Abstract
Primary open-angle glaucoma (POAG) is the most common chronic optic neuropathy worldwide. Epidemiological studies show a robust positive relation between intraocular pressure (IOP) and POAG and modest positive association between IOP and blood pressure (BP), while the relation between BP and POAG is controversial. The International Glaucoma Genetics Consortium (n=27 558), the International Consortium on Blood Pressure (n=69 395), and the National Eye Institute Glaucoma Human Genetics Collaboration Heritable Overall Operational Database (n=37 333), represent genome-wide data sets for IOP, BP traits and POAG, respectively. We formed genome-wide significant variant panels for IOP and diastolic BP and found a strong relation with POAG (odds ratio and 95% confidence interval: 1.18 (1.14–1.21), P=1.8 × 10−27) for the former trait but no association for the latter (P=0.93). Next, we used linkage disequilibrium (LD) score regression, to provide genome-wide estimates of correlation between traits without the need for additional phenotyping. We also compared our genome-wide estimate of heritability between IOP and BP to an estimate based solely on direct measures of these traits in the Erasmus Rucphen Family (ERF; n=2519) study using Sequential Oligogenic Linkage Analysis Routines (SOLAR). LD score regression revealed high genetic correlation between IOP and POAG (48.5%, P=2.1 × 10−5); however, genetic correlation between IOP and diastolic BP (P=0.86) and between diastolic BP and POAG (P=0.42) were negligible. Using SOLAR in the ERF study, we confirmed the minimal heritability between IOP and diastolic BP (P=0.63). Overall, IOP shares genetic basis with POAG, whereas BP has limited shared genetic correlation with IOP or POAG.
Introduction
Primary open-angle glaucoma (POAG) is an intraocular pressure (IOP)-related, chronic optic neuropathy that is a leading cause of blindness worldwide.1 Randomized controlled trials (RCTs) indicate that treatments designed to lower IOP slow disease progression.2 Compared with an IOP of 16 mm Hg or less, an IOP of 35 mm Hg is associated with a 39-fold increased risk of POAG.3 A more modest but consistent positive association between blood pressure (BP) and IOP also exists. Specifically, a meta-analysis of epidemiological surveys found that every 10 mm Hg increase in systolic blood pressure (SBP) or 5 mm Hg increase in diastolic blood pressure (DBP) was associated with 0.26 mm Hg and 0.17 mm Hg increases in IOP, respectively.4
The relationship between BP and POAG is more complex. In pooled analyses, every 10 mm Hg increase in SBP was associated with a 1% increased risk of POAG while every 5 mm Hg increase in DBP was associated a 2% increased risk of POAG.4 However, observational studies report a robust inverse relation between ocular perfusion pressure (OPP) and POAG, where OPP is calculated as either mean arterial pressure (MAP), SBP, or DBP minus IOP.5, 6, 7 Since a lower BP in the context of higher IOP may result in poor optic nerve perfusion, glaucoma drugs are frequently scrutinized for their effects on both IOP and OPP.8, 9 Finally, post hoc analysis of two RCTs found that lower OPP was associated with glaucoma progression.10, 11
Several common gene variants have been identified for IOP12 and BP.13 IOP variants, like TMCO114and CAV115 are also associated with POAG, and a multi-locus IOP genetic panel was associated with POAG.16 Nevertheless, more data regarding IOP genetic variants in relation to POAG subtypes defined by IOP at disease presentation is needed. Furthermore, the relations between BP and IOP single-nucleotide polymorphisms (SNPs) and between BP and POAG SNPs are not known. The International Glaucoma Genetics Consortium (IGGC), the International Consortium of Blood Pressure (ICBP), and the National Eye Institute Glaucoma Human Genetics Collaboration Heritable Overall Operational Database (NEIGHBORHOOD) contain genome-wide data for IOP, BP traits, and POAG, respectively. We use these data sets to explore genetic correlations focusing first on genome-wide significant loci (those with P-value for association <5 × 10−8) for IOP and BP in relation to POAG. Second, we examined the co-heritability between IOP BP traits, and POAG across the genome using linkage disequilibrium (LD) score regression.17, 18, 19 Finally, we compared our genome-wide estimate of heritability between IOP and BP to an estimate based solely on direct measures of these traits and pedigree information in the Erasmus Rucphen Family (ERF) study.
Methods
The NEIGHBORHOOD: a genome-wide association meta-analysis for POAG
The NEIGHBORHOOD data set included 37333 participants of European ancestry (3853 cases and 33480 controls) enrolled as part of eight independent studies from the United States for a genome-wide meta-analysis of POAG. Additional details regarding the cohort composition and analyses performed are described in the original paper.20 All participants had genome-wide genotype data (each contributing data set underwent site-specific genotyping, quality control and imputation of untyped variants using the 1000 Genomes Project reference panel21) and POAG phenotype data. Sample and genotype call rate was ≥95% for each site, and variants with minor allele frequencies (MAF) <5% or imputation quality scores <0.7 were removed. For each study, logistic regression for POAG overall, as well as high-tension glaucoma (HTG) and normal tension glaucoma (NTG) subtypes, adjusted for age, sex, nominally significant principal components of genotype and study-specific covariates was performed. The HTG and NTG subtypes were defined by maximum known IOP ≥22 mm Hg and <22 mm Hg, respectively. Finally, meta-analysis study-specific results were performed using the inverse variance-weighted method and P-values were corrected using genomic control.22
The IGGC and ICBP: genome-wide association studies for IOP and BP
In brief, the IGGC performed a GWAS for IOP that included 35296 multi-ancestry participants from 18 studies;12 however, to reduce the potential impact of population structure, our analysis only considered the meta-analysis results of 27 558 individuals of European ancestry from 14 studies. The ICBP performed a GWAS for BP traits that included 69 395 individuals of European ancestry from 29 studies.13 Participants from both consortia had genome-wide data for ~2.5 million genotyped or imputed SNPs (Supplementary Table 1).
The ERF study: a pedigree with IOP and BP data
The ERF study is an independent family-based cohort containing 2519 participants with simultaneous IOP and BP measurements.23 Details regarding the ERF and how IOP and BP were measured are available in the Supplementary note. We used SOLAR (Sequential Oligogenic Linkage Analysis Routines) to decompose the phenotypic correlation between IOP and BP into genetic and environmental components while accounting for kinship calculated from the pedigree file.24, 25 The genetic correlation between IOP and BP estimated solely from kinship was compared with genome-wide measures of genetic correlation for these traits as described below.
Genetic risk score analyses
For each IOP and DBP locus with MAF >0.05 reported to have at least one genome-wide significant (ie, P<5 × 10−8) associated SNP in the IGGC and ICBP, respectively, we selected the most significant variant based on its P-value for association. We chose to focus on DBP loci for our genetic risk score (GRS) analysis because two studies showed the strongest association between lower diastolic OPP (DBP minus IOP) and POAG as opposed to other OPP parameters (systolic OPP and mean arterial OPP).7, 26 We evaluated the effect of IOP and DBP genome-wide significant SNPs in relation to POAG, HTG and NTG in the NEIGHBORHOOD using GRS’s that aligned alleles associated with increasing IOP and decreasing DBP. GRS’s are conventionally derived using individual level genotype data and then tested for association with the outcome of interest using standard linear or logistic regression. However as proposed by Aschard,27 such a test can be performed using summary statistics data. In brief, one can collect summary statistics for the SNPs that form the GRS and use an inverse variance-weighted sum meta-analysis of individual SNP effect estimates. If 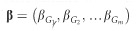 represents the reported effect of the m selected SNP and
represents the reported effect of the m selected SNP and 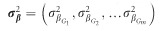 represents the variance of each estimate, under the assumption of independence between the SNPs,
represents the variance of each estimate, under the assumption of independence between the SNPs, 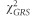 , the χ2 for the GRS effect on POAG over all SNPs Gi,
i=1…m, can be derived as:
, the χ2 for the GRS effect on POAG over all SNPs Gi,
i=1…m, can be derived as:
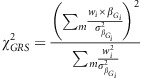 |
where wi is the weight assigned to SNP Gi (commonly, the marginal genetic effect estimate). For unweighted GRSs, as performed in the present study, we set all weights to one. We chose unweighted GRSs for all analyses, as it is difficult to define homogenous weight between the IGGC and ICBP when merging IOP and DBP SNPs, respectively, to form the OPP panel, which represented the combined effect of IOP-increasing gene variants and DBP-decreasing gene variants in relation to POAG.
Genetic correlation and heritability
We used genome-wide summary statistics results across all SNPs for IOP and BP parameters (DBP, SBP, MAP, and pulse pressure) to derive the genetic correlation between those traits and glaucoma phenotypes (POAG, HTG, and NTG) using LD score regression. In brief, given genome-wide summary statistics of two phenotypes, defined as two vectors of z-scores, z1 and z2, and assuming a polygenic model, the expected value of the product z1jz2j for a SNP j equals:17
where ρg is the genetic covariance, M is the number of SNPs analyzed, N1, N2 and Ns are the sample size for phenotype 1, phenotype 2, and the overlapping sample size, respectively; and ρ, is the correlation between the two phenotypes. Finally, lj, the LD score of a variant j is the sum of 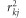 , the squared correlation between the SNP j and all SNPs k=1…M, and is expressed as
, the squared correlation between the SNP j and all SNPs k=1…M, and is expressed as 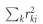 . It follows that the genetic correlation, the parameter of interest, can be estimated using the slope from the regression of z1jz2j on LD Score.
. It follows that the genetic correlation, the parameter of interest, can be estimated using the slope from the regression of z1jz2j on LD Score.
As recommended, we applied the LD score on genetic variants that passed a stringent filter, removing SNPs with poor imputation quality (r2 for imputation<0.9), MAF <5% and SNPs whose effect estimates were derived with <66% of the total sample.18 As this information was only partly available in studies we analyzed, we instead used the systematic filtering proposed for use with the LD score regression software. It specifies analyzing only SNPs that are part of the ~1.2 M common SNPs from the HapMap 3 panel, as these variants have r2 for imputation >0.9 in most studies. Moreover, LD Score of these variants, derived using a European ancestry panel, was readily available as part of the LD Score software. Finally, for binary traits the LD score estimates are based on a liability threshold model (LTM).19In brief, the LTM assumes binary traits are determined by an unobserved normally distributed liability. Individuals whose liability is above a given threshold τ are cases, whereas others are controls. For a population prevalence K, the threshold τ can be derived as τ=CDF−1(1−K), where CDF is the standard normal cumulative distribution function. Hence, this derivation requires the population and sample prevalence of the disease. In this analysis, we assumed that the population prevalence of POAG, HTG, and NTG equaled 0.01, 0.007, and 0.003, respectively, based on the Rotterdam Study,28 which consist of European derived Caucasians.
The LD score regression also allows estimating 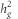 , the genome-wide heritability explained by common variants. However, this estimate might be biased downward when GC correction has been applied to the summary GWAS statistics. As non-GC corrected GWAS was available for POAG and IOP phenotypes, we used these data to estimate
, the genome-wide heritability explained by common variants. However, this estimate might be biased downward when GC correction has been applied to the summary GWAS statistics. As non-GC corrected GWAS was available for POAG and IOP phenotypes, we used these data to estimate 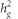 . In brief, the LD score has a linear relationship with SNP
. In brief, the LD score has a linear relationship with SNP 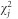 , with a slope proportional to heritability:
, with a slope proportional to heritability:
where N and M are the study sample size and the number of SNPs analyzed and a measures the contribution of confounding biases, such as cryptic relatedness and population stratification. As for the genetic correlation analyses, estimates of heritability were derived using only the ~1.2 M common SNPs from the HapMap 3 panel.
Partitioning POAG and IOP heritability
To gain further insight into the heritability of glaucoma-related traits, we also estimated how heritability partitioned across gene sets expressed in eye tissues. To do so, we leveraged publicly available gene expression data from human eye tissues provided by the National Eye Institute (https://neibank.nei.nih.gov/index.shtml). For each tissue selected, we included genes matching a known gene in the UCSC database with a clone count ≥1. The tissue included ‘ciliary’ (referring to the anterior segment uveal tract responsible for generating aqueous humor, 201 genes), cornea (259 genes), lens (135 genes), optic nerve (349 genes), retina (552 genes), retina pericyte (574 genes), retinal pigment epithelium - choroid (1146 genes) and trabecular meshwork (394 genes). Since a prior report found no relation between hearing loss and POAG29 we included the cochlea as a reference tissue (1797 genes). Using this information we constructed tissue-specific annotation where each SNP within 50kb of the start and end site of the selected genes defined a category. We estimated the proportion of heritability explained by each category using the following formula, as implemented in the LD score software:
where C indexes categories and τC represents the per-SNP contribution to heritability of category C, referred further as the heritability coefficient. As recommended,19 we also included for each analysis a set of 53 (overlapping) baseline annotation regions including a range of functional categories (eg, coding, UTR, promoter and intronic regions and so on) that allow for more accurate estimation of enrichment.
In practice, we performed a single analysis including all tissue-specific and baseline annotation regions, thus estimating all τC jointly. For each category, we reported the fold-enrichment, that is, the ratio of the proportion of total heritability explained to the proportion of SNPs falling into that category, and its associated P-value. We also report the significance of each τC separately. Although the enrichment estimates and its P-value allow for inferences to be made on the contribution of these variants to the heritability for each category, the later P-value addresses the question of whether the category contributes to the outcome after accounting for other functional categories. Data access URL: http://jass.pasteur.fr/RawData.html
Results
IOP and BP genome-wide significant variants in relation to POAG
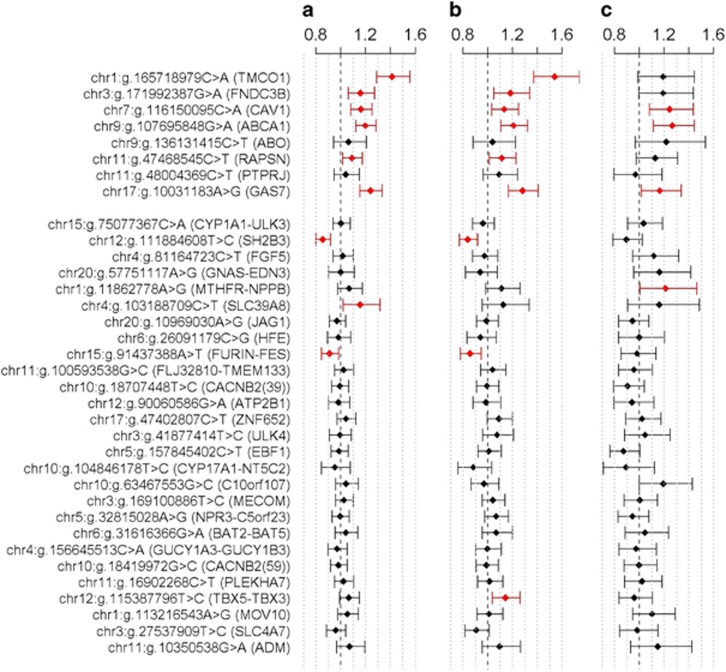
Five of eight genome-wide significant IOP SNPs showed nominally significant positive association with the glaucoma phenotypes (POAG, HTG, and NTG) in NEIGHBORHOOD (Figure 1 and Supplementary Table 2). The strongest association was observed for hg19 chr1:g:165718979C>A (TMCO1; P=5.9 × 10−13, P=1.3 × 10−12, and P=0.065, for POAG, HTG, and NTG, respectively). Many of those associations remained significant after Bonferroni correction accounting for the 105 tests performed (Supplementary Table 2; P=4.76 × 10−4).
Figure 1.
Association of intraocular pressure and diastolic blood pressure gene variants with primary open-angle glaucoma (POAG; n=3853 cases) and the high-tension glaucoma (HTG; n=1774 cases) and normal tension glaucoma (NTG; n=725 cases) subtypes in the NEIGHBORHOOD consortium (n=37 333 total participants). Odds ratios and 95% confidence intervals (indicated with error bars) for association between 8 intraocular pressure loci (upper panel), and 27 diastolic blood pressure loci (bottom panel) and POAG (a), HTG (b), and NTG (c). SNPs with nominally significant associations are highlighted in red. P-values can be found in Supplementary Table 2. The full colour version of this figure is available at European Journal of Human Genetics online.
Five out of the 27 genome-wide DBP SNPs showed nominal significance with at least one glaucoma phenotype but the direction of effects were inconsistent (Figure 1 and Supplementary Table 2) in NEIGHBORHOOD. Specifically, three DBP SNPs (in SLC39A8, MTHFR-NPPB, and TBX5-TBX3) were positively related with at least one glaucoma trait whereas two DBP SNPs (in SH2B3 and FURIN-FES) were negatively related with glaucoma traits. None of these associations remain significant after Bonferroni correction with the exception of rs3184504 (SH2B3), which displayed strong negative associations with POAG and HTG (Supplementary Table 2; P=6.2 × 10−6 and P=1.2 × 10−4, respectively). Interestingly, the SH2B3 locus is in high LD with hg19 chr12:g.111932800C>T (ATXN2, r2=0.90 in CEU samples from the 1000 genomes project),21 which was associated with POAG in a recent report.20 Only one DBP SNP was nominally associated with IOP (MOV10, P=0.018, Supplementary Table 3).
The GRS of IOP-increasing SNPs showed strongly significant positive correlation with POAG and its subtypes (OR=1.18, P=1.8 × 10−27; OR=1.20, P=1.1 × 10−20; and OR=1.18; P=6.2 × 10−9, for POAG, HTG, and NTG, respectively; Table 1). However, there was no relation between the GRS for DBP-decreasing SNPs and POAG or its subtypes (P=0.93, P=0.97, and P=0.96, for POAG, HTG, and NTG, respectively). The significance of the positive association between the GRS of OPP-lowering alleles (the combined effect of IOP-increasing alleles and DBP-lowering alleles) and the various glaucoma phenotypes (OR=1.04, P=7.4 × 10−7; OR=1.04, P=1.6 × 10−5; and OR=1.04, P=6.1 × 10−3 for POAG, HTG, and NTG, respectively) was markedly attenuated when compared to the association between the GRS for IOP-increasing alleles and glaucoma phenotypes.
Table 1. Polygenetic risk scores for intraocular pressure (IOP), diastolic blood pressure (DBP) and the combined ocular perfusion pressure (OPP) panels in relation to primary open-angle glaucoma (POAG; n=3 853 cases) and its high-tension glaucoma (HTG; n=1 774 cases) and normal tension glaucoma (NTG; n=725 cases) subtypes in the NEIGHBORHOOD consortium (n=37 333).
| Association studied | Odds Ratio | 95% lower bound | 95% upper bound | P-value |
|---|---|---|---|---|
| IOP SNPs–POAG | 1.18 | 1.14 | 1.21 | 1.8 × 10−27 |
| DBP SNPs–POAG | 1 | 0.98 | 1.01 | 0.93 |
| OPP SNPs–POAG | 1.04 | 1.02 | 1.05 | 7.4 × 10−7 |
| IOP SNPs–HTG | 1.2 | 1.15 | 1.25 | 1.1 × 10−20 |
| DBP DBP–HTG | 1 | 0.98 | 1.02 | 0.97 |
| OPP SNPs–HTG | 1.04 | 1.02 | 1.06 | 1.6 × 10−5 |
| IOP SNPs–NTG | 1.18 | 1.12 | 1.25 | 6.2 × 10−9 |
| DBP SNPs–NTG | 1 | 0.97 | 1.03 | 0.96 |
| OPP SNPs–NTG | 1.04 | 1.01 | 1.07 | 0.0061 |
Abbreviation: SNPs, single-nucleotide polymorphisms. NB: DBP allele variants coded as allele associated with lower diastolic blood pressure and IOP allele variants coded as allele with higher intraocular pressure so as to mimic the OPP term. Polygenetic risk scores are unweighted. Bold values represent p-values<0.05.
Genetic correlations and heritability between IOP, BP traits and glaucoma traits using data from the IGGC, ICBP, and NEIGHBORHOOD
The genetic correlation between BP phenotypes using LD score regression was very high (eg, the genetic correlation between DBP and SBP was 0.884, P<10−300), in agreement with previous studies (Table 2).30 IOP showed strong genetic correlation with POAG (0.485; SE=0.110; P=2.1 × 10−5) and HTG (0.548; SE=0.116; P=9.6 × 10−4). Possibly owing to a smaller number of documented NTG cases (n=725), the genetic correlations between IOP and NTG (0.210; SE=0.198; P=0.28) and between HTG and NTG (0.241; SE=0.290; P=0.41) were not statistically significant. Conversely, we did not observe any genetic correlations between IOP and BP phenotypes, nor between BP phenotypes and POAG (P≥0.32). We also reported estimates of 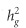 for our glaucoma phenotypes explained by common SNPs. Applying the LD score regression we obtained
for our glaucoma phenotypes explained by common SNPs. Applying the LD score regression we obtained 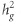 of 0.102 (SE=0.021), 0.099 (SE=0.033), 0.165 (SE=0.082), and 0.116 (SE=0.018) for POAG, HTG, NTG, and IOP, respectively. In sensitivity analysis, doubling POAG prevalence from 1 to 2% did not appreciably alter
of 0.102 (SE=0.021), 0.099 (SE=0.033), 0.165 (SE=0.082), and 0.116 (SE=0.018) for POAG, HTG, NTG, and IOP, respectively. In sensitivity analysis, doubling POAG prevalence from 1 to 2% did not appreciably alter 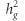 estimates (eg,
estimates (eg, 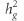 increased from 0.102 to 0.122 for POAG).
increased from 0.102 to 0.122 for POAG).
Table 2. Genetic correlation matrix for the various glaucoma and blood pressure traits using summary data from NEIGHBORHOOD (n=37 333), the International Glaucoma Genetics Consortium (n=27 558) and the International Consortium of Blood Pressure (n=69 395).
| HTG | NTG | IOP | DBP | SBP | MAP | PP | |
|---|---|---|---|---|---|---|---|
| POAG | 0.820 (0.071) P=2.5 × 10−30 | 0.593 (0.150) P=1.2 × 10−4 | 0.485 (0.110) P=2.1 × 10−5 | 0.023 (0.028) P=0.42 | 0.008 (0.027) P=0.77 | 0.012 (0.031) P=0.67 | 0.011 (0.032) P=0.73 |
| HTG | − | 0.241 (0.290) P=0.41 | 0.548 (0.166) P=9.6 × 10−4 | 0.036 (0.036) P=0.32 | −0.015 (0.034) P=0.67 | −0.004 (0.035) P=0.99 | −0.027 (0.040) P=0.49 |
| NTG | − | − | 0.210 (0.198) P=0.28 | 0.023 (0.055) P=0.67 | 0.039 (0.058) P=0.50 | 0.052 (0.058) P=0.37 | 0.031 (0.062) P=0.62 |
| IOP | − | − | − | 0.004 (0.021) P=0.86 | −0.009 (0.019) P=0.65 | −0.011 (0.021) P=0.58 | 0.005 (0.025) P=0.83 |
| DBP | − | − | − | − | 0.884 (0.012) p ~ 0 | 0.95 (0.009) p ~ 0 | 0.71(0.022) p ~ 0 |
| SBP | − | − | − | − | − | 0.93 (0.01) p ~ 0 | 0.883(0.015) p ~ 0 |
| MAP | − | − | − | − | − | − | 0.760 (0.022) p ~ 0 |
Abbreviations: BP, systolic blood pressure; DBP, diastolic blood pressure; HTG, high-tension glaucoma (1774 cases); IOP, intraocular pressure; MAP, mean arterial pressure; NTG, normal tension glaucoma (725 cases); POAG, primary open-angle glaucoma (3853 cases); PP, pulse pressure; SBP, systolic blood pressure. The off-diagonal indicates the genetic correlation (standard error) and the corresponding P-value.
Statistically significant values are in bold and italicized.
p ~ 0 refers to P<10–300 power.
As IOP and BP traits were not measured in the ICBP and IGGC respectively, we sought to validate our estimate of shared heritability of these traits in the ERF study where IOP and BP were directly measured in defined pedigrees (see Supplementary note). Shared heritability estimates using purely family-based data were not significantly different from zero between IOP and the four BP phenotypes (P≥0.63; Supplementary Table 4), supporting our LD score regression results.
Partitioning of glaucoma trait heritability with ocular tissue genes using data from the NEIGBORHOOD and IGGC
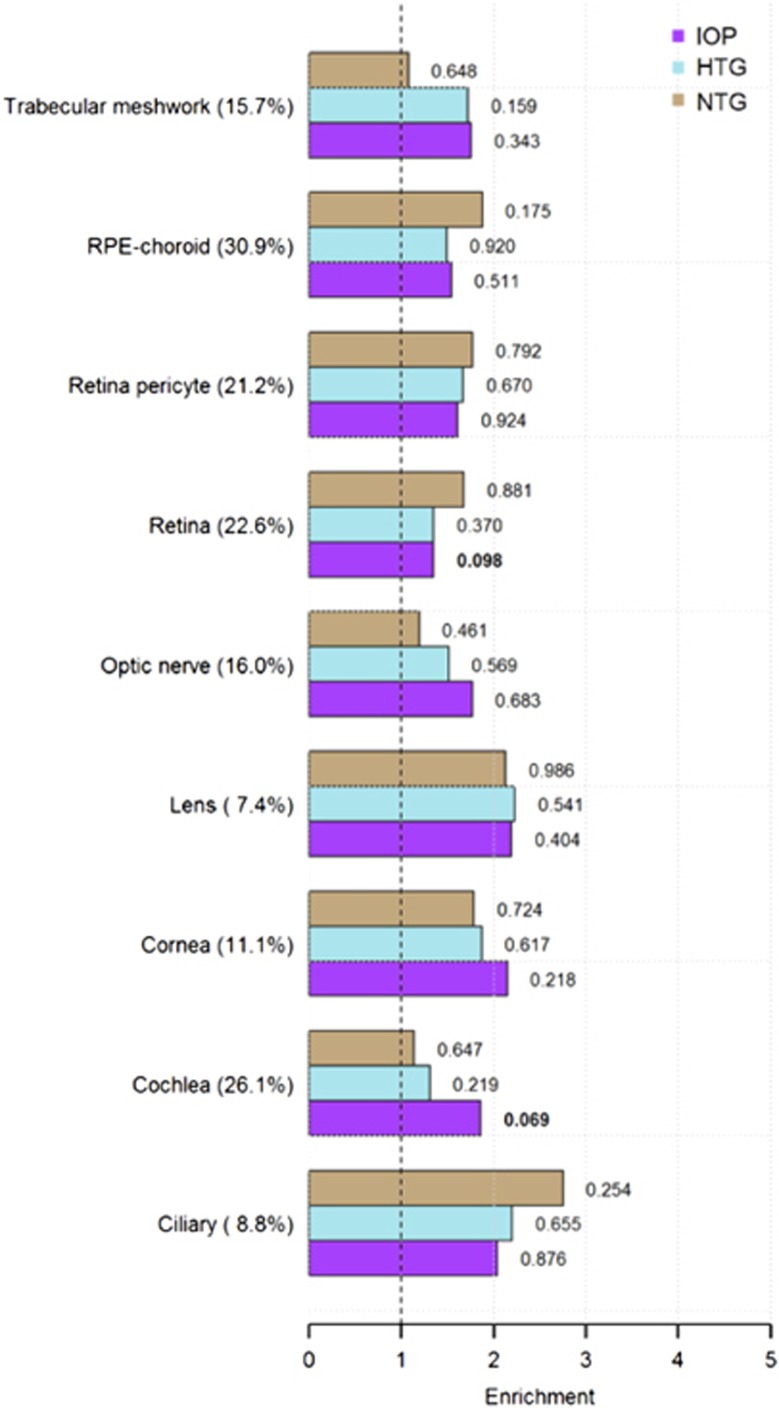
Overall, after accounting for the strong overlap between the various ocular tissue SNP sets and the heterogeneous representation of these SNP collections in the available GWAS (7.4–30.9%), we find no significant enrichment of heritability for IOP and glaucoma traits in any ocular tissue gene set (Figure 2). Supplementary Table 5 provides further detail regarding heritability partitioning between IOP, glaucoma traits and ocular tissue genes.
Figure 2.
Heritability enrichment of selected glaucoma-related traits in various ocular tissues and the cochlea in NEIHBORHOOD and the International Glaucoma Genetics Consortium. See text for how tissue-specific SNP sets were assembled. Tissue categories with percent available GWAS SNPs are represented on the vertical axis. Heritability enrichment (E) estimates are presented on the horizontal axis along with tissue-specific heritability P-values. These P-values, as opposed to the E P-values, are provided here because they represent a more conservative estimate of significance that simultaneously accounts for all tissue categories. E estimates for IOP are derived from the IGGC, whereas similar estimates for HTG and NTG are from the NEIHBORHOOD. For a complete list of tissue-specific E values, E P-values and tissue-specific heritability P-values for all glaucoma traits, see Supplementary Table 5. NB: Ciliary refers to ciliary body. RPE=retinal pigment epithelium. IOP=intraocular pressure, HTG=high-tension glaucoma, NTG=normal tension glaucoma.
Discussion
In this large multi-cohort study of European ancestry we found strong associations between genome-wide significant IOP SNPs and POAG, as expected for an IOP-related optic neuropathy. In fact, even the association between the IOP GRS and NTG, where the highest known IOP was <22 mmHg, was highly significant (OR=1.18; P=6.2 × 10−9; Table 1). Overall, significant variants from this analysis were already known to be associated with POAG phenotypes. chr7:g.116150095C>A (CAV1/CAV2 intergenic region) is a known POAG SNP;15 chr1:g.165718979C>A (TMCO1), chr17:g.10031183A>G (GAS7), and chr9:g.107695848G>A (ABCA1) were identified by the NEIGHBORHOOD GWAS;20 and chr3:g.171992387G>A (FNDC3B) has been associated with POAG as part of studies that used NEIGHBORHOOD data.12, 31 However, we found weak, inconsistent relations between genome-wide significant DBP SNPs and POAG. Finally, we found minimal genome-wide shared genetic heritability between IOP and BP traits and these results were replicated in the ERF study.
We formed a SNP panel that combined DBP and IOP variants in an effort to mimic OPP (a measure of DBP minus IOP), a popularly derived biomarker felt to reflect optic nerve perfusion.5, 6, 7 We found that the relation between OPP and POAG appeared to be driven by the IOP variants. The only BP SNP showing an association signal with POAG after correction for multiple comparisons (see Table 1 and Supplementary Table 2) was chr2:g.111884608T>C (SH2B3). Interestingly, this variant encodes a missense amino-acid change and has already been found associated with other outcomes such as type 1 diabetes32 and autoimmune hepatitis type 1,ref.33 suggesting that this association is driven by a pleiotropic biological mechanism independent of the other variants.
The shared heritability across the genome between IOP and POAG was high (0.49; SE=0.11; P=2.1 × 10−5; Table 2), but between IOP and BP parameters, (≤0.005; P≥0.58; Table 2) or between BP parameters and POAG (≤0.023; P≥0.42; Table 2) the shared genome-wide heritability was minimal. Taken collectively, our work suggests that, unlike IOP, BP is not in the causal pathway of POAG from a genetic perspective. Furthermore, the genetic correlation between IOP and HTG across the genome was also high (0.55±0.16; P=9.6 × 10−4 Table 2). Conversely, Although an IOP GRS was significantly associated with NTG, an estimate of genetic correlation between IOP and NTG across the genome suggests a lower heritability (0.21±0.20; P=0.28; Table 2), though this estimate is unstable due to the smaller sample size (n=725 NTG cases). Finally, across the genome, there was non-significant shared heritably between NTG and HTG (0.24±0.29; P=0.41; Table 2). To date, only the CDKN2BAS region unequivocally appears to be shared between these two glaucoma phenotypes,34, 35 underscoring the need to carefully phenotype POAG for known maximum IOP and to perform additional high-throughput genotyping, particularly for NTG.
Investigators have pointed out that it is impossible to dis-entangle the BP and IOP components of OPP when considering the latter term in relation to POAG.36 In fact, in the Rotterdam Study, the relation between OPP and incident open-angle glaucoma was null when adjustment for baseline IOP was made.37 Our agnostic genomic approach is consistent with epidemiologic data, suggesting that IOP is more important than BP in predicting POAG risk across a range of presenting IOPs. Epidemiological research is clear that there is a modest positive relation between BP and IOP4 but our data suggests that this is not mediated by the examined genetic factors. The mechanism by which higher BP might be associated with higher IOP is unknown but could represent the pleotropic effects of multiple environmental influences.
Our estimate for heritability of POAG is consistent with a prior classic twin study (h2=0.11)38 but lower than a contemporary estimate using GWAS data (h2=0.42).39 The sample size for the classic twin study was small. In the latter study, cases and controls were drawn from different sources and the number of cases (1105) was considerably less than in our data set (3853). On the other hand, IOP has a reported heritability of 0.5640 but our estimate from the IGGC based on common variants is 0.12. The missing IOP heritability could be partially related to imprecision in phenotyping as IOP measurements vary by time of day,41 season of year,42 central corneal thickness (CCT),43 variation in IOP measurement methodology44 and other factors. CCT, a highly heritable trait (h2=0.95),45 was collected in IGGC but there is no agreed upon algorithm to adjust IOP based upon the CCT.46 In the IGGC, the other variables effecting IOP variability were not systemically adjusted for in the collection of this glaucoma-related trait.
Study limitations include our candidate SNP analyses, which focused on selected genome-wide significant variants. These variants together capture only a small proportion of the total genetic component of those traits, and more extensive panels might show different characteristics. For example future work might compare SNP effects for variants showing only suggestive genome-wide significance or for variants annotated for functional characteristics. A recent meta-analysis47 uncovered an additional 31 BP loci but many of these are rare variants and assessing the more common ones in relation to IOP and glaucoma traits will not change our results about shared genetic correlation across the genome. Nonetheless, it is still possible that there is a small subset of BP genetic markers related to IOP and glaucoma traits. Next, for co-heritability estimates, whereas we did not observe genetic correlation between BP traits and glaucoma traits, isolated shared genetic loci between BP parameters and glaucoma traits may exist when very large data sets are considered. Furthermore, our findings are derived from individuals of European ancestry and it is not clear if they apply to other ethnicities. Finally, as previously discussed, the LD score regression approach is sensitive to GC correction,17 which was applied to the BP meta-analysis. This may result in a slight underestimation of genetic correlation between BP and POAG traits, though our replication analysis in the ERF study confirmed that if any correlation exists, its magnitude is likely very low. The LD score regression also relies on the assumption that the single SNP effect sizes are normally distributed. While violation of this assumption does not bias the regression, it would increase the standard error, making heritability and co-heritability estimates unstable.
In summary, using the largest data sets available to date for IOP, BP, and POAG we confirm a strong genetic link between IOP and POAG but we cannot detect any substantial shared genetic effect between BP and IOP, nor between BP and POAG. Thus, if BP contributes to POAG by altering optic nerve perfusion, it does so via non-genetic effects or genetic influences we could not detect.
Acknowledgments
NIH EY015473 supports this work. The numerous other grants that support this work are listed as Supplementary material. Web Resources: The LD score regression analyses were performed using the implementation of the LD score method provided at: https://github.com/bulik/ldsc.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY: Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 2014; 121: 2081–2090. [DOI] [PubMed] [Google Scholar]
- Boland MV, Ervin AM, Friedman DS et al: Comparative effectiveness of treatments for open-angle glaucoma: a systematic review for the U.S. Preventive Services Task Force. Ann Intern Med 2013; 158: 271–279. [DOI] [PubMed] [Google Scholar]
- Sommer A, Tielsch JM, Katz J et al: Relationship between intraocular pressure and primary open angle glaucoma among white and black Americans. The Baltimore Eye Survey. Arch Ophthalmol 1991; 109: 1090–1095. [DOI] [PubMed] [Google Scholar]
- Zhao D, Cho J, Kim MH, Guallar E: The association of blood pressure and primary open-angle glaucoma: a meta-analysis. Am J Ophthalmol 2014; 158: 615–627. [DOI] [PubMed] [Google Scholar]
- Leske MC, Wu SY, Hennis A, Honkanen R, Nemesure B: Risk factors for incident open-angle glaucoma: the Barbados Eye Studies. Ophthalmology 2008; 115: 85–93. [DOI] [PubMed] [Google Scholar]
- Topouzis F, Wilson MR, Harris et al: Association of open-angle glaucoma with perfusion pressure status in the Thessaloniki Eye Study. Am J Ophthalmol 2013; 155: 843–851. [DOI] [PubMed] [Google Scholar]
- Tielsch JM, Katz J, Sommer A, Quigley HA, Javitt JC: Hypertension, perfusion pressure, and primary open-angle glaucoma. A population-based assessment. Arch Ophthalmol 1995; 113: 216–221. [DOI] [PubMed] [Google Scholar]
- Rossetti L, Sacchi M, Karabatsas CH et al: Comparison of the effects of bimatoprost and a fixed combination of latanoprost and timolol on 24-hour blood and ocular perfusion pressures: the results of a randomized trial. BMC Ophthalmol 2015; 15: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januleviciene I, Siaudvytyte L, Diliene V et al: Comparison of intraocular pressure, blood pressure, ocular perfusion pressure and blood flow fluctuations during dorzolamide versus timolol add-on therapy in prostaglandin analogue treated glaucoma subjects. Pharmaceuticals (Basel) 2012; 5: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leske MC, Heijl A, Hyman L, Bengtsson B, Dong L, Yang Z: Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology 2007; 114: 1965–1972. [DOI] [PubMed] [Google Scholar]
- De Moraes CG, Liebmann JM, Greenfield DS, Gardiner SK, Ritch R, Krupin T: Risk factors for visual field progression in the low-pressure glaucoma treatment study. Am J Ophthalmol 2012; 154: 702–711. [DOI] [PubMed] [Google Scholar]
- Hysi PG, Cheng CY, Springelkamp H et al: Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet 2014; 46: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret GB, Munroe PB, Rice KM et al: Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 2011; 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon KP, Macgregor S, Hewitt AW et al: Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet 2011; 43: 574–578. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Hewitt AW et al: Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet 2010; 42: 906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham YC, Liao J, Vithana EN et al: Aggregate effects of intraocular pressure and cup-to-disc ratio genetic variants on glaucoma in a multiethnic asian population. Ophthalmology 2015; 122: 1149–1157. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V et al: An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev et al: Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015; 47: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK et al: LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JN, Loomis SJ, Kang JH et al: Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet 2016; 48: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium1000 Genomes Project Consortium, Abecasis GR 1000 Genomes Project Consortium, Auton et al: An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roder K: Genomic control for association studies. Biometrics 1999; 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- Henneman P, Aulchenko YS, Frents RR et al: Prevalence and heritability of th emetabolic syndrome and its individual components in a Dutch osilate: the Erasmus Rucphen Family Study. J Med Genet 2008; 45: 572–577. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J: Mulitpoint qunatitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998; 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J: Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 1997; 14: 953–958. [DOI] [PubMed] [Google Scholar]
- Bonomi L, MarchiniG, Marraffa M et al: Vascular risk factors for primary open angle glaucoma: the Egna-Neumarkt Study. Ophthalmology 2000; 107: 1287–1293. [DOI] [PubMed] [Google Scholar]
- Aschard H: A perspective on interaction effects in genetic association studies. Genet Epidemiol 2016; 40: 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dielemens I, Vingerling JR, Wolfs RC, Hofman A, Grobbee DE, deJong PT: The prevalence of primary open-angle glaucoma in a population-based study the Netherlands. The Rotterdam Study. Ophthalmology 1994; 10: 1851–1855. [DOI] [PubMed] [Google Scholar]
- Tryggvason G, Jonasson F, Cotch MF et al: Hearing in older adults with exfoliation syndrome/exfoliation glaucoma or primary open-angle glaucoma. Acta Ophthalmol 2016; 94: 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn MJ, Schut AF, Aulchenko YS et al: Heritability of blood pressure traits and the genetic contribution to blood pressure variance explained by four blood-pressure-related genes. J Hypertens 2007; 25: 565–570. [DOI] [PubMed] [Google Scholar]
- Lu Y, Vitart V, Burdon KP et al: Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet 2013; 45: 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P et al: Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009; 41: 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer YS, van Gerven NM, Zwiers et al: Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology 2014; 147: 443–452. [DOI] [PubMed] [Google Scholar]
- Wiggs JL, Yaspan BL, Hauser MA et al: Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degenerationin glaucoma. PLoS Genet 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hughes G, Chen X et al: Genetic variants associated with different risks for high tension glaucoma and normal tension glaucoma in a Chinese population. Invest Ophthalmol Vis Sci 2015; 56: 2595–2600. [DOI] [PubMed] [Google Scholar]
- Khawaja A, Crabb DP, Jansonius NM: Time to abandon over-simplified surrogates of ocular perfusion pressure in glaucoma research. Acta Ophthalmologica 2015; 93: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas WD, Wolfs RC, Hofman A, deJOng PT, Vingerling JR, Jansonius WM: Ocular perfusion pressure and the incidence of glaucoma: real effect or artifact. The Rotterdam Study. Invest Ophthalmol Vis Sci 2011; 52: 6875–6881. [DOI] [PubMed] [Google Scholar]
- Tekiara JM: Genetic factors in open-angle (simple and capsular) glaucoma. A population-based twin study. Acta Ophthalmol (Copenh) 1987; 65: 715–720. [DOI] [PubMed] [Google Scholar]
- Cuellar-Partida G, Craig JE, Burdon KP et al: Assessment of polygenic effects links primary open-angle glaucoma and age-related macular degeneration. Sci Rep 2016; 6: 26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo PG, Hewitt AN, Hammond CJ, Mackey DA: The heritability of ocular traits. Surv Ophthalmol 2010; 44: 561–583. [DOI] [PubMed] [Google Scholar]
- Sit AJ: Intraocular pressure variations: causes and clinical significance. Can J Ophthalmol 2014; 49: 484–488. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Xi XR, Lu HJ, Wu XD, Huang YB, Shiarkar E: Effect of seasons upon intraocular pressure in healthy population of China. Korean J Ophthalmol 1996; 10: 29–33. [DOI] [PubMed] [Google Scholar]
- Doughty MJ, Zaman ML: Human corneal thickness and its impact on intraocular pressure measures: a review and meta-analysis approach. Surv Ophthalmol 2000; 44: 367–408. [DOI] [PubMed] [Google Scholar]
- Lamparter J, Hoffmann EM: Measuring intraocular pressure by different methods. Ophthalmologe 2009; 106: 676–682. [DOI] [PubMed] [Google Scholar]
- Dimasi DP, Burdon KP, Craig JE: The genetics of central corneal thickness. Br J Ophthalmol 2010; 94: 971–976. [DOI] [PubMed] [Google Scholar]
- Park SJ, Ang GS, Nicholas S, Wells AP: The effect of thin, thick and normal corneas on Goldmann intraocular pressure measurements and correction formulae in individual eyes. Ophthalmology 2012; 119: 443–449. [DOI] [PubMed] [Google Scholar]
- Liu C, Kraja A, Smith JA et al: Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet 2016; 48: 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.