Abstract
Background
The bacteria of the genus Bartonella are obligate parasites of vertebrates. Their distribution range covers almost the entire world from America, Europe, Asia to Africa and Australia. Some species of Bartonella are pathogenic for humans. Their main vectors are blood-sucking arthropods such as fleas, ticks and blood-feeding flies. One such dipteran able to transfer vector-borne pathogens is the deer ked (Lipoptena cervi) of the family Hippoboscidae. This species acts as a transmitter of Bartonella spp. in cervid hosts in Europe.
Methods
In the present study, 217 specimens of deer ked (Lipoptena cervi) were collected from 26 red deer (Cervus elaphus) hunted in January 2014. A short fragment (333 bp) of the rpoB gene was used as a marker to identify Bartonella spp. in deer ked tissue by PCR test. A longer fragment (850 bp) of the rpoB gene was amplified from 21 of the positive samples, sequenced and used for phylogenetic analysis.
Results
The overall prevalence of Lipoptena cervi infection with Bartonella spp. was 75.12% (163/217); 86.67% (104/120) of females and 60.82% (59/97) of males collected from red deer hunted in the Strzałowo Forest District in Poland (53°45′57.03″N, 21°25′17.79″E) were infected. The nucleotide sequences from 14 isolates (Bartonella sp. 1) showed close similarity to Bartonella schoenbuchensis isolated from moose blood from Sweden (GenBank: KB915628) and human blood from France (GenBank: HG977196); Bartonella sp. 2 (5 isolates) and Bartonella sp. 3 (one isolate) were similar to Bartonella sp. from Japanese sika deer (GenBank: AB703149), and Bartonella sp. 4 (one isolate) was almost identical to Bartonella sp. isolated from Japanese sika deer from Japan (GenBank: AB703146).
Conclusions
To the best of our knowledge, this is the first report to confirm the presence of Bartonella spp. in deer keds (Lipoptena cervi) in Poland by molecular methods. Bartonella sp. 1 isolates were most closely related to B. schoenbuchensis isolated from moose from Sweden and human blood from France. The rest of our isolates (Bartonella spp. 2–4) were similar to Bartonella spp. isolated from Japanese sika deer from Japan.
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2413-0) contains supplementary material, which is available to authorized users.
Keywords: Bartonella spp., Bartonella schoenbuchensis, Lipoptena cervi, Cervus elaphus
Background
The genus Bartonella comprises small, Gram-negative bacteria which act as obligate intracellular parasites of vertebrates. About 30 species, as well as three subspecies, have been described [1]. The zoonotic reservoir for some Bartonella species (B. schoenbuchensis, B. baciliformis and B. quintana), is composed of wild mammals, which usually only possess asymptomatic bacteraemia. The prevalence of bacteremia in wild animals is often very high, ranging from 50 to 95% in selected rodent and ruminant populations [2, 3] and strongly depends on the season [3]. As the reservoir hosts include a wide range of wild mammals, including typically ruminants, rodents and carnivores, the infections are easily spread and have been noted all over the world [2]. A good example is that of ruminant infections: Bartonella infection has been recorded in cattle in three different countries (Thailand, Guatemala and Georgia) and in buffalo (Bubalus bubalis) from Thailand [4]. The prevalence of Bartonella infection in these regions varies between 10 and 90%. Similar investigations in Japan found no infection in cattle, but recorded a prevalence of Bartonella sp., 67.5% in Honshu sika deer (Cervus nippon centralis) and 50% in Yezo sika deer (Cervus nippon yesoensis) [5].
Many Bartonella species are considered as human pathogens and causative agents of zoonotic diseases: B. bacilliformis, the agent of Carrion’s disease and Verruga peruna is the chronic delayed stage of infection; B. quintana, the agent of trench fever and bacillary angiomatosis; B. henselae, the agent of cat scratch disease and of bacillary angiomatosis; B. clarridgeiae, B. elizabethae, B. vinsonii subsp. arupensis, B. vinsonii subsp. berkhoffii, B. alsatica, B. koehlerae and B. washoensis all cause endocarditis [6–8]. The symptoms of human bartonellosis vary with regard to the bacterial species and general condition of the patient; however, Bartonella infection most often manifests as various cardiovascular, neurological and rheumatologic conditions [8, 9].
Bartonellosis has been observed in humans throughout Europe, Asia and North America. One of the most important species is Bartonella schoenbuchensis, which is found in cattle, wild animals (such as the cervids) and humans [10–12]. It has been identified in wild living roe deer (Capreolus capreolus) in Germany [10], in cattle in France [11] and in humans; B. schoenbuchensis was first detected in France [12].
Bartonella are transmitted by blood-sucking arthropods, such as lice, flies, fleas and ticks, many of which are emerging pathogens of humans and animals [2, 13–15]. Dehio et al. [16] identified B. schoenbuchensis in Lipoptena cervi collected from red deer and roe deer. Halos et al. [14] reported the presence of Bartonella spp. in hippoboscide flies, and suggested that flies can be a vector for pathogens. Other authors [14, 17] have found Bartonella spp. in wingless adult deer keds.
Lipoptena cervi are blood-sucking parasites which belong to a highly specialized family of louse flies (Diptera: Hippoboscidae) [18, 19]. This species has a Palaearctic distribution and occurs in Europe, Asia and North America; it is known to be present in many countries, including Sweden, Norway, Japan, the USA and Finland [20, 21]. In America, it is regarded as an invasive species, believed to have been transported with deer from Europe in the 1800s. This species was identified in the USA for the first time during the Second World War [18]. In Europe, the most common blood-sucking ectoparasites of mammals belonging to this family are louse fly (Hippobosca equina), parasitizing cows and horses, deer keds (Lipoptena cervi and Lipoptena fortisetosa), parasitizing cervids, and sheep ked (Melophagus ovinus), which ectoparasites of sheep [22, 23]. Lipoptena fortisetosa in Europe could be introduced with Japanese sika deer (Cervus nippon). This cervids were introduced to England in 1860, and their number and range have since increased [24]. From England, the Japanese sika deer were late introduced to Poland, where two species of Lipoptena have been identified: L. cervi and L. fortisetosa [25, 26].
Although the dominant hosts for two species of Lipoptena (L. cervi and L. fortisetosa) are cervids (Alces alces, Cervus elaphus, Cervus elaphus maral, Cervus nippon, Dama dama, Capreolus capreolus and Rangifer tarandus), the insects can attack a wide range of animals, including bovids (Ovis aries musimon, Bison bonasus, cattle, Capra aegagrus hircus and Ovis aries), carnivores (domestic dogs, Vulpes vulpes and Meles meles) and suids (Sus scrofa) [27]. The most important hosts of Lipoptena cervi in Europe are red deer (Cervus elaphus), roe deer (Capreolus capreolus), moose (Alces alces) and sika deer (Cervus nippon), while Japanese sika deer (Cervus nippon) are the predominate host in Japan, and white-tailed deer (Odocoileus virginianus), moose (Alces alces) and cattle (Bos taurus) in North America [21, 28, 29].
Generally, Lipoptena cervi is found swarming in large numbers on the host. Deer keds most frequently bite animal hosts; they can land on humans, but rarely bite in this case. Nevertheless, some authors have reported ked bites on humans [30, 31].
The life-cycle of Lipoptena cervi begins with free-ranging, winged adult deer keds that search for a suitable host (cervids). The winged deer keds usually suck blood from the host and mate; however, after landing on a host, they can crawl into the fur, shed their wings and become permanently associated with the host. The larvae develop up to stage 3 (L3) in the oviduct and are then deposited in the cervid fur as a white prepupa, which immediately starts to pupate. The fully developed pupa drops to the ground and remains there until August-September, when a new generation of winged adult deer keds can appear [29, 32, 33]. The female can produce to 20–25 larvae per year. Lipoptena cervi can overwinter on the host, and most deer keds can live to one year [34].
The winged adult Lipoptena cervi are attracted to large moving dark-colored objects when actively searching for a host [35]. Lipoptena cervi only fly for short distances and frequently attack accidental hosts such as humans or dogs [32].
Lipoptena cervi is a potential vector for other zoonotic pathogens, such as Anaplasma, Borrelia and Rickettsia species [36]. Korhonen et al. [37] showed for the first time transstadial transmission of Bartonella spp. in all development stages of deer ked (pupa, unfed adult winged deer ked). These data show vector competence for transmission of Bartonella spp. by Lipoptena cervi. In America, Bartonella sp. has been reported in Lipoptena cervi from white-tailed deer in Massachusetts [28], and in Lipoptena mazamae from white-tailed deer (Odocoileus virginianus) in Georgia and South Carolina (USA) [21]. Additionaly, it has also been suggested that wingless adults of L. mazamae, could be transmitted mechanically from female white-tailed deer to their offsprings [38]. Bartonella sp. has also been found in ticks (Ixodes ricinus) parasitizing roe deer in Poland [39].
The main goal of the study was detection of Bartonella spp. DNA in Lipoptena cervi by PCR test and verification of prevalence of this infection in adult wingless males and females. Molecular characterization of specimens of Bartonella spp. was done by analysis of partial (850 bp) rpoB gene sequences.
Methods
Sample collection
Living deer keds (Lipoptena cervi) were collected from 26 red deer (Cervus elaphus) hunted in January 2014 in the Strzałowo Forest District (53°45′57.03″N, 21°25′17.79″E). After collection, all insects were preserved in 70% ethanol for further morphological and molecular processing. Identification of deer ked (L. cervi) species and sex were conducted using appropriate identification key [34] and molecular methods to confirm vector species.
PCR and sequence analyses
In the laboratory, the flies were removed from 70% alcohol and air-dried. The DNA was isolated from whole insect body using the AX Tissue Mini kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s protocol.
For PCR test the primers rpoR and rpoF were used. These primers amplify a 333 bp fragment of the rpoB gene of Bartonella spp. PCR reactions were conducted according to Paziewska et al. [40]. 21 of 163 positive samples were used to obtain a longer (850 bp) fragment of the rpoB gene with second set of primers (1400F and 2300R) [41] (Table 1).
Table 1.
List of primers sequences used in this study
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| L700F | AAAGTTTAACCTGCCCACTGAT | This study |
| L1213R | CTGAACTCAGATCACGTAAGAAT | This study |
| rpoR | CGCATTATGGTCGTATTTGTCC | Paziewska et al. [40] |
| rpoF | GCACGATTYGCATCATCATTTTCC | Paziewska et al. [40] |
| 1400F | CGCATTGGCTTACTTCGTATG | Renesto et al. [41] |
| 2300R | GTAGACTGATTAGAACGCTG | Renesto et al. [41] |
PCR reactions were conducted in a 50 μl reaction mixture containing 2 μl of DNA template, 0.5 U (0.1 μl) of RUN Taq polymerase (A&A Biotechnology, Gdynia, Poland), 1 μl of dNTPs (10 mM), 0.5 μl of each primer (20 mM), and 5 μl of 10× Taq DNA polymerase buffer (pH 8.6, 25 mM MgCl2). In the negative control, nuclease-free water was added to the PCR mix instead of the tested DNA.
DNA amplification (1400F/2300R) was performed using the DNA Engine PTC-200 Thermal Cycler (BioRad, Hercules, USA) using the following program: initial denaturation was performed at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s and extension at 72 °C for 60 s. The final extension was performed at 72 °C for 7 min and then kept at 10 °C in a thermocycler.
The PCR products were visualized on a 1.0% agarose gel stained with ethidium bromide. Visualization was performed using ChemiDoc, MP Lab software (Imagine, BioRad, Hercules, USA). The resulting product was compared using the Nova 100 bp DNA Ladder Novazym (Poznań, Poland). The PCR amplicons were purified using a QIAEX II Gel Extraction Kit (Qiagen, Hilden, Germany), sequenced in both directions by Genomed (Poland) and contiguous sequences assembled using ContigExpress, Vector NTI Advance 11.0 (Invitrogen Life Technologies, New York, USA). The derived sequences were submitted to the GenBank database under the accession numbers MF580655–MF580675.
To confirm the morphological species determination of Lipoptena cervi, DNA from four specimens (positive in Bartonella spp. test and used in Bartonella isolate analysis), L700F and L1213R primers were used to amplify and sequence 412 bp fragments of 16S rDNA (Table 1).
In order to perform PCR amplification, the following mixture reaction was used: 4 μl of DNA extract was added to 46 μl of reaction mixture consisting of 0.1 μl of Allegro Taq Polymerase DNA (5 U/μl) (Poznań, Poland), 0.5 μl dNTPs (10 mM), 1 μl of each primer (20 mM), 5 μl of 10× Taq DNA polymerase buffer (pH 8.6, 25 mM MgCl2), and 38.4 μl of deionized water.
DNA amplification (L700F/L1213R) was performed using the DNA Engine PTC-200 Thermal Cycler (BioRad, Hercules, USA) using the following program:an initial denaturation was performed at 92 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 10 s and extension at 72 °C for 30 s. The final extension was performed at 72 °C for 5 min and then kept at 12 °C in a thermocycler.
Phylogenetic analyses
We used Bayesian inference (BI) analysis with MrBayes version 3.2 [42]. Analysis of partial rpoB gene sequence data was based on an alignment of 804 bp (268 amino acids) using the GTR + I + G model. The GTR models were chosen on the basis of jModelTest version 2.1.4 [43, 44] using the Akaike information criterion.
Results
In total, 217 deer keds were collected from 26 red deer (Cervus elaphus). All insects were identified as Lipoptena cervi using morphological features [34]. The prevalence of Bartonella spp. infection was 75.12% (163/217) by PCR test. In the tested group of L. cervi, a greater proportion of females (86.67%) was found to be positive for Bartonella spp. than males (60.82%).
PCR, sequence and molecular analyses
Fourteen sequences (Bartonella sp. 1: MF580662–MF580675) obtained in this study share over 99% similarity with B. schoenbuchensis isolated from moose blood from Sweden (GenBank: KB915628) and from human blood from France (GenBank: HG977196). Five sequences Bartonella sp. 2 (MF580657–MF580661) showed 94.6% similarity with Bartonella sp. 3, and 94.4% with Bartonella sp. from Japanese sika deer from Japan (AB703149). Bartonella sp. 3 (MF580656) showed 99.7% similarity with another samples isolated from Japanese sika deer from Wakayama Prefecture Japan (AB703149). Bartonella sp. 4 (MF580655) was 99,7% similar with Bartonella sp. isolated from Japanese sika deer (Cervus nippon centralis) from Nara Prefecture Japan (AB703146). These results are summarized in Table 2 and Additional file 1: Table S1.
Table 2.
Bartonella spp. used in the phylogenetic analysis
| Isolate/sequence ID | Species | Host | Source | Country of isolation |
|---|---|---|---|---|
| MF580662–MF580675 | Bartonella sp. 1 | Lipoptena cervi | whole body | Poland |
| MF580657–MF580661 | Bartonella sp. 2 | Lipoptena cervi | whole body | Poland |
| MF580656 | Bartonella sp. 3 | Lipoptena cervi | whole body | Poland |
| MF580655 | Bartonella sp. 4 | Lipoptena cervi | whole body | Poland |
| DQ356077 | Bartonella bovis | bovine | blood | Italy |
| EF432062 | B. bovis | cow | valve (heart) | France |
| KR733195 | B. bovis | cattle | blood | Malaysia |
| KR733194 | B. bovis | cattle | blood | Malaysia |
| KF218224 | B. bovis | water buffalo | blood | Thailand |
| KF218220 | B. bovis | cattle | blood | Thailand |
| KF218218 | B. bovis | cattle | blood | Guatemala |
| KF218217 | B. bovis | cattle | blood | France |
| HM167505 | Bartonella capreoli | moose | blood | USA |
| AB703143 | B. capreoli | Japanese sika deer | blood | Japan |
| AB703142 | B. capreoli | Japanese sika deer | blood | Japan |
| AB703149 | Bartonella sp. | Japanese sika deer | blood | Japan |
| AB703146 | Bartonella sp. | Japanese sika deer | blood | Japan |
| AB703145 | Bartonella sp. | Japanese sika deer | blood | Japan |
| KB915628 | Bartonella schoenbuchensis | moose | blood | Sweden |
| KM215709 | Bartonella chomelii | cattle | blood | Spain |
| KM215710 | Bartonella chomelii | cattle | blood | Spain |
| JN646664 | Bartonella chomelii | cattle | blood | New Caledonia |
| KJ909808 | Bartonella bovis | cattle | blood | Israel |
| AB703148 | Bartonella sp. | Japanese sika deer | blood | Japan |
| AB703144 | Bartonella sp. | Japanese sika deer | blood | Japan |
| HG977196 | Bartonella schoenbuchensis | human | blood | France |
| CP019789 | Bartonella schoenbuchensis | European roe deer | blood | Germany |
Four 16S rDNA sequences of deer ked were obtained, which were identical with Lipoptena cervi collected in the Czech Republic (AF322437). The sequences derived during this study were submitted to the GenBank database under the accession numbers MF541726–MF541729.
Discussion
Our findings indicate the presence of Bartonella species in deer keds (Lipoptena cervi) obtained from red deer (Cervus elaphus). Dehio et al. [16] noted that L. cervi appears to be a natural reservoir supporting the transmission of Bartonella schoenbuchensis. The largest group of isolates (Bartonella sp. 1) showed closed similarity with Bartonella schoenbuchensis. Our data indicate a high prevalence of Bartonella spp. A prevalence of 75.12% was recorded in the whole tested group, a result similar to global data; however, higher values were observed in Finland (90%) and France (94%) [14, 37].
To the best of our knowledge, the present paper is the first report in Poland to identify differences in the prevalence of Bartonella spp. infection in male and female deer keds. Females were more frequently infected than males; 86.67% of wingless females were infected by Bartonella, similar to a study from Hungary (76.0%), while only 60.82% of wingless males were infected, also similar to a result in Hungary (58%) [45].
The reason for such a difference in infection prevalence is not clear; however, it might be associated with the multiple bites needed for larvae production (20–25 larvae per year) [34]. De Bruin et al. [45] suggested that Bartonella species are able to colonize or survive more efficiently as females than males, but the molecular mechanism for this remains unknown. The same authors [45] suggested that Bartonella spp. infection in female deer keds might lead to more female offspring than uninfected females, possibly resulting in the observed asymmetry in the female:male ratio of infected individuals.
By transmitting pathogens, Lipoptena cervi can be potentially dangerous for animals and humans. A study conducted in Finland found that L. cervi can transmit some pathogens by biting, and this is associated with hair-loss epizootics in moose [17]. Since the 1970s, in Finland an increasing number of cases of recurrent, and occasionally long-lasting, dermatitis associated with deer keds bites has been observed [46].
Fourteen of the sequences (Bartonella sp. 1, see Table 2.) isolated from deer keds demonstrated 99.2% homology with a Bartonella schoenbuchensis sample isolated from moose in Sweden (KB915628). Bartonella schoenbuchensis could be found in vectors, such as L. cervi or L. mazamae, or in definitive hosts like cattle, wild ruminants and humans [10, 11, 16]. It is difficult to see how the relationship between B. schoenbuchensis and deer ectoparasites may relate to similar cases in humans. Lipoptena cervi has fed on humans under experimental conditions and in the natural environment [18, 47]. As humans may be infected with B. schoenbuchensis during occasional biting by L. cervi, hunters, forestry workers and cross-country runners, among others, are at increased risk of infection [16]. Although L. cervi has not yet been definitively demonstrated to transmit B. schoenbuchensis through bites, it is possible that hunters are at risk to infection of Bartonella schoenbuchensis or Anaplasma phagocytophilum following exposure to deer blood [48].
In Massachusetts, a Bartonella sp. similar to B. schoenbuchensis has been found in Lipoptena mazamae [21], which suggests that Lipoptena species can extend the range of B. schoenbuchensis. Our phylogenetic analysis of the partial rpoB gene sequences found that our samples were in the same clade as other B. schoenbuchensis isolates from moose (KB915628) and human (HG977196). Bartonella schoenbuchensis (strain closely related to B. schoenbuchensis strain R1–86.6%, gltA gene) was previously reported from roe deer (C. capreolus) in Poland [49].
Bartonella sp. 2 (5 isolates) and Bartonella sp. 3 (MF580656) demonstrated high similarity to sequences obtained from Japanese sika deer from Japan (AB703149, 94.6% and 99.7% similarity, respectively. These Bartonella isolates are related with the B. bovis clade in our phylogenetic analysis (Fig. 1).
Fig. 1.
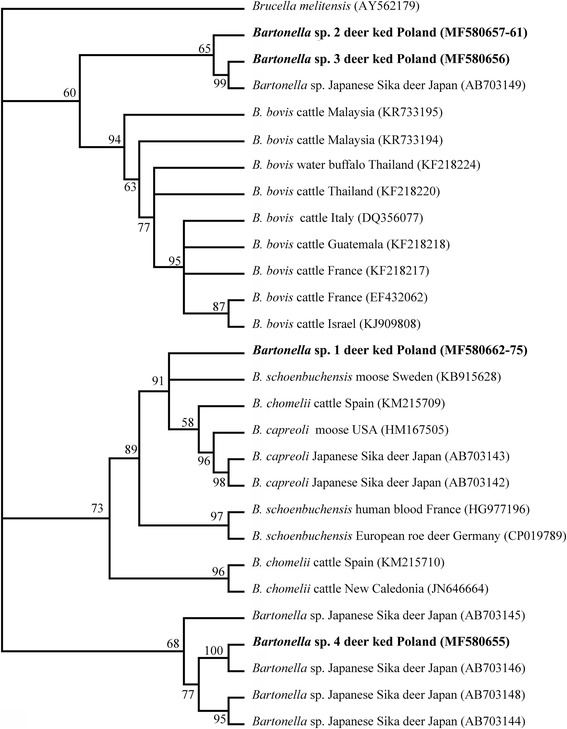
Phylogenetic tree of Bartonella spp., constructed by Bayesian inference (BI) analysis using MrBayes version 3.2. For BI codon analysis (nucmodel = codon), the GTR + I + G model was chosen based on jModelTest version 2.1.4 [43, 44] using Akaike Information Criterion. Analysis was run for 8,000,000 generations, with 2,000,000 generations discarded as ‘burn-in’. Hosts, country and GenBank accession numbers of origin are shown. Nodal support is indicated as Bayesian posterior probabilities. Sequence from Brucella melitensis (AY562179) was used as outgroup. Sequences generated in this study are shown in bold
Bartonella sp. 4 (MF580655) is very similar (99.7%) to Bartonella sp. isolates collected from Japanese sika deer (AB703146, see Fig. 1) and in our phylogenetic analyses these isolates are associated with other Bartonella sp. isolates from Japanese sika deer and formed a distinct clade with Bartonella spp. isolated from Japanese sika deer. Probably “japanese” Bartonella isolates were introducted to Poland. This observation suggests that the strain of Bartonella sp. bacteria identified in the present study is derived from Japanese sika deer introduced to Europe by vectors to environment, indicating that this Asian strains could be spread by L. cervi to European red deer (Cervus elaphus).
Some Japanese Bartonella sp. samples, isolated from different prefectures, comprise a group distinct from other samples of Bartonella on GenBank; however, further testing is required utilizing other genes, including gltA and ftsZ, to confirm whether they could be regarded as a distinct species.
Conclusions
Our data confirm that Bartonella spp. can be transmitted by deer ked in central Europe, and the prevalence of this pathogen is very high. In this area, Lipoptena cervi are infected by various species of the genus Bartonella.
Additional files
Pairwise comparison of partial (804 bp) mitochondrial gene rpoB DNA and amino acid sequences variability among species/sequences of Bartonella used in phylogenetical analysis. Above diagonal: number of variable sites in the 268 amino acid gene rpoB sequences. Below diagonal: number of variable sites in the nucleotides gene rpoB sequences. The percentage of variable sites for each gene fragment between 2 species/sequences is given in parenthesis. Each species isolates/sequence information are provided in Table 2. (DOCX 23 kb)
Acknowledgements
The authors would like to express their gratitude to Marek Bogdaszewski, Head of the Research Station in Kosewo Górne (W. Stefański Institute of Parasitology), for enabling us to collect the samples from red deer.
Funding
No funding was received.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional file. The sequences were submitted to the GenBank database under the accession numbers MF580655–MF580675 (Uncultured Bartonella sp.) and MF541726–MF541729 (Lipoptena cervi).
Authors’ contributions
TS planned and organized the study. JW and ŻSB collected samples. TS and JW extracted DNA. ZL performed PCR, sequencing and analyzed sequence data. TS, WJ and ZL drafted the manuscript, and wrote the final version together with GK. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2413-0) contains supplementary material, which is available to authorized users.
Contributor Information
Tomasz Szewczyk, Email: t.szewczyk86@gmail.com.
Joanna Werszko, Email: joanna.werszko@gmail.com.
Żaneta Steiner-Bogdaszewska, Email: zaneta.steiner@gmail.com.
Witold Jeżewski, Email: jezw@twarda.pan.pl.
Zdzisław Laskowski, Email: laskowz@twarda.pan.pl.
Grzegorz Karbowiak, Email: grzgrz@twarda.pan.pl.
References
- 1.Bai Y, Kandaurov A, Imnadze P, Kosoy M. Molecular survey of bacterial zoonotic agents in bats from the country of Georgia (Caucasus) PLoS One. 2017;12(1):e0171175. doi: 10.1371/journal.pone.0171175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev. 2000;13:428–438. doi: 10.1128/CMR.13.3.428-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karbowiak G, Rychlik L, Nowakowski W, Wita I. Natural infections of small mammals with blood parasites on the borderland of boreal and temperate forest zones. Acta Theriol. 2005;50:31–42. doi: 10.1007/BF03192616. [DOI] [Google Scholar]
- 4.Bai Y, Malania L, Alvarez Castillo D, Moran D, Boonmar S, Chanlun A, et al. Global distribution of Bartonella infection in domestic bovine and characterization of Bartonella bovis strain using multi-locus sequence typing. PLoS One. 2013;8(11):e80894. doi: 10.1371/journal.pone.0080894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato S, Kabeya H, Yamazaki M, Takeno S, Suzuki K, Kobayashi S, et al. Prevalence and genetic diversity of Bartonella species in sika deer (Cervus nippon) in Japan. Comp Immunol Microbiol Infect Dis. 2012;35(6):575–581. doi: 10.1016/j.cimid.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Boulouis HJ, Piemont Y. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.) Int J Syst Evol Microbiol. 2000;50:1973–1979. doi: 10.1099/00207713-50-6-1973. [DOI] [PubMed] [Google Scholar]
- 7.Chomel B. Emerging bacterial zoonoses. Point Veterinaire. 2000;31:15–22. [Google Scholar]
- 8.Breitschwerdt EB, Maggi RG, Chomel BB, Lapin MR. Bartonellosis: an emerging infectious diseases of zoonotic importance to animal and humans being. J Vet Emerg Crit Care (San Antonio) 2010;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 9.Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, Correa M, Breitschwerdt EB. Bartonella spp. bacteremia and rheumatic symptoms in patient from Lyme disease-endemic region. Emerg Infect Dis. 2012;18:783–791. doi: 10.3201/eid1811.121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehio C, Lanz C, Pohl R, Behrens P, Bermond D, Piemont Y, Pelz K, Sander A. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int J Syst Evol Microbiol. 2001;51:1557–1565. doi: 10.1099/00207713-51-4-1557. [DOI] [PubMed] [Google Scholar]
- 11.Rolain JM, Rousset E, La Scola B, Duquesnel R, Raoult D. Bartonella schoenbuchensis isolated from the blood of a French cow. Ann NY Acad Sci. 2003;990:236–8. [DOI] [PubMed]
- 12.Vayssier-Taussat M, Moutailler S, Fèmènia F, Raymond P, Croce O, La Scola B, et al. Identification of novel zoonotic activity of Bartonella spp., France. Emerg Infect Dis. 2016;22:457–462. doi: 10.3201/eid2203.150269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller A, Reiter M, Schötta AM, Stockinger H, Stanek G. Detection of Bartonella spp. in Ixodes ricinus ticks and Bartonella seroprevalence in human population. Ticks Tick Borne Dis. 2016;7:763–767. doi: 10.1016/j.ttbdis.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, et al. Role of Hippoboscidae flies as potential vector of Bartonella spp. infecting wild domestic ruminants. Appl Environ Microbiol. 2004;70(10):6302–6305. doi: 10.1128/AEM.70.10.6302-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sytykiewicz H, Karbowiak G, Werszko J, Czerniewicz P, Sprawka I, Mitrus J. Molecular screening for Bartonella henselae and Borrelia burgdorferi sensu lato co-existence within Ixodes ricinus populations in central and eastern parts of Poland. Ann Agric Environ Med. 2012;19(3):451–456. [PubMed] [Google Scholar]
- 16.Dehio C, Sauder U, Hiestand R. Isolation of Bartonella schoenbuchensis in Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J Clin Microbiol. 2004;42(11):5320–5323. doi: 10.1128/JCM.42.11.5320-5323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madslien K, Ytrehus B, Vikøren T, Malmsten J, Isaksen K, Hygen HO, Solberg EJ. Hair-loss epizootic in moose (Alces alces) associated with massive deer ked (Lipoptena cervi) infestation. J Wild Dis. 2011;47(4):893–906. doi: 10.7589/0090-3558-47.4.893. [DOI] [PubMed] [Google Scholar]
- 18.Bequaert JC. Monograph of the Melophaginae of ked-flies of sheep, goats, deer and antelopes (Diptera, Hippoboscidae) Bull Brooklyn Ent Soc. 1942;22:1–220. [Google Scholar]
- 19.Maa TCA. synopsis of the Lipopteninae (Diptera: Hippoboscidae) J Med Entomol. 1965;2:233–248. doi: 10.1093/jmedent/2.3.233. [DOI] [PubMed] [Google Scholar]
- 20.Valimaki P, Madslien K, Malmsten J, Harkonen I, Harkonen S, Kaitala A, et al. Fennoscandian distribution of an important parasite of cervids, the deer ked (Lipoptena cervi), revisted. Parasitol Res. 2010;107:117–125. doi: 10.1007/s00436-010-1845-7. [DOI] [PubMed] [Google Scholar]
- 21.Reeves WK, Nelder MP, Cobb KD, Dasch GA. Bartonella spp. in deer keds, Lipoptena mazamae (Diptera: Hippoboscidae), from Georgia and South Carolina, USA. J Wildl Dis. 2006;42(2):391–396. doi: 10.7589/0090-3558-42.2.391. [DOI] [PubMed] [Google Scholar]
- 22.Kettle DS. Medical and veterinary entomology. London, UK: Croom Helm. 1984:285–92.
- 23.Haigh JC, Mackintosh C, Griffin F. Viral, parasitic and prion diseases of farmed deer and bison. Rev Sci Tech (Int Off Epizoot) 2002;21:219. doi: 10.20506/rst.21.2.1331. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Espona S, Pemberton JM, Putman R. Red and sika deer in the British Isles, current management issues and management policy. Mamm Biol. 2009;74:247–262. doi: 10.1016/j.mambio.2009.01.003. [DOI] [Google Scholar]
- 25.Sokół R, Gałęcki R. Prevalence of keds on city dogs in central Poland. Med Vet Entomol. 2017;31:114–116. doi: 10.1111/mve.12209. [DOI] [PubMed] [Google Scholar]
- 26.Kowal J, Nosal P, Rościszewska M, Matysek M. New records of Lipoptena fortisetosa MAA, 1965 (Diptera: Hippoboscidae) in Poland. Dipteron. 2009;25:27–29. [Google Scholar]
- 27.Kowal J, Nosal P, Kornaś S, Wajdzik M, Matysek M, Basiaga M, Różnorodność i. znaczenie muchówek z rodziny narzępikowatych - pasożytów jeleniowatych. Med Weter. 2016;72:745–749. [Google Scholar]
- 28.Matsumoto K, Berrada ZL, Klinger E, Goethert HK, Telford SR. Molecular detection of Bartonella schoenbuchensis from ectoparasites of deer in Massachusetts. Vector Borne Zoonotic Dis. 2008;8(4):549–554. doi: 10.1089/vbz.2007.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chistyakov AF. Skin lesions in people due to bites of Lipoptena cervi. Vestn Dermatol Venerol. 1968;42:59–62. [PubMed] [Google Scholar]
- 30.Rantanen T, Reuanala T, Vuojolahti P, Hackman W. Presistent pruritic papules from deer ked bites. Acta Derm Venereol. 1982;62:307–311. [PubMed] [Google Scholar]
- 31.Haarlov N. Life cycle and distribution pattern of Lipoptena cervi (L.) (Dipt. Hippobosc.) on Danish deer. Oikos. 1964;15:93–129. doi: 10.2307/3564750. [DOI] [Google Scholar]
- 32.Madslien K, Ytrehus B, Vilijugrein H, Solberg EJ, Braten KR, Mysterud AA. factors affecting deer ked (Lipoptena cervi) prevalence and intensity in moose (Alces alces) in Norway. Parasit Vectors. 2012;5:251. doi: 10.1186/1756-3305-5-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mysterud A, Madslien K, Herland A, Viljugrein H, Ytrehus B. Phenology of deer ked (Lipoptena cervi) host-seeking flight activity and its relationship with prevailing autumn weather. Parasit Vectors. 2016;9:95. doi: 10.1186/s13071-016-1387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borowiec L. [Wpleszczowate Hippoboscidae. Klucze do oznaczania owadów Polski.] Część XXVIII/77 77. Wrocław 1984 (In Polish).
- 35.Kortet R, Härkönen L, Hokkanen P, Härkönen S, Kaitala A, Kaunisto S, et al. Experiments on the ectoparasitic deer ked that often attacks humans; preferences for body parts, colour and temperature. Bull Entomol Res. 2010;100:279–285. doi: 10.1017/S0007485309990277. [DOI] [PubMed] [Google Scholar]
- 36.Buss M, Case L, Kearney B, Coleman C, Henning JD. Detection of Lyme disease and anaplasmosis pathogens via PCR in Pennsylvania deer ked. J Vector Ecol. 2016;41:292–294. doi: 10.1111/jvec.12225. [DOI] [PubMed] [Google Scholar]
- 37.Korhonen EM, Pèrez Vera C, Pulliainen AT, Sironen T, Aaltonen K, Kortet R, et al. Molecular detection of Bartonella spp. in deer ked pupae, adult keds and moose blood in Finland. Epidemiol Infect. 2014;143:578–585. doi: 10.1017/S0950268814001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuel W, Trainer D. Lipoptena mazamae Rondani, 1878 (Diptera: Hippoboscidae) on white-tailed deer in southern Texas. J Med Entomol. 1972;9:104–106. doi: 10.1093/jmedent/9.1.104. [DOI] [PubMed] [Google Scholar]
- 39.Adamska M. Infestation of game animals from north-western Poland by common tick (Ixodes ricinus) Wiadi Parazytol. 2008;54:31–36. [PubMed] [Google Scholar]
- 40.Paziewska A, Harris PD, Zwolińska L, Bajer A, Siński E. Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb Ecol. 2011;61:134–45. [DOI] [PMC free article] [PubMed]
- 41.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;3(2):430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel OA. simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and paralel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Bruin A, van Leeuwen AD, Jahfari S, Takken W, Földvári M, Dremmel L, et al. Vertical transmission of Bartonella schoenbuchensis in Lipoptena cervi. Parasit Vectors. 2015;8:176. doi: 10.1186/s13071-015-0764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Härkönen S, Laine M, Vornanen M, Reunala T. Deer ked (Lipoptena cervi ) dermatitis in humans - an increasing nuisance in Finland. Alces. 2009;45:73–79. [Google Scholar]
- 47.Starzyk J. Strzyżak sarni, Lipoptena cervi (L.) (Diptera, Hippoboscidae) - pasożyt zewnętrzny parzystokopytnych. Prz. Zool. 1975;17:446–448. [Google Scholar]
- 48.Telford SR, III, Bakken JS, Dumler JS. Risk for acquiring human granulocytic ehrlichiosis: exposure to deer blood or deer ticks? Clin Infect Dis. 1997;24:531–533. doi: 10.1093/clinids/24.3.531. [DOI] [PubMed] [Google Scholar]
- 49.Welc-Falęciak R, Werszko J, Cydzik K, Bajer A, Michalik J, Behnke JM. Co-infection and genetic diversity of tick-borne pathogens in roe deer from Poland. Vector Borne Zoonotic Dis. 2013;13:277–288. doi: 10.1089/vbz.2012.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pairwise comparison of partial (804 bp) mitochondrial gene rpoB DNA and amino acid sequences variability among species/sequences of Bartonella used in phylogenetical analysis. Above diagonal: number of variable sites in the 268 amino acid gene rpoB sequences. Below diagonal: number of variable sites in the nucleotides gene rpoB sequences. The percentage of variable sites for each gene fragment between 2 species/sequences is given in parenthesis. Each species isolates/sequence information are provided in Table 2. (DOCX 23 kb)
Data Availability Statement
The data supporting the conclusions of this article are included within the article and its additional file. The sequences were submitted to the GenBank database under the accession numbers MF580655–MF580675 (Uncultured Bartonella sp.) and MF541726–MF541729 (Lipoptena cervi).


