type of mutation. In view of the therapeutic potential of mutant allele-specific gene silencing, we have compared and integrated two major systems of HTT haplotype definition, combining data from 74 sequence variants to identify the most frequent disease-associated and control chromosome backbones and revealing that there is potential for additional resolution of HD haplotypes. We have used the large collection of 4078 heterozygous HD subjects analyzed in our recent genome-wide association study of HD age at onset to estimate the frequency of these haplotypes in European subjects, finding that common genetic variation at HTT can distinguish the normal and CAG-expanded chromosomes for more than 95% of European HD individuals. As a resource for the HD research community, we have also determined the haplotypes present in a series of publicly available HD subject-derived fibroblasts, induced pluripotent cells, and embryonic stem cells in order to facilitate efforts to develop inclusive methods of allele-specific HTT silencing applicable to most HD patients. Our data providing genetic guidance for therapeutic gene-based targeting will significantly contribute to the developments of rational treatments and implementation of precision medicine in HD.
Introduction
Huntington’s disease (HD) [MIM 143100] is a progressive neurodegenerative disorder caused by expansion of a CAG repeat in huntingtin (HTT) exon 1 that lengthens a normally polymorphic polyglutamine tract in HTT protein1 and produces characteristic motor disturbances, along with cognitive and psychiatric manifestations.2 Both the age at onset and the age at death of HD subjects are inversely correlated with the length of their CAG repeat, while the duration from onset to death, typically 15–20 years is largely independent of the mutation size.3, 4 Currently, there is no treatment to either delay the onset or slow the progression of HD, but the recent discovery of genetic modifiers of age at onset establishes that the rate of HD pathogenesis can be altered before symptoms appear.5 Genetic analysis of large HD cohorts has demonstrated that HD is inherited as a complete dominant where a single mutant HTT allele determines the timing of disease onset, with no discernible impact of either the normal HTT allele or, when present, a second mutant HTT allele.4 Consequently, suppression of the expression of mutant HTT is an appealing therapeutic strategy which, if achieved in an allele-specific manner,6 could avoid any potential negative consequences attributable to deficiency of normal huntingtin activity.
HTT allele-specific gene silencing strategies can either directly target the expanded CAG repeat or aim at other genetic variants in the surrounding haplotype.7, 8, 9 While the former is an attractive target that would be applicable in all HD subjects, establishing allele specificity in individuals where the second HTT CAG repeat is high in the normal range, and limiting the effect to HTT when there are other expressed CAG repeats in the human genome may be technically challenging. However, targeting genetic variants on the mutant HTT haplotype can achieve allele-specificity only in those HD individuals who are heterozygous for those variants.10, 11, 12 A multiplicity of HTT haplotypes, with normal and expanded repeats, have been observed in HD individuals of European ancestry.13, 14, 15, 16, 17 We have previously delineated the eight most common HTT haplotypes bearing expanded alleles based upon 21 genetic variants14, 18 while others have described three major haplogroups,16 which they recently resolved into subtypes using 63 variants.11 The two marker sets, which are only partially overlapping, have each been used to define sites that are most frequently heterozygous in HD subjects as potential targets for allele-specific HTT silencing.6, 11 In order to facilitate research and development towards this goal, we have compared and integrated the two haplotype systems, better estimated HTT haplotype frequencies on normal and disease chromosomes in Europeans, and delineated the HTT haplotypes present in publicly available cell line resources available to the HD research community.
Materials and methods
Definitions of HTT haplotypes
Selection of variants, mainly single-nucleotide polymorphisms (SNPs), and samples initially used to characterize HTT haplotypes on HD expanded chromosomes and normal chromosomes were described elsewhere.14 Briefly, 20 SNPs and one 3 bp insertion-deletion (indel) that showed significant association with HD in either (1) comparison of all HD vs controls, or (2) comparison of those HD individuals lacking the major disease haplotype vs controls, were used for haplotype phasing.14 The HTT CAG repeat sizes in HD individuals were coded as bi-allelic genotypes (expanded and normal), each person being a heterozygote. In contrast, each control individual was coded as homozygous normal for the HTT CAG repeat. Haplotype phasing of SNP genotypes was performed by the MaCH program,19 and the 10 most frequent haplotypes on each of expanded chromosomes and normal chromosomes were identified. As four haplotypes overlapped between both disease and normal, the union set comprised 16 distinct haplotypes. Definitions of haplotypes described previously (hap.01~hap.07)14 are same as those in this study. The phylogeny tree of haplotypes was obtained by the MEGA5 program (neighbor-joining method, P-distance model; http://www.megasoftware.net/).
Haplotype-specific SNP sites for mutant allele-selective silencing
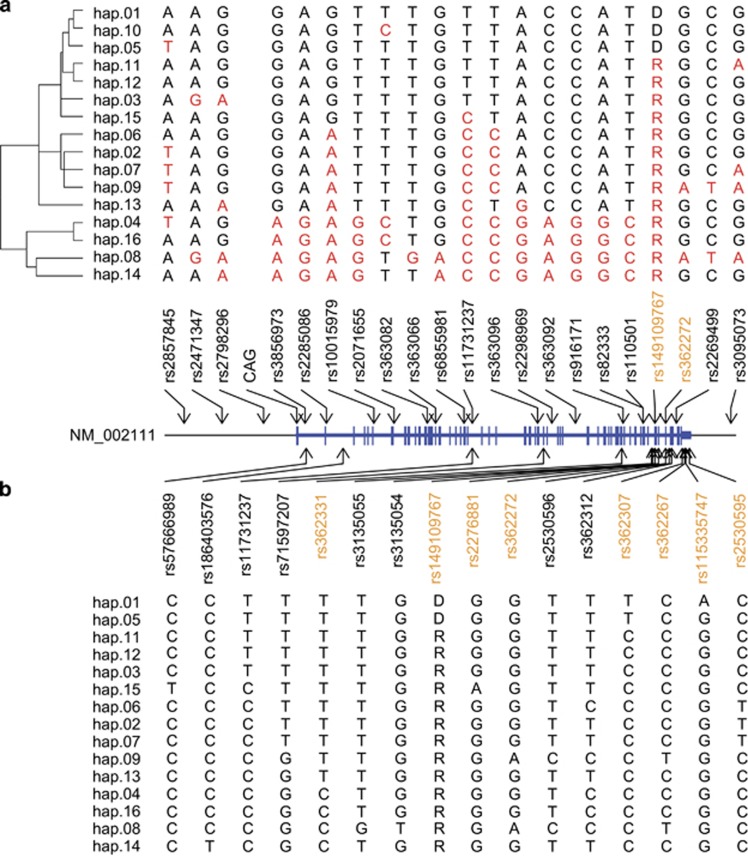
Previously, based on cumulative heterozygosity analyses of HD subjects with European ancestry, we revealed 20 SNP sites that can be targeted for mutant allele-specific HTT silencing/lowering.18 In order to relate alleles of target SNPs to haplotypes, we determined consensus alleles of those 20 SNPs (10 exon SNPs and 10 intron SNPs) for each haplotype. Briefly, for a given haplotype, we extracted chromosomes from 1000 Genomes Project data (Phase 1; http://www.internationalgenome.org/data/) to determine consensus alleles by taking the most frequent allele of each of 20 target SNP sites. Some of SNP sites are not variable among 16 haplotypes, and consensus alleles of variable SNPs are indicated in Figure 1b together with 2 exon SNPs used to define haplotypes. In 1000 Genomes data, hap.10 is not present, and therefore excluded in this analysis.
Figure 1.
Definitions and sequence relationships of HTT haplotypes. (a) Twenty SNPs, one 3 bp indel (rs149109767, alleles R-reference and D-deletion) and the CAG repeat polymorphism are shown at their genomic locations relative to that of the HTT RefSeq transcript (NM_002111). Genotype at each marker on each of 16 HTT haplotypes, defined in the text, is shown above the marker. Haplotypes are ordered based upon a neighbor-joining method (p-distance model) in a dendrogram with two main branches, each with different sizes of sub-clusters. Alleles in red represent differences from hap.01, the most frequent haplotype on CAG-expanded HD chromosomes. (b) Consensus alleles of 10 exon SNPs and 10 intron SNPs that showed the biggest cumulative heterozygosity were determined for each haplotype based on 1000 Genomes Project data. A consensus allele for a given SNP site represents the most frequent allele among a collection of chromosomes with same haplotype. Since hap.10 is not present in 1000 Genomes data (Phase 1), hap.10 was excluded in this analysis. Subsequently, alleles of SNPs that show variable alleles in 15 haplotypes and alleles of two exon SNPs that were used to define the haplotypes are indicated. SNPs in orange and black font colors represent SNPs on exons and introns of RefSeq NM_002111, respectively.
Haplotypes of publicly available cell lines
We assembled genotypes of 21 tagging SNPs, either from genome-wide association (GWA) data5 or from specific TaqMan assays applied to DNA from blood, lymphoblasts, fibroblasts, induced pluripotent stem cells or derived neural progenitor cells. Those cell line data described in this study represent 59 individuals whose fibroblast cell lines are available in public repositories and 7 human embryonic stem cell lines from Genea Biocells Inc. (http://geneabiocells.com/). HTT CAG repeat length was also determined as described previously.20 Cell line genotype data and the HTT CAG repeat genotype coded as a bi-allelic system (expanded or normal) were combined for haplotype phasing in order to identify haplotype carrying expanded CAG or normal repeat. Genotype data for HD and control subjects that were used to define haplotypes14 were also included to increase the accuracy of computational population phasing by the MaCH program.19 Familial relationships (Supplementary Table 1) were further considered when the relationships between CAG repeats and haplotypes were ambiguous to determine the phase of CAG repeats and haplotypes (eg, control subjects).
Frequencies of haplotypes and haplogroups in control samples
Fully phased 1000 Genomes Project data (Phase 1) were used to estimate population frequencies of HTT haplotypes defined in this study and haplogroups described by Kay et al.11 Each chromosome was classified into haplotypes based on 21 SNPs, and further summarized for each population group (ie, Europeans, Asians, Africans, and Ad Mixed Americans). The haplogroup of each chromosome was determined similarly based on 63 SNPs, permitting direct delineation of correspondence between haplotype systems on normal chromosomes.
Genotype imputation of HD samples
Genotypes on chromosome 4 were imputed for HD samples with European ancestry used in a recent onset-modifier GWA study (4082 Europeans)5 and control samples (1676 Europeans)21 using the Michigan Imputation Server.22 Pre-phasing was performed by Eagle223 and imputation was performed by Minimac3 using 1000 Genomes Phase 1 as a reference panel (all populations).24 A set of SNPs used for haplotype and/or haplogroup analysis were then extracted from imputed data to determine relationships between haplotypes and haplogroups.
Determination of the relationship between haplotypes and haplogroups
Twenty-one genetic variations from our study14 and 63 tagging SNPs from Kay and colleagues11 were used to classify haplotypes of samples used in the HD modifier GWA study.5 There were 10 shared SNPs between the two haplotype systems (rs2798296, GRCh37 chr4:g.3062165A>G; rs3856973, GRCh37 chr4:g.3080173G>A; rs2285086, GRCh37 chr4:g.3089259A>G; rs10015979, GRCh37 chr4:g.3109442A>G; rs11731237, GRCh37 chr4:g.3151813C>T; rs363096, GRCh37 chr4:g.3180021T>C; rs2298969, GRCh37 chr4:g.3186244A>G; rs363092, GRCh37 chr4:g.3196029A>C; rs916171, GRCh37 chr4:g.3216815C>G; and rs362272, GRCh37 chr4:g.3234980G>A) so genotypes for a total of 74 variants were extracted from the imputed data. Then the recoded bi-allelic HTT CAG repeat length genotype (expanded or normal) was added to the imputed genotype data, and haplotype phasing was performed for the 75 variant sites in the HD samples (4082 Europeans),5 control samples (1676 Europeans),21 and 1000 Genomes Phase 1 samples (379 Europeans, 181 Ad Mixed Americans, 246 Africans, 286 Asians)24 by the Beagle program.25 Subsequently, the CAG-expanded and normal chromosomes from each HD heterozygous subject (4078 Europeans) were named based on (1) our haplotype definitions, and (2) haplotype definitions used by Kay and colleagues11 in order to delineate the relationships between the two haplotype systems.
Description of SNPs, website, and public access
Detailed description of SNPs used in this study can be found in Supplementary Table 2. In addition, description of SNPs, definition of haplotypes, and genotype data are available at chgr.partners.org/htt.haplotype.html. The genotype data set is also available at the European Variation Archive (http://www.ebi.ac.uk/eva/) (accession number: PRJEB20817).
Results
Common SNP-based haplotypes
We previously defined the eight most frequent haplotypes (hap.01 to hap.08) on HD disease-causing chromosomes using 21 common genetic variants, including 20 SNPs and one 3 bp indel, genotyped in 699 unrelated HD subjects and 1676 population controls of European ancestry.14, 18 Approximate locations and alleles of DNA variations that were used for haplotype analysis are summarized in Figure 1a. Here, we extend the definitions in that data set to the most frequent 10 HD and the 10 most frequent normal European HTT haplotypes. Four haplotypes were shared between the two groups, so the union created a single set of 16 different haplotypes. These were named based first upon decreasing frequency on CAG-expanded disease chromosomes in this initial HD data set (hap.01 through hap.10) and then, after excluding the four shared haplotypes (hap.08, hap.02, hap.03 and hap.01, in order of normal frequency), based upon decreasing frequency on normal chromosomes (hap.11 through hap.16); all other rare haplotypes were grouped as ‘hap.other’. A comparison of the potential relationships between these 16 haplotypes was achieved by phylogeny analysis using a neighbor-joining algorithm. The result is a dendrogram with two main branches containing different-sized sub-clusters (Figure 1a). For example, hap.01, the most common haplotype on the HD disease chromosomes forms a cluster with hap.05 and hap.10, whereas hap.08, the most common haplotype on normal chromosomes is a part of a cluster of haplotypes involving hap.04, hap.16, and hap.14. Divergence of related haplotypes could potentially be explained by a single marker allele change in some cases (eg, hap.01, hap.05, and hap.10; hap.11 and hap.12; hap.02 and hap.07; hap.04 and hap.16), by insertion/deletion of a simple repeat (eg, hap.01 and hap.12), and by combinations of various genetic events including local recombination or gene conversion. The two main branches of the dendrogram suggest at least two different ancestral origins of de novo CAG expansion mutation. However, it is likely that the haplotype diversity within the subclusters reflects the occurrence of many more de novo expansions rather than being the result of haplotype decay since we have previously demonstrated de novo CAG expansion on both hap.01 and hap.05.18
Haplotype-specific target SNP sites for allele-specific silencing
Initial selection of SNPs for haplotyping was based on comparisons between HD subjects and normal control individuals, and therefore did not represent the combination of disease chromosomes and normal chromosomes in HD subjects.14 Subsequently, we performed iterative heterozygosity analyses aiming at revealing a minimal number of SNPs covering the maximum proportion of HD patients in allele-specific gene targeting therapies.18 Cumulatively, 10 exon SNPs and 10 intron SNPs covered 93.8 and 97% of HD subjects, respectively, indicating that the vast majority of HD subjects with European ancestry carry at least one heterozygous SNP site among 20 nominated targetable locations.18 However, the heterozygosity analysis did not immediately show mutant alleles to target. Here, we determined consensus alleles of 20 targetable SNP sites on each of haplotypes based on 1000 Genomes Project data, and mapped variable alleles on each haplotype. As summarized in Figure 1b, target SNP sites for each diplotype can be selected immediately by comparing two haplotypes (assuming one is mutant chromosome and the other is normal chromosome). For example, if a HD individual carries mutant hap.01 and normal hap.08 chromosomes, there are 12 SNP sites that can be used to distinguish mutant allele from normal allele (Figure 1b).
Haplotypes of publicly available cell line resources
Results of mutant allele-specific gene silencing studies have produced promising results in animal models,10 encouraging the application of this approach to human HD. In this context, cell lines derived from HD subjects provide valuable tools to test the specificity and efficacy of allele-specific silencing reagents in pre-clinical experiments. Thus, we performed haplotype analysis using our haplotype system for HD cell lines readily available from various public repositories. Supplementary Table 6 gives the HTT haplotypes for 59 fibroblast lines available from the NIGMS Repository at the Coriell Institute (https://catalog.coriell.org/1/NIGMS) or the NINDS Human Cell and Data Repository at RUCDR Infinite Biologics (https://nindsgenetics.org/). These include 43 lines representing individuals (from 26 families) with an expanded HTT repeat, whose allele lengths range from 38 to 180 CAGs. The remaining 16 lines from 10 families represent control individuals with CAG repeat lengths 33 or shorter. Where possible the phase of the CAG repeat with respect to the HTT haplotype was confirmed from family relationships (Supplementary Table 1). In the remaining instances (unrelated subjects; noted by * on the sample ID in Supplementary Table 6), the phase of the expanded repeat was assigned probabilistically using MaCH program (see Methods) or the phase of distinguishable normal alleles was assigned arbitrarily for control individuals. As expected from HD population data, the most frequent haplotype on the disease and normal chromosomes in these families are hap.01 and hap.08, respectively, and this most common HD diplotype, hap.01/hap.08, is present in multiple lines from independent families. However, many other HD haplotypes and diplotypes are also represented. Only five of the HD individuals are homozygous for the same haplotype, and four of these, two of which are also homozygous for an expanded CAG repeat, derive from the large Venezuela HD kindreds in which the disease segregates with hap.03 haplotype.
Induced pluripotent stem cell lines are already available to the research community from the above repositories or from the Cedars-Sinai iPSC Core (https://www.cedars-sinai.edu/Research/Research-Cores/Induced-Pluripotent-Stem-Cell-Core-/) for 11 of the subjects with expanded repeat fibroblast lines and 5 of the normals, as noted in Supplementary Table 6. In addition, we have performed haplotyping for 7 human embryonic stem cell lines, with expanded CAG alleles ranging from 40 to 48 repeats, available from Genea Biocells, as shown in Supplementary Table 7. The HD mutation in these lines resides either on hap.01 (four independent lines) or hap.02 (three lines from the same family).
Haplogroup definition of HD chromosomes
A different set of genetic markers (Supplementary Table 2) has been used by others to define haplogroups A, B, and C, each of which represents a cluster of similar haplotypes.13, 16, 17 Recently, Kay et al performed a more detailed analysis of the haplogroup system in 738 European reference haplotypes from the 1000 Genomes Project and 2364 haplotypes from HD patients and relatives in Canada and Europe to define individual subtypes within each haplogroup based upon 63 genetic variants across HTT.11 Across the Canadian and European HD subjects, selected subtypes from the A haplogroup accounted for 86% of all CAG-expanded chromosomes, but the remaining HD chromosomes fell into haplogroup B or C subtypes, or rarely, into none of the three major haplogroups (‘Other’).
Comparison and integration of the two HTT haplotype systems
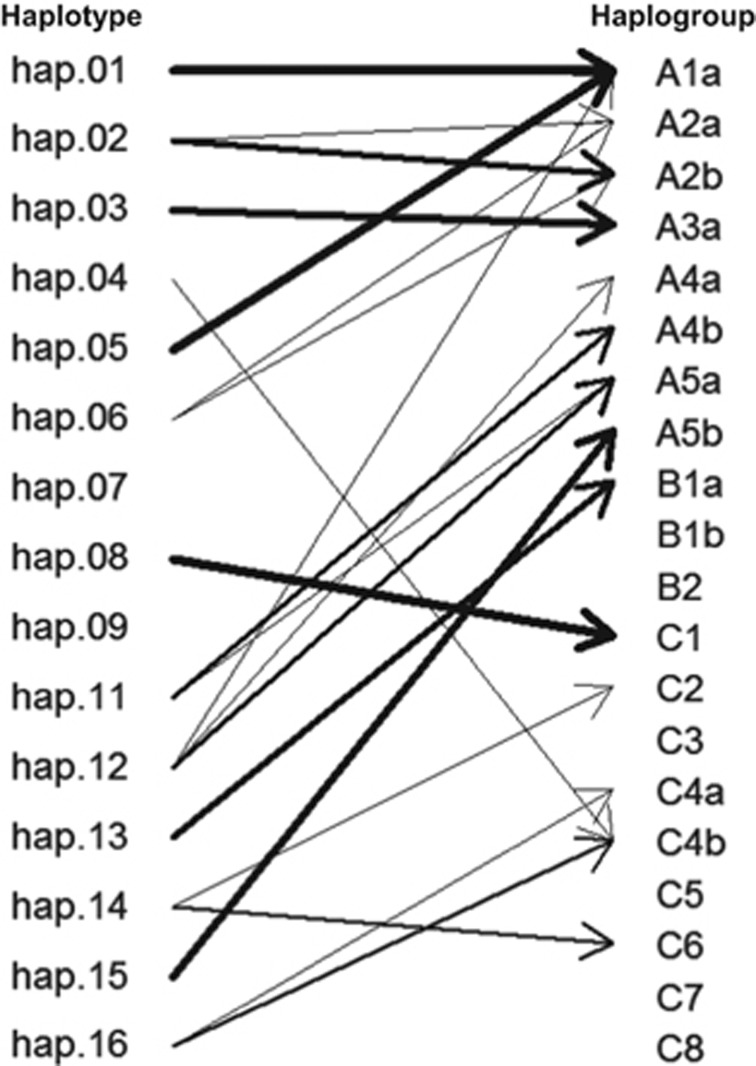
Between the 21 markers used in our haplotype system and the 63 markers used in the recent subdividing of A, B, and C haplogroups, only 10 markers are overlapping (Supplementary Table 2). In order to maximize the utility of both haplotype systems, we have directly compared them by examining the fully phased 1000 Genomes Project haplotype data (Phase 1). To extend the analysis across all available populations rather than only Europeans, we analyzed a total of 1092 control individuals (2184 normal chromosomes) consisting of Africans (ASW, LWK, and YRI), Ad Mixed Americans (CLM, MXL, and PUR), East Asians (CHB, CHS, and JPT), and Europeans (CEU, FIN, GBR, IBS, and TSI). Each 1000 Genomes chromosome was independently classified into our haplotypes using 21 variant sites and haplogroup subtypes using 63 variant sites. The hap.01–hap.16 designations encompassed almost 76% of European chromosomes and more than 60% of Ad Mixed American and Asian chromosomes, but only 23% of African chromosomes, which display far greater genetic complexity (Supplementary Table 3). A similar pattern was evident using haplogroup subtypes which accounted for almost 67% of European chromosomes, about half of Ad Mixed American and Asian chromosomes, and only about 7% of African chromosomes (Supplementary Table 3). Subsequently, we delineated the relationships between the two haplotype systems by calculating the percentage of chromosomes with each haplotype defined in our system that distributed to each haplotype defined in the haplogroup system (Supplementary Table 4; Figure 2) and, vice versa (Supplementary Table 5). For example, 93.6 and 100% of chromosomes defined as bearing the related hap.01 or hap.05 haplotypes are classified as haplogroup subtype A1a (Supplementary Table 4; Figure 2). Among only Europeans, the same correspondence is 100% for both haplotypes. The third related member of this haplotype subcluster from Figure 1, hap.10, was originally defined from HD chromosomes but was not seen on any 1000 Genomes Project chromosomes and so is not reflected in the Tables. The haplotype most common on European normal chromosomes, hap.08, corresponds 95.1% of the time with the C1 subtype designation in the haplogroup system (Supplementary Table 4). However for some other haplotypes, the correspondence is not so direct, as some hap.02 chromosomes (25.6%) are classified as haplogroup subtype A2a while others (61.0%) are classified as A2b. Similarly, hap.06 also divides between these two related A2 subtypes, but is primarily assigned to the ‘Other’ class, not being classified as haplogroup A, B or C. Interestingly, haplotypes hap.04, hap.07, and hap.09, which were named by decreasing order of their frequency on HD disease chromosomes in our original study all correspond to the ‘Other’ class of haplogroups except 12.5% of the hap.04 group, which are designated as C4b.
Figure 2.
Correspondences of haplotypes and haplogroups. Based on Supplementary Table 4, correspondences of haplotypes to haplogroups were summarized. ‘hap.other’ and ‘Other’ were excluded to focus on distinct haplotypes. Thickness of an arrow represents relative proportion of a specific haplotype-haplogroup correspondence for a given haplotype. For example, most of hap.02 is classified as haplogroup A2b, and a small portion of hap.02 is classified as haplogroup A2a. Actual haplotype-haplogroup correspondence data can be found in Supplementary Table 4.
Considering the reverse comparison of chromosomes named by the haplogroup system to our haplotypes (Supplementary Table 5), the correspondence is similar to the above, with the A1, A2 and A3 designations, which are the most common on European HD chromosomes, corresponding largely to hap.01+hap.05, hap.02+hap.06 and hap.03, respectively, encompassing most of the haplotypes seen frequently on European HD chromosomes. Those haplogroup subtypes rarely seen on HD chromosomes, such as A4a, A4b, A5a, A5b, B1a, C2, C4, and C6 correspond largely with haplotypes seen on the normal chromosomes in our HD data set (hap.12, hap.11, hap.12, hap.15, hap.13, hap.14, hap.16, and hap.14, respectively), or in the cases of B1b, B2, C3, C5, C7, and C8, among the mixed hap.other group of less frequent normal haplotypes.
Overall, these comparisons indicate that the haplotypes most frequently associated with HD disease chromosomes (ie, hap.01, hap.02, and hap.03) correspond in general with haplogroup subtypes A1, A2, and A3. However, there is the potential for additional resolution in both systems, as illustrated by the fact that the subtypes of A2 (A2a and A2b) subdivide the hap.02 chromosomes, but each subtype (A2a and A2b) is also classified into either hap.02 or hap.06. Similarly, the lack of strong correspondence with haplogroup subtypes of some of the rarer haplotypes identified on HD chromosomes in our studies suggests yet greater diversity among disease chromosomes, and predicts that additional genetic variants can further subdivide the defined haplotypes and haplogroups, particularly in non-European populations.
HTT haplotype frequencies on CAG-expanded and normal chromosomes
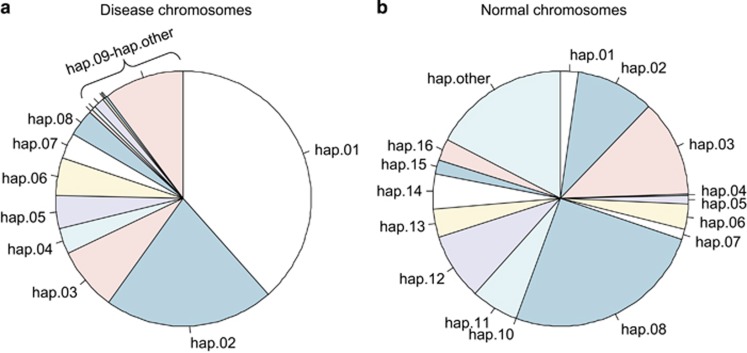
The comparisons in Supplementary Tables 4 and 5 relied on fully phased control chromosomes to define haplotype/haplogroup relationships in samples with various ancestries. To estimate the frequency of these groupings on HD chromosomes of European ancestry, we examined the imputed genotypes of 4078 heterozygous HD subjects recently studied in a GWA study of HD modifiers.5 We extracted a union set of 74 SNPs (representing 21 SNPs used to define our HTT haplotypes and the 53 non-overlapping variant sites used by Kay et al), and performed probabilistic phasing of the marker alleles using the Beagle program.25 Each of 74-SNP haplotypes (either expanded CAG or normal CAG chromosome) was assigned to both a haplotype and a haplogroup subtype, generating a data set that permitted assessment of the frequency of each haplotype/haplogroup subtype combination. When focusing on our haplotypes defined in this study (Figure 1), frequencies of haplotypes of the expanded and normal chromosomes based on a large collection of HD subjects with European ancestry revealed that HD expansion mutation sits on diverse haplotypes that are also present in normal chromosomes (Figure 3). In addition, comparisons of haplotype frequencies revealed overrepresented and underrepresented haplotypes in HD. For example, hap.01 and hap.08 are enriched in disease and normal chromosomes, respectively (Figure 3). Frequency data predicted that the most common diplotype in heterozygous HD subjects would be expanded CAG repeat on hap.01 and normal CAG repeat on hap.08. When comparing our haplotypes to haplogroups, overall, 78 and 71% of European HD and normal chromosomes, respectively, were assignable to discrete ‘super’-haplotype backbones that combined discrete haplotypes and haplogroup subtypes, excluding the uncertain hap.other and haplogroup ‘Other’ catch-all categories (Table 1). As expected, the most frequent HD chromosome backbone was hap.01/A1a and comprised over 38% of European HD chromosomes from the GWA study. Similarly, the most frequent control backbone hap.08/C1 accounted for about 25% of normal chromosomes. Examination of diplotypes of the 4078 European HD individuals revealed that 56% possessed HD and normal chromosomes that could both be assigned to a fully defined haplotype/haplogroup backbone, without the uncertainty of the hap.other and haplogroup ‘Other’ categories (Supplementary Table 8). Notably, less than 5% of these HD subjects had fully defined chromosomal backbones that were identical on disease and normal chromosomes, being homozygous for all tagging markers. If all 4078 heterozygous HD subjects were analyzed, 4.9% of them carry identical alleles for 74 SNPs, suggesting that the majority of HD subjects of European descent are eligible for allele-specific gene targeting strategies. Our previous full sequence analysis of HD hap.01 chromosomes suggests that many of the individuals with the same haplotype backbone on the normal and disease chromosomes could harbor heterozygous variants not considered in the current haplotypes/haplogroups,18 further implying an additional likelihood of allele discrimination.
Figure 3.
Frequencies of haplotypes in HD disease and normal chromosomes. HD subjects carrying one expanded and one normal chromosome were included in this analysis to estimate overall frequencies of haplotypes. From haplotypes probabilistically determined based on a union set of 74 SNPs, we used our haplotype definitions to classify each chromosome. Subsequently, frequencies of our haplotypes in HD disease chromosomes (a) and normal chromosomes in HD subjects (b) were calculated and summarized.
Table 1. Frequency of combined haplotype/haplogroup system backbones on CAG-expanded and normal chromosomes in European HD subjects.
| Haplotype | Haplogroup | # HD normal chromosomes | Percent |
|---|---|---|---|
| 'Super'-haplotype of CAG-expanded chromosomes | |||
| hap.01 | A1a | 1556 | 38.16% |
| hap.02 | A2b | 553 | 13.56% |
| hap.03 | A3a | 323 | 7.92% |
| hap.02 | A2a | 291 | 7.14% |
| hap.05 | A1a | 164 | 4.02% |
| hap.08 | C1 | 136 | 3.33% |
| hap.06 | A2b | 107 | 2.62% |
| hap.06 | A2a | 60 | 1.47% |
| hap.11 | A4b | 3 | 0.07% |
| hap.12 | A5a | 3 | 0.07% |
| hap.15 | A5b | 2 | 0.05% |
| hap.12 | A1a | 1 | 0.02% |
| Sum | 78.45% | ||
| Haplotype ‘hap.other’ or haplogroup ‘Other’ categories of CAG-expanded chromosomes | |||
| hap.other | Other | 380 | 9.32% |
| hap.04 | Other | 134 | 3.29% |
| hap.07 | Other | 134 | 3.29% |
| hap.12 | Other | 51 | 1.25% |
| hap.02 | Other | 33 | 0.81% |
| hap.06 | Other | 30 | 0.74% |
| hap.other | B2 | 22 | 0.54% |
| hap.09 | Other | 18 | 0.44% |
| hap.11 | Other | 17 | 0.42% |
| hap.14 | Other | 14 | 0.34% |
| hap.01 | Other | 13 | 0.32% |
| hap.16 | Other | 10 | 0.25% |
| hap.other | C5 | 7 | 0.17% |
| hap.05 | Other | 4 | 0.10% |
| hap.03 | Other | 3 | 0.07% |
| hap.other | B1b | 3 | 0.07% |
| hap.other | A2b | 2 | 0.05% |
| hap.other | A5a | 2 | 0.05% |
| hap.08 | Other | 1 | 0.02% |
| hap.other | A1a | 1 | 0.02% |
| Sum | 21.55% | ||
| 'Super'-haplotype of normal chromosomes | |||
| hap.08 | C1 | 1034 | 25.36% |
| hap.03 | A3a | 479 | 11.75% |
| hap.02 | A2b | 242 | 5.93% |
| hap.11 | A4b | 197 | 4.83% |
| hap.02 | A2a | 153 | 3.75% |
| hap.13 | B1a | 141 | 3.46% |
| hap.01 | A1a | 82 | 2.01% |
| hap.12 | A5a | 81 | 1.99% |
| hap.12 | A4a | 76 | 1.86% |
| hap.15 | A5b | 71 | 1.74% |
| hap.06 | A2a | 63 | 1.54% |
| hap.14 | C6 | 61 | 1.50% |
| hap.06 | A2b | 53 | 1.30% |
| hap.16 | C4b | 51 | 1.25% |
| hap.16 | C4a | 47 | 1.15% |
| hap.05 | A1a | 42 | 1.03% |
| hap.14 | C2 | 26 | 0.64% |
| hap.10 | A1a | 1 | 0.02% |
| hap.12 | A1a | 1 | 0.02% |
| Sum | 71.14% | ||
| Haplotype ‘hap.other’ or haplogroup ‘Other’ categories of normal chromosomes | |||
| hap.other | Other | 559 | 13.71% |
| hap.12 | Other | 187 | 4.59% |
| hap.14 | Other | 96 | 2.35% |
| hap.other | B1b | 73 | 1.79% |
| hap.07 | Other | 54 | 1.32% |
| hap.11 | Other | 47 | 1.15% |
| hap.other | C8 | 36 | 0.88% |
| hap.03 | Other | 21 | 0.51% |
| hap.06 | Other | 16 | 0.39% |
| hap.other | C5 | 14 | 0.34% |
| hap.16 | Other | 11 | 0.27% |
| hap.04 | Other | 10 | 0.25% |
| hap.other | C7 | 10 | 0.25% |
| hap.02 | Other | 9 | 0.22% |
| hap.01 | Other | 8 | 0.20% |
| hap.other | C1 | 6 | 0.15% |
| hap.08 | Other | 4 | 0.10% |
| hap.13 | Other | 3 | 0.07% |
| hap.other | B1a | 3 | 0.07% |
| hap.15 | Other | 2 | 0.05% |
| hap.other | A5a | 2 | 0.05% |
| hap.other | B2 | 2 | 0.05% |
| hap.other | A2a | 1 | 0.02% |
| hap.other | A4b | 1 | 0.02% |
| hap.other | A5b | 1 | 0.02% |
| Sum | 28.84% | ||
Phased haplotypes of subjects (4078 heterozygous HD) were grouped into HD disease chromosomes and normal chromosomes. Subsequently, the frequency of each combined haplotype/haplogroup (ie, ‘super’-haplotype) was calculated for HD disease and normal chromosomes. Frequency and corresponding percentage value of each ‘super’-haplotype were based on (1) haplotypes not involving ‘hap.other’ or ‘Other’ and (2) haplotypes involving ‘hap.other’ or ‘Other’.
Discussion
HTT shows evolutionarily conserved structural characteristics, and deficiency or hypomorphism of huntingtin are associated with pleiotropic effects involving a number of critical biological processes,26 suggesting that HTT silencing approaches to treat HD may need to be specific to the mutant allele. Allele-specific silencing of HTT can be achieved either by directly targeting the CAG repeats or, alternatively, by targeting polymorphisms in linkage disequilibrium with the CAG expansion.8, 10 Because the HD mutation can occur across a wide range of pathogenic sizes, and CAG repeats are found in many other genes, directly targeting the CAG expansion could result in variable levels of allele selectivity and off-target effects. Previous studies have demonstrated the feasibility of silencing the expression of the expanded allele by targeting a variation on the expanded chromosome.10, 27, 28, 29 Recently, SNP heterozygosity analysis has revealed that the disease chromosome can be distinguished from the normal chromosome in most HD subjects of European ancestry.11, 16, 18, 28, 30 Therapeutic strategy leveraging a SNP-targeting approach is therefore possible (Figure 1b), but would require knowledge about presence of target SNP sites, haplotype phasing, and preferably additional exon SNP sites for outcome measurements (ie, levels of mutant HTT) for a given HD individual. Still, analytical pipelines to identify variant alleles on the CAG-expanded chromosome of an HD individual are yet to be developed because simple genotyping assays do not differentiate allelic phase unless family members are also analyzed. This limitation can be overcome by computational haplotype phasing approaches, because haplotype phasing with a large collection of HD data allows relatively accurate inference of the disease and normal chromosome. Results described here on haplotype phasing of large populus of HD individuals can help populate attributes on HD patient database, and inform where patient groups enriched for targeting SNP can be sought. Subsequent sequence analysis of representative common HD haplotypes and pair-wise comparisons then provide a comprehensive list of targetable sites for each diplotype. In addition, development of allele-specific HTT quantification assays to assess the efficacy and allele specificity of silencing reagents require knowledge of variations and their relationships to the expanded chromosomes. Therefore, haplotypes of expanded chromosomes, individual-level diplotype data, and our analytical pipelines provide guidance for identifying targets for mutant allele-specific HTT lowering strategies and a route to developing allele-specific readouts to assess specificity of silencing reagents. In addition, genome-wide genotyping assays for HD subjects in a large observational study is ongoing (ie, ENROLL-HD), and our pipelines can efficiently identify each individual’s expanded and normal chromosomes. Such individual level diplotype data will be critically important in stratifying subjects to identify optimal study populations in clinical trials.
In summary, we performed individual level haplotype analyses on a large cohort of HD subjects to evaluate the power of haplotype-based genetics in stratifying HD subjects. Our haplotypes based on a relatively small number of SNPs were able to distinguish mutant chromosomes from their normal counterparts, and confirmed that the majority of HD subjects carry two different haplotypes, further supporting the conclusion from population-based SNP analysis that most HD individuals could be eligible for allele-specific gene silencing18 and demonstrating the efficiency of haplotype-based approaches. By providing the HD haplotypes of commonly used publicly available cell lines and haplotype conversion tables for the comparable haplogroup classification strategy, we hope to promote and facilitate the use of these resources to accelerate pre-clinical allele-specific gene silencing studies and a true precision medicine approach to HD.
Acknowledgments
We would like to thank all HD patients and their families who generously participated in this study. The full list of clinical investigators contributing samples to the generation of genetic data sets used in this study can be found at PMC4524551. This work was supported by the CHDI Foundation, by grants U01NS082079, R01NS091161, R01HG002449 and P50NS016367 from the National Institutes of Health (USA), and by grants G0801418 and MR/L010305/1 from the Medical Research Council (UK).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- The Huntington's Disease Collaborative Research Group: a novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993; 72: 971–983. [DOI] [PubMed] [Google Scholar]
- Bates GP, Dorsey R, Gusella JF et al: Huntington disease. Nat Rev Dis Primers 2015; 15005. [DOI] [PubMed]
- Keum JW, Shin A, Gillis T et al: The HTT CAG-expansion mutation determines age at death but not disease duration in Huntington disease. Am J Hum Genet 2016; 98: 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Ramos EM, Lee JH et al: CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology 2012; 78: 690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium: identification of genetic factors that modify clinical onset of Huntington's disease. Cell 2015; 162: 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Kim KH, Chao MJ et al: Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 2016; 25: 4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT et al: Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol 2009; 27: 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MS, Kordasiewicz HB, McBride JL: Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington's disease and spinocerebellar ataxia. Hum Mol Genet 2015; 25 (R1): R53–R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J et al: Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 2012; 150: 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL et al: Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther 2011; 19: 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay C, Collins JA, Skotte NH et al: Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in Huntington disease patients of European ancestry. Mol Ther 2015; 23: 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell AL, Skotte NH, Kordasiewicz HB et al: In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther 2014; 22: 2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baine FK, Kay C, Ketelaar ME et al: Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur J Hum Genet 2013; 21: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Gillis T, Mysore JS et al: Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet 2012; 90: 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EM, Gillis T, Mysore JS et al: Prevalence of Huntington's disease gene CAG trinucleotide repeat alleles in patients with bipolar disorder. Bipolar Disord 2015; 17: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR et al: CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet 2009; 84: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Visscher H, Collins JA et al: HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet 2011; 19: 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Kim KH, Shin A et al: Sequence-level analysis of the major European Huntington disease haplotype. Am J Hum Genet 2015; 97: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR: MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Mysore J et al: Prevalence of incompletely penetrant Huntington's disease alleles among individuals with major depressive disorder. Am J Psychiatry 2010; 167: 574–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myocardial Infarction Genetics ConsortiumMyocardial Infarction Genetics Consortium, Kathiresan S, Myocardial Infarction Genetics Consortium, Voight BF et al: Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009; 41: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S et al: Next-generation genotype imputation service and methods. Nat Genet 2016; 48: 1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Danecek P, Palamara PF et al: Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016; 48: 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium: An integrated map of genetic variation from 1092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL: Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan LH, Cohen J, Fatemi A et al: A novel neurodevelopmental disorder associated with compound heterozygous variants in the huntingtin gene. Eur J Hum Genet 2016; 24: 1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard ME, Southwell AL, Kordasiewicz H et al: Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res 2013; 41: 9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J et al: Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol 2009; 19: 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Engelman J, Friedlander RM: Allele-specific silencing of mutant Huntington's disease gene. J Neurochem 2009; 108: 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C et al: A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol 2009; 217: 312–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.