Abstract
Massive parallel sequencing (MPS) can accurately quantify mitochondrial DNA (mtDNA) single nucleotide variants (SNVs), but no MPS methods are currently validated to simultaneously and accurately establish the breakpoints and frequency of large deletions at low heteroplasmic loads. Here we present the thorough validation of an MPS protocol to quantify the load of very low frequency, large mtDNA deletions in bulk DNA and single cells, along with SNV calling by standard methods. We used a set of well-characterized DNA samples, DNA mixes and single cells to thoroughly control the study. We developed a custom script for the detection of mtDNA rearrangements that proved to be more accurate in detecting and quantifying deletions than pre-existing tools. We also show that PCR conditions and primersets must be carefully chosen to avoid biases in the retrieved variants and an increase in background noise, and established a lower detection limit of 0.5% heteroplasmic load for large deletions, and 1.5 and 2% for SNVs, for bulk DNA and single cells, respectively. Finally, the analysis of different single cells provided novel insights into mtDNA cellular mosaicism.
Introduction
Depending on the cell type, each human cell contains several hundreds to thousands of mitochondria, with each mitochondrion holding numerous copies of mitochondrial DNA (mtDNA).1 The mtDNA can contain small insertions and deletions (indels), single nucleotide variants (SNV) and large rearrangements that can exist in a homoplasmic or heteroplasmic state, with variable loads.2 mtDNA changes maternally transmitted at a high load may result in hereditary disease.3 It is typical for these mtDNA diseases to be caused by a single or a small number of changes that are present in the majority of the mtDNA molecules.4 Mitochondrial variants have also been associated with a number of different pathologies, such as Parkinson’s disease,5 multiple sclerosis6 and Huntington’s disease,7 and it has been shown recently that relatively low variant loads can deteriorate the health of the individuals and their offspring.8
In genetics, new discoveries have often come hand in hand with the development of novel technologies. In this sense, the in-depth study of large sets of genomes has become reality with the advent of massive parallel sequencing (MPS). This also holds true for the mtDNA and MPS at a sufficiently high depth allows the identification of SNVs and the simultaneous determination of their loads. Several groups have shown that thorough control of the experimental setup allows for a detection of SNVs at heteroplasmy levels as low as 1–1.6%,9, 10, 11, 12 0.2%13 and even 0.1% by using mother–child pairs.14 Their work has revealed, for instance, that individuals of the general population often carry mtDNA variants at low frequencies13, 14 and there are previously unsuspected levels of mtDNA diversity amongst cells of the same origin.11
Currently, there are a number of very good bioinformatic tools specifically designed for the SNV analysis of mtDNA MPS data, including MToolBox,15 MitoSeek16 and mtDNA-server.17 Conversely, the quantification of mtDNA rearrangements using MPS data has been, up to date, limited. In samples with high variant loads, mtDNA deletions may be visualized as gaps in the depth of sequencing.9, 12, 18 This approach, though, is not useful in samples with multiple deletions or rearrangements present at low individual loads. A few groups have used breakpoint mapping by identifying chimeric reads, which indeed enables the detection of lower frequency events, while simultaneously establishing the breakpoints and the load.19, 20 Conversely, their work did not include a thorough validation of this approach, leaving open questions such as what is the lower detection limit of this method, what is the false positive rate and what are the biases associated with the different methodological setup.
In this work, we optimized a protocol including both wet lab and bioinformatics procedures to detect mtDNA changes, with a focus on the detection of large rearrangements with a low heteroplasmic load. We tested several bioinformatics approaches to identify large mtDNA deletions in MPS data sets and found that a custom-made script, based on chimeric read identification, was the most effective in establishing the breakpoints and loads of very low frequency events. By studying the same region of the mtDNA using two different primersets, we identified PCR-specific artifacts, including false mtDNA rearrangements and biases in the retrieved frequency, depending on the primer set used and the PCR conditions. Finally, the protocol was downscaled for its use in single-cell analysis and validated on single fibroblasts and single muscle fibers.
Materials and methods
Fibroblast culture and single-cell isolation
Fibroblasts were cultured in F-12 Nutrient Mix Ham (Life Technologies, Thermo Fisher, Waltham, MA, USA) supplemented with 20% fetal calf serum, 0.5% penicillin/streptomycin and 1% glutamine. The cells were harvested at passage 4 and individually collected and lysed as previously described.21 Briefly, the cells were washed in individual PBS droplets by mouth-controlled pipetting. The cells were collected in reaction tubes containing 2.5 μL lysis solution (200 mM NaOH and 50 MM dithiothreitol). The cells were stored at −20 °C until PCR amplification. Directly before PCR analysis, the cells were lysed by incubation for 10 min at 65 °C. Single-muscle fibers, negatively stained for cytochrome c oxidase activity, were isolated using laser-capture microdissection (PixCell II laser-capture microscope, Arcturus, Thermo Fisher, Waltham, MA, USA) as previously described.22 A single-fiber section was captured on LCM transfer film on a CapSure cap and the non-captured material was removed with CapSure pads (Arcturus, Thermo Fisher). The cells were frozen at −20 °C in 10 μL of Pico Pure DNA Extraction solution (Arcturus, Thermo Fisher).
Bulk DNA extraction
DNA from blood was extracted from 7 mL whole blood with a Chemagic DNA blood kit special (7 mL) on a Chemagic Prepito-D instrument (PerkinElmer, Baesweiler, Germany), following the manufacturer’s instruction. DNA from fibroblast culture and muscle tissue was isolated by proteinase K–SDS lysis, followed by phenol–chloroform extraction and ethanol precipitation.
mtDNA enrichment
To obtain a selective and high coverage of the mitochondrial genome, we chose a PCR-based enrichment, which is an approach that can be reliably downscaled to the single-cell level. We tested two overlapping primer sets, designed to provide a specific amplification of the mtDNA and broad coverage. Set 1 generates an amplicon of 12.96 kb (5042f-1424r) and set 2 amplifies a region of 8.7 kb (8286f-421r). The sequences for primer set 1 were: set1f 5′-AGCAGTTCTACCGTACAAC-3′ and set1r 5′-ATCCACCTTCGACCCTTAAG-3′. For set 2, they were 5′-TCTAGAGCCCACTGTAAAGCTAACTT-3′ for set2f and 5′-AGTGCATACCGCCAAAAGATAAAATT-3′ for set2r. Fifty nanograms of DNA were amplified with the LongAmp Taq DNA Polymerase kit (New England Biolabs, Ipswich, MA, USA)) in a 50 μL reaction including 10 μL of 5 × LongAmp Buffer, 1.5 μL of 10 mM dNTPs, 2 μL of each primer (10 μM) and 2 μL (5 units) of DNA polymerase. The amplification protocol for set 1 included an initial denaturation of 30 s at 94 °C, followed by a touchdown step of 8 cycles with 15 s denaturation at 94 °C, 30 s of annealing at a starting temperature of 64 °C and subsequent decrease of 0.4 °C per cycle, and an elongation time of 11 min at 65 °C. After the touchdown step, 22 additional cycles were performed with 15 s denaturation at 94 °C, 30 s annealing at 61 °C and 11 min at 65 °C; the final step was an elongation of 11 min at 65 °C. For set 2, the elongation time was reduced to 7 min 25 s due to the shorter length of the amplicon. Absence of amplification of nuclear DNA was assessed for both sets using DNA extracted from mitochondria-lacking RhoZero cells.
Single-cell PCR
Tricine (2.5 μL of 200 mM; Sigma-Aldrich, Darmstadt, Germany) was added to the PCR reaction mix to buffer the lysis solution. The mix was prepared under a dedicated vertical flow to avoid contamination. After the lysis step, the mix was added directly to the tubes; the samples were then subjected to the same touchdown amplification protocol listed above but with 45 total cycles instead of 30.
Massive parallel sequencing
Long-range PCR products were sheared with a Covaris M220 sonicator (Life Technologies) to obtain an average fragment size of 350 bp. After the shearing, the samples were size-selected for fragments between 200 and 900 bp. The fragmented products underwent end repair, adenylation and paired-end adapters ligation with the TruSeq DNA PCR-Free Library Preparation Kit (Illumina, San Diego, CA, USA). The samples were pooled and sequenced on a Miseq with the MiSeq Reagent Micro Kit, v2 (Illumina). MPS was performed with paired-end reads of 2 × 150 bp, with an average sequencing error for the Miseq phiX internal control of 0.41%. Finally, we aimed for an average sequencing depth of around 6000 × (average in the experiments presented in this study: 7100 ×), to be able to accurately analyze low frequency events.
Data processing: variant calling and identification of large rearrangements
For the calling of SNVs and small indels, the files were first aligned to the mitochondrial revised Cambridge Reference sequence (rCRS, NC 012920.1) using BWA-MEM and sorted. This is followed by GATK23 realignment around indels and recalibration. Finally, variant calling itself was done using CLC Genomics Workbench (CLC Bio-Qiagen, Aarhus, Denmark). The frequency was set at 0.5% and SNVs were considered when having a quality >20, a forward to reverse ratio of >0.1 and a frequency >1%.24
For the quantification and identification of the exact breakpoints of large deletions present at low frequencies, we tested four tools designed for breakpoint recognition in the nuclear genome, namely Pindel version 0.2.5,25 Delly version 0.7.2,26 Platypus version 0.8.1,27 and GATK haplotype caller version 3.5,23 and compared it with the performance of a custom script. All pre-existing tools were used with default parameters with rCRS NC_012920.1 as the reference genome. Pindel variants were filtered to exclude deletions smaller than 150 bp. The allele frequency for deleted sequences was calculated as the variant allele divided by the sum of variant and reference alleles. Our custom script uses a local installation of Blastn28 to align all reads. Next, all reads mapping to two different regions of the mtDNA (ie, chimeric reads) are retrieved. Those with identical breakpoints are pooled. Next, the script adds all the incomplete alignments with discordant paired reads to the list of deletions if they match the location of breakpoints detected in the first processing. The final processing involves the calculation of the frequency of the deletions. This is calculated by dividing the number of chimeric reads containing the breakpoints of the deletion by the total number of reads of the position of the breakpoint. A detailed description of the script and the script code in Perl can be found in Supplementary Materials and Methods section. All variants have been submitted to the MITOMAP database (www.mitomap.org).
Results
Analysis of mixed DNA samples
To test the different tools for the detection of mtDNA deletions, along with SNV analysis, we performed experiments with samples that consisted of the mix of the DNA of two well-characterized individuals at different ratios (an overview of the characteristics of the different samples used in this study can be found in Supplementary Table 1). We mixed DNA1, a 100% full-length mitochondrial genome,24 with DNA2, carrying a large deletion with a load of approximately 80% (the sequencing results for this patient can be found in a section below). Both samples also carried different homoplasmic SNVs and DNA1 carried the SNV m.12071T>C at a load of 9–13%, the estimated load depending on the primer set used for the analysis. We generated mixes of 50, 25, 12.5, 5, 1, 0.5 and 0.1% of DNA2 in DNA1. For the SNV analysis, we used nine variants belonging to the haplogroup of DNA2 that were located outside of the deleted region. The load was calculated as the average loads of the nine SNVs. As DNA2 contained 80% of deleted molecules, the expected heteroplasmic loads for the deletion were of 40, 20, 10, 4, 0.8, 0.4 and 0.08%.
We analyzed these samples in three independent experiments, using two overlapping primer sets, to evaluate whether different PCR setups can lead to different results in identical samples. In two experiments we carried out 30 PCR cycles and in one we carried out 35 cycles. This was done to assess the PCR-induced bias in favor of mtDNA molecules carrying a large deletion, based on the notion that there could be a preferential amplification of shorter molecules. We evaluated the quantification of the large deletion using Delly, Platypus, Pindel, the GATK haplotype caller and our custom script. Only Pindel and our custom script were able to identify the deletion and give a quantitative report of the frequencies. The other tools only detected deletions in the initial 250 bp of the amplicon, which we later identified to be PCR artifacts (see below).
For both the SNVs and the deletions, the results show that both sets perform well in terms of relative quantification, as there is a very good correlation between the expected and observed frequency (Figures 1a and b). There is also a good correspondence between the SNV call and the deletion load, albeit that the method under-called the deletion loads in some cases. For both sets, we observe no drastic PCR-induced bias in the experiment using 35 cycles versus 30 cycles. Conversely, in terms of absolute quantification, set 2 overestimates the frequency of the mutated molecules and shows inconsistency between experiments. For both primer sets, we found that deletions spiked in at loads as low as 0.08% and SNVs at loads of 1% could be reliably detected, although in the case of set 2 these are overestimated. Finally, a detailed comparison of the SNVs found in the test samples with the SNVs found in the two DNA samples used to create the mixes identified three additional SNVs in two of the 24 sequenced mixes (data not shown). These SNVs were present at frequencies below 1.1% hence, we chose to increase the threshold for SNV detection to 1.5%. Finally, a second set of mixes was studied, using DNA6, which contained a larger deletion than DNA 2. The results were very similar to those in the previous experiments, suggesting that the size of the deletion has little impact on the quantification efficiency by this method (all results shown in Supplementary Figure 1).
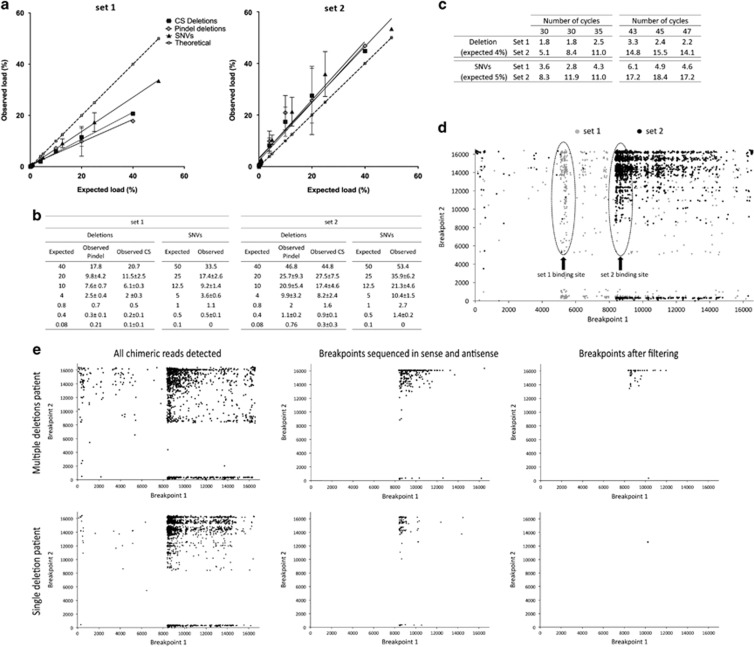
Figure 1.
(a,b) Results of the analysis of the spiked-in DNA samples with the two primer sets. The data points correspond to the analysis of DNA samples carrying 50, 25, 12.5, 5, 1, 0.5 and 0.1% of a set of SNVs and of 40, 20, 10%, 4, 0.8, 0.4 and 0.08% of a single large deletion. In a, the axes show the expected versus the observed frequency (in %), as obtained by the SNV analysis and the deletion analysis using Pindel and the custom script (CS). The error bars reflect the variability between independent experiments. (b) The tables show the data of all sequencing experiments. The values are all percentages and the average frequencies are shown with their SD (n=1 for 1% and 50% samples, n=3 for all the other data points). (c) Impact of the increase in number of PCR cycles on the quantification of the DNA mix containing 4% of the deletion and 5% of the SNVs (same sample as in a,b). All values are percentages. The results show that increasing the number of cycles results in a bias in the quantification of the deletion when using primer set 2. (d) Plotting of all the chimeric reads retrieved by the initial step of the custom script, from one DNA sample (4% mix). The breakpoints are plotted as x,y coordinates, in grey breakpoints obtained with set 1 and black the one found with set 2. The majority of breakpoints cluster in the regions within the first 250 bp of each amplicon (dotted circles). This, along with the fact that the deletions identified in set 2 are not found in set 1 strongly suggests that they are artifacts. Further data processing as illustrated in e filters these out. (e) Examples of the results of the sequential processing of the chimeric reads by the custom script in samples of patients with single and multiple deletions (DNA samples 2 and 3, respectively). The script initially retrieves a wide variety of chimeric reads (left side). Next, only breakpoints that are sequenced in sense and antisense are considered (middle). In the last step, several thresholds are applied that result in the detection of bona fide deletion sequences (right side).
Next, we downscaled the procedure to the single-cell level. First, we tested for PCR-induced amplification bias by amplifying 10 pg of DNA, roughly corresponding to the content of a single cell, of the 5% mixture DNA for 43, 45 and 47 cycles (Figure 1c). Although both primer sets showed a tendency towards higher frequency calls, both for the deletion and the SNVs, the strongest bias appeared after using primer set 2 when using higher numbers of PCR cycles, in line with the previous experiments. Set 2 showed an increase in the frequency of the deletion from 8.2 to 14.8% (80% increase) with the higher number of cycles (43 to 45) and set 1 from 2.01 to 2.64% (30% increase), when compared with 30–35 cycles. When considering SNVs, the increase in the higher cycles is from 10.4% to 17.6% (70% increase) for set 2 and from 3.6 to 5.2% (46% increase) for set 1. Overall, despite the observed increase, set 1 still showed an absolute quantification close to the expected value, whereas set 2 showed a stronger tendency towards bias in the results with the increase of cycles. Regarding the appearance of false SNVs, we identified two additional variants in one sample (Supplementary Table 2), both present at a frequency below 1.5%, leading us to increase the threshold to 2% for a reliable SNV detection in single cells.
False mtDNA rearrangements
Next to the expected deletion in the mixed DNA samples, we observed also molecules resembling bona fide deletions, albeit at low frequencies. The breakpoints of these low-frequency putative deletions significantly differ in the same sample depending on the primers used for the PCR. In fact, most of the very low-frequency deletions cluster to the initial 250 bp of the amplicon, in a primer set-dependent manner (Figure 1d). Furthermore, we did not detect the same deletion breakpoints for the overlapping regions of the primer sets. These observations led us to suspect that these deletions could be library preparation and/or PCR generated.
The first hypothesis was that DNA molecules resembling deletions are generated after the DNA shearing step and the ligation during the library preparation. Furthermore, if the library preparation procedure includes a PCR step, these artifacts can be exponentially amplified, appearing in the final data as low frequency deletions. We tested for this type of artifact generation by sequencing a pUC19. We found that the TruSeq DNA PCR-Free Sample Preparation (Illumina) resulted in minimal levels of false rearrangements. Supplementary Table 3 lists all putative rearrangements detected after the sequencing of the pUC19 with this protocol, in two independent experiments. As all detected (false) rearrangements were single events, sequenced in one sense only, we included a step in our custom script to remove all putative rearrangements sequenced only once or in one sense.
The second potential source of false deletions is the long-range PCR used to enrich for mtDNA sequences. To test this hypothesis we carried out single-molecule PCR. In this setup, the PCR was carried out on a DNA dilution where the template was theoretically one molecule. In this way, each PCR product represented the amplification of one molecule only, thus making low-frequency rearrangements impossible. We performed this experiment on two DNA samples and we pooled multiple single-molecule reactions from the same DNA (12 amplified products for DNA5 and 7 for DNA1) to obtain the required amount of DNA for the library preparation. Both experiments showed deletions at very low frequencies (below 0.05%), strongly suggesting they are PCR artifacts (Supplementary Table 4).
Consequently, although the experiments on the different DNA mixtures indicated that we could detect deletions down to frequencies of 0.1%, to minimize the risk of calling false positives we set the lower limit of detection for deletions at 0.5%. Furthermore, due to the clustering of the majority of the artifacts to the first and last 250 bp of the amplicons, along with a very strong imbalance in heavy/light strand coverage in that region, we decided to exclude these regions from further analysis. Figure 1e shows an example of the outcome of each different step in the processing of the data, starting with the full range of detected chimeric reads, which are then filtered by only considering breakpoints sequenced in sense and antisense, and finishing by applying the thresholds described above.
DNA samples of patients with large mtDNA deletions
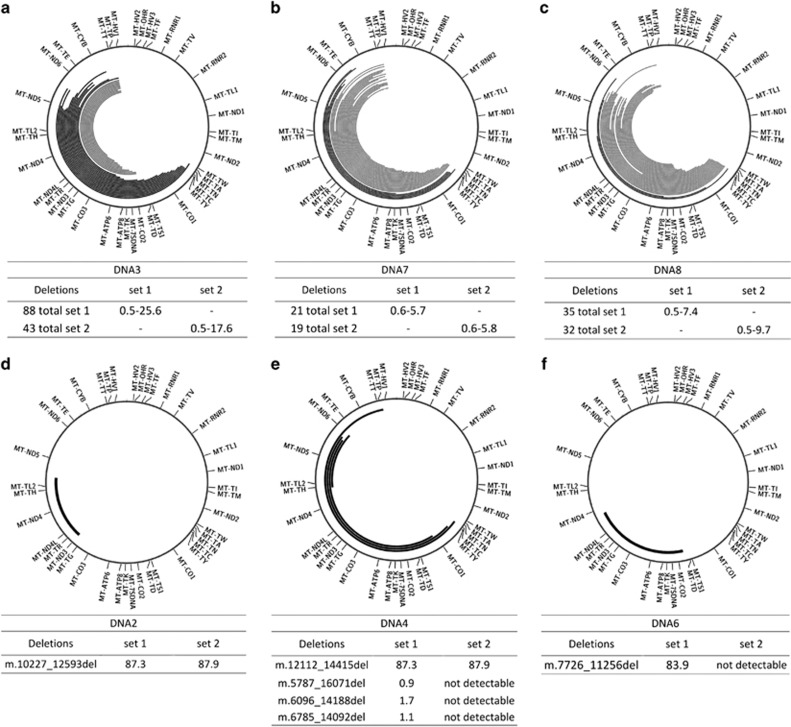
As a proof-of-principle, we analyzed, using both primer sets, six DNA samples extracted from muscle homogenates of patients known to carry deletions in their mtDNA as established by Southern blot analysis and some from patients suspected of mitochondrial disease (details on the patients can be found in the Supplementary Table 1). The results are summarized in the circos plots29 of Figure 2. In DNA samples with single or few deletions, both the custom script and Pindel reliably identified the same breakpoints. Conversely, in DNA3, DNA7 and DNA8, containing multiple mtDNA deletions, we found discrepancies between the two analysis methods. Mainly, Pindel was not able to detect deletions with one breakpoint in the hypervariable region (Figure 2a–c grey deletions). This is possibly due to the design of the Pindel algorithm to detect simple deletions in large genomes. For this reason we discontinued the use of Pindel in the later experiments, and focused our work on the use of our custom script. With this approach, we found various deletions with frequencies ranging from 0.5 to 25.6% in DNA3, from 0.5 to 5.7% in DNA7 and from 0.5 to 9.6% for DNA8 (Figures 2a–c). DNA2 showed a major single deletion of 2367 bp (m.10227_12593del) in around 80% of the molecules, DNA6 showed a larger deletion of 3531bp in 84% of the molecules and DNA4 showed a major single deletion of 2304 bp (m.12112_14415del) at a frequency of 90%, with three additional deletions found at very low frequency (Figure 4d–f). The data sets from both primer pairs identified the same breakpoints for DNA2 and DNA4, giving a comparable load of the deleted molecules. DNA6 could not be analyzed with set 2. The patients carrying multiple deletions also showed a good correspondence between the two primer sets, although with some differences in the estimated frequencies (Supplementary Tables 5, 6 and 7).
Figure 2.
Graphical representation of the deletions found in six patients with known or suspected mitochondrial disease. In black, deletions identified by both Pindel and the custom script. In grey, breakpoints identified only with the custom script. (a) Sequencing of a patient (DNA3) carrying the mutation in the Twinkle gene, known to be associated with the formation of multiple deletions. The plot shows the 30 deletions with highest frequency, the full dataset can be found in Supplementary Table 5. (b,c) sequencing results of two other patients (DNA7 and DNA8) with a suspicion of mitochondrial disease found to carry multiple deletions at low frequency. Extensive data can be found in Supplementary Tables 6 and 7. (d–f) show the results of patients carrying single deletions (DNA2, DNA4 and DNA6). The tables show the breakpoints and the retrieved frequencies with the different primer sets. All values are percentages. ‘Not detectable’: outside the amplicon of set 2.
Single-cell analysis
First, we studied 11 single fibroblasts from the donor of DNA1 and matched the results with the bulk DNA extracted from a fibroblast culture of the same individual. We amplified eight fibroblasts with primerset1 and three with primer set 2. The results show an overall good correspondence between the SNVs called using the two primersets on the bulk DNA of the fibroblast culture. The results are summarized in Table 1 and the complete data on single cells is shown in Supplementary Table 8. When looking at the individual cells, the data showed interesting variation in terms of the segregation of SNVs, illustrating a form of somatic mosaicism. First, the variant m.12071T>C, which is present in the blood at a heteroplasmy of 11–13%, and of 9% in the bulk DNA of the fibroblast culture, appears only in two of the 11 cells and this at a frequency of 100%, whereas the other cells were homoplasmic for the reference allele. Second, we observed two other SNVs (m.12850A>G and m.9276G>A), only present in the fibroblast culture and not in the blood at 15 and 3%, respectively. These SNVs also appear to segregate in a similar way as the m.12071T>C variant, being present at a higher level in some cells while absent in the majority of them. Furthermore, they appear to be linked, co-segregating at similar frequencies in two distinct single cells (Table 1, cell 4 and 9). The variant m.15617G>A was also found in only one out of 11 cells at higher frequency compared to the bulk (Table 1, cell 8). In terms of deletions, none were observed in the bulk DNA or in the single cells.
Table 1. Frequencies of the variants found in fibroblast bulk DNA versus single cells, after the analysis using primer set 1 and primer set 2.
| Single fibroblasts | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bulk DNA fibroblasts | set 1 | set 2 | |||||||||||
| Variants | set 1 | set 2 | Cell 1 | Cell 2 | Cell 3 | Cell 4 | Cell 5 | Cell 6 | Cell 7 | Cell 8 | Cell 9 | Cell 10 | Cell 11 |
| m.414T>G | 6.2 | Not detectable | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.6933T>C | 2.4 | Not detectable | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.8188A>G | 2.5 | Not detectable | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.9276G>A | 3.1 | 3.1 | 0 | 0 | 0 | 14.9 | 0 | 0 | 0 | 0 | 20.1 | 0 | 0 |
| m.9947G>A | 3.1 | 2.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.9987T>C | 1.7 | 1.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.12071T>Ca | 9.9 | 8.0 | 97.8 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| m.12850A>G | 15.6 | 15.8 | 0 | 0 | 0 | 99.9 | 0 | 0 | 0 | 0 | 96.1 | 0 | 0 |
| m.13552G>A | 1.0 | 3.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.14033G>A | 2.3 | 1.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| m.15617G>A | 4.2 | 6.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40.9 | 0 | 0 | 0 |
| m.16166A>G | 3.0 | 2.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
All the variants detected in the bulk are indicated and matched with the single cells. The complete list of variants in the single cells can be found in Supplementary Table 8. All values are percentages. ‘Not detectable’, outside the amplicon of set 2.
Only variant retrieved in blood, at a frequency of 10.8% with set 1 and 9.6% with set 2.
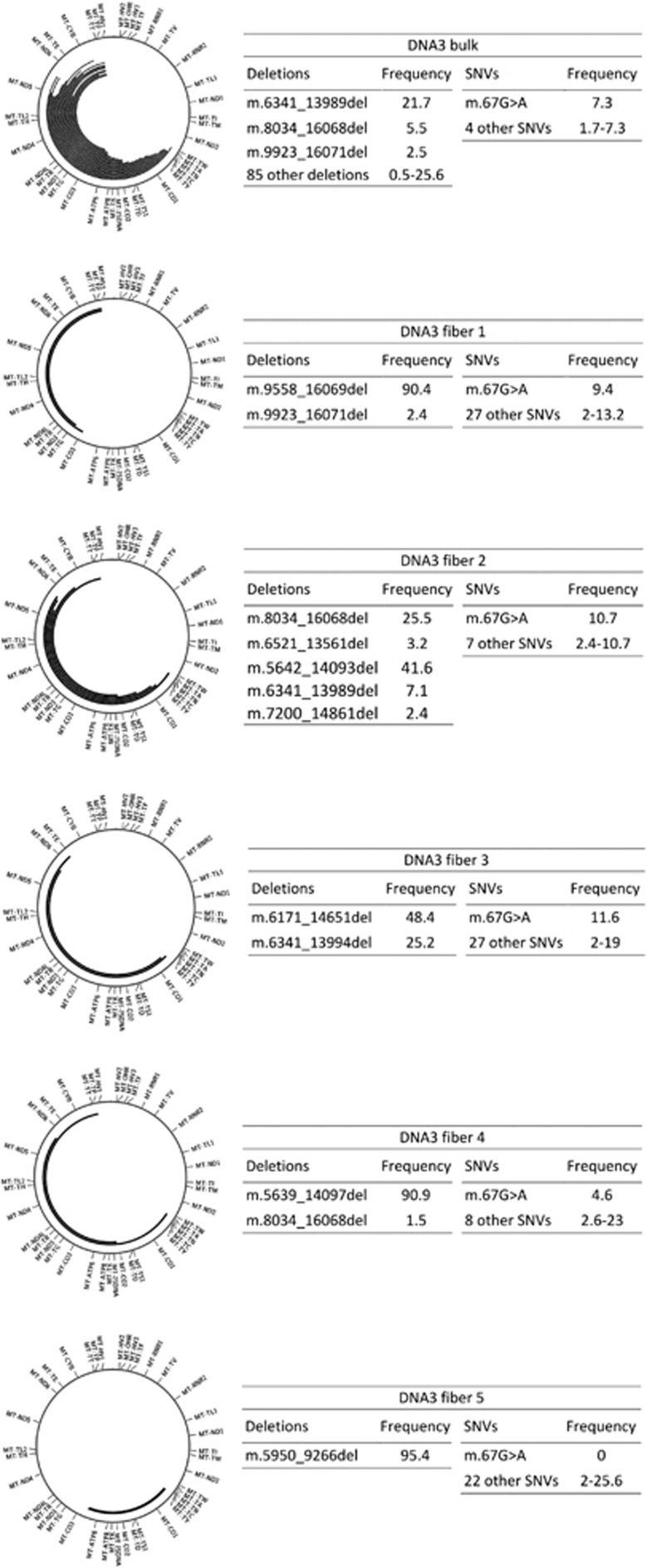
Next, to study large deletions at the single-cell level, we compared the data of the whole muscle DNA of the patient containing multiple deletions (DNA3) with the data obtained from five individual COX− muscle fibers with set 1. We excluded from SNV analysis the deleted regions that showed coverage depth below 1000. In the single cells, we observed a higher number of variants with low heteroplasmic loads as compared to the bulk muscle sample, where only five variants were detected above the threshold of 1.5% (Figure 3 and Supplementary Table 9). Variant m.67G>A is present in bulk DNA and most of the single cells in a heteroplasmic status (Figure 3). In each of the individual muscle fibers we observed a small number of deletions, in three out of five cases with frequencies over 90% (Figure 3). The deletions detected in the single muscle fibers were not identical, but some overlap was found with the deletions identified in the bulk muscle DNA of the same patient and among the different fibers. The frequencies retrieved in bulk muscle (Supplementary Table 5) were always lower than the ones found in single cells (Figure 3), again illustrating the somatic mosaicism as found in fibroblasts.
Figure 3.
Overview of the deletions and SNVs found in five single COX− muscle fibers, as compared with the analysis of the bulk DNA extracted from muscle homogenate. The large deletions are represented in the circos plots and their frequencies, along with that of the SNVs are in the adjacent boxes (all values are percentages). We have highlighted the variant m.67A>G, present in most cells and in the bulk DNA.
Discussion
In this work, we tested whether tools currently available for the detection of rearrangements in the nuclear genome could perform the same task on data obtained by ultra-deep sequencing of the mtDNA and tested a custom-made bioinformatics approach. We controlled different aspects of the full analysis process, including PCR error rates, different primer sets performance and MPS-induced artifacts, to set reliable detection thresholds. Finally, we evaluated our setup in the analysis of single cells.
During the optimization of the mtDNA enrichment step, we found that SNV detection is affected by the number of PCR cycles used for the enrichment of the mtDNA in the samples. When using 30 PCR cycles, as in the case of the analysis of bulk DNA, we found a 1.5% threshold to be most reliable, which is in line with the works of Zhang et al9 and He et al,11 in which the authors set the lower threshold to 1.3 and 1.6%, respectively. Conversely, the use of a higher number of cycles as required for the analysis of minute DNA samples (such as 10 pg) or single cells, increases the threshold to 2% to avoid false positive calling.
For the detection of large deletions, we found that from the different published tools we tested only Pindel was capable of identifying single deletions at very low frequencies, but it was unreliable in the study of samples with multiple deletions, and particularly in deletions with breakpoints occurring towards the end of the mtDNA sequence. On the other hand, our custom pipeline did not show this limitation, and allowed for the detection of events appearing in single MPS reads. However, this type of analysis resulted in the detection of a high number of deletions, which strongly resembled those reported by Williams et al20 in human putamen by using a similar bioinformatics approach (i.e. the identification of incompletely aligned MPS reads). In this report, the authors did not further control for the reliability of their approach, and we speculate that together with the “true” rearrangements, they were detecting the same artifacts found by us, strongly resembling bona fide deletions at low individual frequencies. We experimentally addressed the nature of the rearrangements, and found that they are likely artifacts generated during the PCR amplification. These artifacts are characterized by breakpoints falling in regions very close to either ends of the amplicon, and with a very strong bias in the sense/antisense ratio, depending on which end of the amplicon is closest. Furthermore, of the two primersets used, only one proved to yield consistent and reliable results, while the other showed an overestimation of the deletions called, and an increase in the number and frequency of the artifacts. A possible explanation for this is an amplification bias by this primer set in favor of shorter fragments (ie, the deleted molecules). Taken together, these findings highlight the critical importance of the PCR setup and the use of controls to reliably establish the thresholds for variant detection. Future work should address the use of multiple robust primers, such as locked or biotinylated oligonucleotides, for an accurate validation of deletions and an improvement in the detection of rare events.
From a biological point of view, the analysis of single cells carried out in this study illustrates the type of insight that single-cell work can provide into the mechanisms of mtDNA segregation and cellular diversity. For instance, in one of our control individuals, we found one SNV present at constant frequency (~10–13%) in both bulk DNA sources we analyzed: blood and fibroblast culture. However, the analysis of single fibroblasts of the same individual showed that the frequency detected in bulk DNA was actually an average, resulting from the fact that the individual had one cell homoplasmic for the SNV for every eight–nine cells homoplasmic for the wild-type nucleotide. Other groups have made analogous findings by other methods and in other cell types, such as blood cells,30, 31 neurons, glia32 and single muscle fibers.33 Regarding large mtDNA rearrangements, our analysis of the patient with a mutation in the Twinkle gene showed another type of mosaicism, in this case disease-linked.34 The comparison of the deletions retrieved in whole muscle homogenates and in single COX-deficient muscle fibers showed that each single cell contained a few deletions at a high frequency, while the full tissue sample showed an average of all the different deletions in the single cells. These findings are in line with the work of others, albeit with different methodological approaches, showing that individual muscle fibers carry clonally expanded deletions, each with unique breakpoints.33, 19
In conclusion, our study presents a controlled setup for the detection and quantification of large deletions with frequencies as rare as 0.5% in MPS data sets, in both bulk DNA samples and single cells. We demonstrate that in order to achieve a comprehensive and reliable analysis, the setup of the experiments must be thoroughly controlled and validated. Especially in the initial PCR enrichment, suboptimally designed primers can cause selective amplification and generate additional artifacts resembling bona fide mtDNA deletions, biasing frequency calculation for events at low frequencies. Finally, the data we report also provides interesting information on the nature of mtDNA heteroplasmy, with different examples of cell-to-cell mosaicism and variant segregation in different cells and tissues.
Data access
The SRA accession number for the data reported in this paper is SRA: SRP090128.
Acknowledgments
This work was supported by the wetenschappelijk fonds Willy Gepts of the University Hospital UZ Brussel and the Methusalem grant to Karen Sermon of the Research Council of the Vrije Universiteit Brussel. FZ is co-funded by the Methusalem grant and S.I.S.Me.R. reproductive medicine unit, Bologna. DB is supported by the Flemish Institute for Biotechnology (VIB).
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Shokolenko IN, Alexeyev MF: Mitochondrial DNA: a disposable genome? Biochim Biophys Acta 2015; 1852: 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Chinnery PF: The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 2015; 16: 530–542. [DOI] [PubMed] [Google Scholar]
- Schapira AHV: Mitochondrial diseases. Lancet 2012; 379: 1825–1834. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S: An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature 1989; 339: 309–311. [DOI] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM et al: High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 2006; 38: 515–517. [DOI] [PubMed] [Google Scholar]
- Campbell GR, Kraytsberg Y, Krishnan KJ et al: Clonally expanded mitochondrial DNA deletions within the choroid plexus in multiple sclerosis. Acta Neuropathol 2012; 124: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Moody JP, Edgerly CK et al: Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet 2010; 19: 3919–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JM, Stewart JB, Hagström E et al: Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature 2013; 501: 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cui H, Wong L-JC: Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin Chem 2012; 58: 1322–1331. [DOI] [PubMed] [Google Scholar]
- Rebolledo-Jaramillo B, MS-W Su, Stoler N et al: Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc Natl Acad Sci 2014; 111: 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC et al: Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010; 464: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Li F, Chen D et al: Comprehensive next-generation sequence analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med 2013; 15: 388–394. [DOI] [PubMed] [Google Scholar]
- Payne BaI, Wilson IJ, Yu-Wai-Man P et al: Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 2013; 22: 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li C-I, Sheng Q et al: Very low-level heteroplasmy mtDNA variations are inherited in humans. J Genet Genomics 2013; 40: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C, Simone D, Diroma MA et al: MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics 2014; 30: 3115–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li J, Li C-I, Shyr Y, Samuels DC: MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics 2013; 29: 1210–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissensteiner H, Forer L, Fuchsberger C et al: mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic Acids Res 2016; 44: gkw247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneca S, Vancampenhout K, Van Coster R et al: Analysis of the whole mitochondrial genome: translation of the Ion Torrent Personal Genome Machine system to the diagnostic bench? Eur J Hum Genet 2015; 23: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygiel Ka, Tuppen Ha, Grady JP et al: Complex mitochondrial DNA rearrangements in individual cells from patients with sporadic inclusion body myositis. Nucleic Acids Res 2016; pp 1–17. [DOI] [PMC free article] [PubMed]
- Williams SL, Mash DC, Züchner S, Moraes CT: Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet 2013; 9: e1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C, Le Caignec C, De Rycke M et al: Whole-genome multiple displacement amplification from single cells. Nat Protoc 2006; 1: 1965–1970. [DOI] [PubMed] [Google Scholar]
- Cao Z, Wanagat J, McKiernan SH, Aiken JM: Mitochondrial DNA deletion mutations are concomitant with ragged red regions of individual, aged muscle fibers: analysis by laser-capture microdissection. Nucleic Acids Res 2001; 29: 4502–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancampenhout K, Caljon B, Spits C et al: A bumpy ride on the diagnostic bench of massive parallel sequencing, the case of the mitochondrial genome. PLoS One 2014; 9: e112950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z: Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009; 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO: DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012; 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I et al: Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 2014; 46: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS et al: BLAST: a more efficient report with usability improvements. Nucleic Acids Res 2013; 41: W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I et al: Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Ogasawara Y, Kajigaya S et al: Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood 2007; 109: 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y, Nakayama K, Tarnowka M et al: Mitochondrial DNA spectra of single human CD34+ cells, T cells, B cells, and granulocytes. Blood 2005; 106: 3271–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Lin MT, Zheng K et al: Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging 2005; 26: 1343–1355. [DOI] [PubMed] [Google Scholar]
- Payne BaI, Wilson IJ, Hateley Ca et al: Mitochondrial aging is accelerated by anti-retroviral therapy through the clonal expansion of mtDNA mutations. Nat Genet 2011; 43: 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN: Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet 2009; 18: 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.