Abstract
Background & objectives:
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) has been instrumental in revolutionizing microbiological identification, especially in high-throughput laboratories. It has enabled the identification of organisms like non-fermenting Gram-negative bacilli (NFGNB), which has been a challenging task using conventional methods alone. In this study an attempt was made to validate MALDI-TOF MS for the identification of clinical isolates of each of the three most common NFGNB, other than Pseudomonas spp., taking molecular methods as the gold standard.
Methods:
One hundred and fifty clinical isolates of NFGNB, confirmed by molecular methods such as Acinetobacter baumannii[oxa-51 polymerase chain reaction (PCR)], Burkholderia cepacia complex (expanded multilocus sequence typing) and Stenotrophomonas maltophilia (species-specific PCR), were taken. Isolated colonies from fresh cultures of all 150 isolates were smeared onto ground steel plate, with and without formic acid extraction step. The identification was carried out using MALDI-TOF MS Biotyper database.
Results:
A concordance of 100 and 73.33 per cent was found between the molecular techniques and MALDI-TOF MS system in the identification of these isolates up to genus and species levels, respectively. Using a cut-off of 1.9 for reliable identification, rate of species identification rose to 82.66 per cent. Principal component analysis dendrogram and cluster analysis further increased discrimination of isolates.
Interpretation & conclusions:
Our findings showed MALDI-TOF MS-based identification of NFGNB as a good, robust method for high-throughput laboratories.
Key words: Acinetobacter baumannii, Burkholderia cepacia complex, matrix-assisted laser desorption ionization time-of-flight-mass spectrometry, MLST, non-fermenting Gram-negative bacilli, Stenotrophomonas maltophilia
Non-fermenting Gram-negative bacilli (NFGNB) cause a multitude of severe and disabling infections. Though many varied pathogenic organisms come under the term NFGNB, the majority of serious infections are caused by Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia and Burkholderia cepacia complex (Bcc)1. Unlike P. aeruginosa, the rest of the NFGNBs lack easily discernable phenotypic characteristics. Their contrasting susceptibilities, high intrinsic resistance and remarkable ability to develop resistance to essentially all commonly used antibiotics including the anti-pseudomonal drugs2 limit the therapeutic options, causing a delay in patient management. Bcc is intrinsically resistant to aminoglycosides and polymyxins while S. maltophilia is intrinsically resistant to aminoglycosides and commonly used carbapenems3. A. baumannii is increasingly becoming resistant to the carbapenems, leaving polymyxins as the only option4,5. Consequently, their correct identification is essential as no single drug is effective against all, which hinders the initiation of appropriate empirical treatment resulting in increased morbidity and mortality.
It takes several days for a routine laboratory to identify these organisms due to their inert biochemical activities and difficulty in interpretation of phenotypic characteristics. These analyses require expertise and are time-consuming and often give unreliable and overlapping results. The automated systems are not much discriminatory6, and molecular methods are cost-and labour-intensive. Thus, a simple, rapid and reliable technique is the need of the hour. Matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF MS) is emerging as an efficient, accurate and cost-effective alternative tool in microbial identification7.
In this study the identification accuracy of MALDI-TOF MS was compared with standard molecular methods for identifying clinical isolates of NFGNB other than P. aeruginosa.
Material & Methods
The study was conducted in the department of Medical Microbiology, Post Graduate Institute of Medical Education and Research, Chandigarh, India, between August 2013 and October 2014. The isolates were obtained from clinical samples over the years (2005-2013) and were well preserved by stocking up in 20 per cent glycerol vials and reviving from time to time. These were isolated from various sources such as blood, endotracheal aspirates, cerebrospinal fluid, wounds and drain fluids.
It was noted that in literature most studies though have quoted 100 per cent genus identification for NFGNBs, but correct species identification was as low as 72 per cent also8. Thus, we took the best possible result of 100 and 72 per cent given by the available data for species concordance, giving an average of 86 per cent with a margin of error of 14 per cent. Using the standard formula [(Z-score)2×SD×(1−SD)/(margin of error)2], the sample size was calculated to be 49 taking a confidence level of 95 per cent (corresponding to Z-score of 1.96), standard deviation of 0.5 and margin of error of 14 per cent. Hence, 150 clinical isolates of NFGNB (50 isolates each of A. baumannii, S. maltophilia and Bcc) were included in the study.
Criteria of selection of isolates
All isolates that could be revived from stock cultures that gave pure colony growth and fulfilled the molecular identification for their respective organisms were included. Those isolates which did not revive in pure form, showed any contamination or were misidentified by molecular gold standards were excluded.
Identification of isolates
These organisms were presumptively identified by conventional techniques such as colony morphology and a set of standardized biochemical reactions9. The identity was further confirmed using standard molecular techniques for each as, oxa-51 polymerase chain reaction (PCR) for confirming A. baumannii10, species-specific PCR for S. maltophilia11 and expanded multilocus sequence typing (E-MLST) for Bcc12. In Bcc isolates, identification was also carried out by subjecting the amplicon of recA-PCR to sequencing using Big Dye Terminator Cycle Sequencing Kit, version 3.1 (Applied Biosystems, USA). The nucleotide sequences were analyzed on ABI 3130 Genetic Analyzer (Applied Biosystems) and compared with sequences available on the internet (http://www.ncbi.nlm.nih.gov/BLAST/).
MALDI-TOF MS
The Microflex LT MALDI-TOF MS (Bruker Daltonics, Germany) with a 60-Hz nitrogen laser was used to analyze spectra over a mass range of 2000-20000 Da. All specimens were processed as per the manufacturer's instructions. Pre-analytic preparation of samples was performed by using a sterile wooden tip to pick an isolated bacterial colony freshly grown on defined agar medium and then smearing a thin film in duplicate onto a ground steel MALDI biotarget 96 plate (Direct Transfer procedure). One of the microbial films was overlaid directly with 1.0 μl α-cyano-4-hydroxycinnamic acid (MALDI TOF HCCA) matrix solution while the other pair was first treated with 1 μl 100 per cent formic acid (FA) for on-plate extraction13. The sample-matrix mixture was dried at room temperature and subsequently inserted into the system for data acquisition. A sum spectrum was acquired by summing the laser shots. Quality controls were internally calibrated using Escherichia coli DH5α supplied by Bruker Daltonics, following the same procedure. The data were processed automatically by the instrument software and the spectra were compared with reference libraries for bacterial identification matching.
Spectra were analyzed using MALDI Biotyper database version 3 (Bruker Daltonics, Germany). Manufacturer-recommended score cut-offs were used to determine genus level (1.7000 to 1.999) or species level (≥2.000) of the organism. A score of <1.7 was considered unreliable for genus identification. For this study, a Biotyper score cut-off of ≥2.0 was considered for reliable species identification when analyzing under automatic mode. Any isolate getting lower score was reanalyzed before labelling an isolate as ‘Not reliable identification’. The NFGNB were also analyzed taking a cut-off score of ≥1.9 for species-level identification, instead of ≥2.0 as also suggested by Ford and Burnham13. An attempt to harness the potential of MALDI-TOF MS as a typing technique was also carried out by creating main spectra for all the isolates and studying them by principal component analysis (PCA) dendrogram. The correlation distance from peaks measured by the software was used to construct dendrogram on the basis of ribonucleic protein profile of the isolates. In cases of misidentification, cluster analysis using Biotyper version 3 was also carried out.
Statistical analysis
Statistical analysis was performed using the Mann–Whitney Wilcoxon test with GraphPad software (GraphPad software Inc., California, USA).
Results
Acinetobacter baumannii
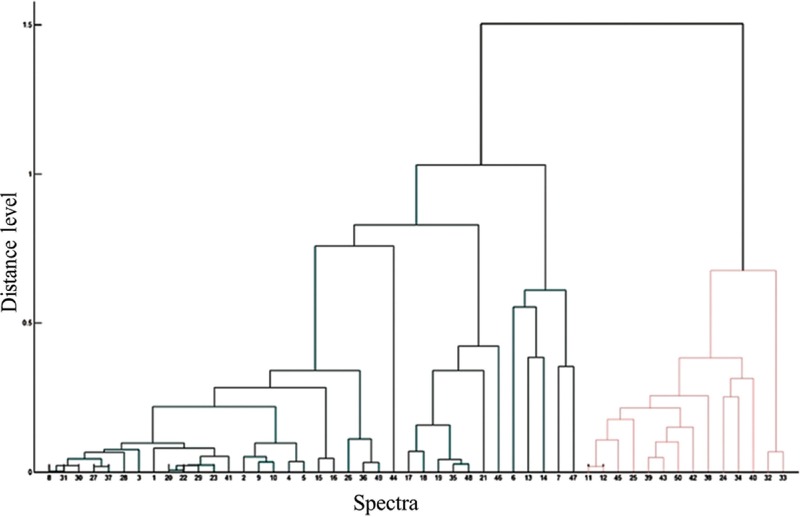
Oxa-51 PCR confirmed all 50 isolates presumptively identified as A. baumannii. Using manufacturer's cut-offs, the concordance rate of MALDI-TOF MS with oxa-51 PCR for correct identification up to genus level was obtained in 100 per cent isolates, while higher discriminatory resolution up to species level was noted in 88 per cent (44/50). A four per cent increase in the rate of species identification was obtained when the cut-off of ≥1.9 was taken instead of ≥2. Direct transfer procedure could identify all the isolates, whereas two per cent remained unidentified with FA on-plate extraction method. There were no misidentifications (Table I). On PCA dendrogram, all 50 isolates fell into two distinct groups without any outliers (Fig. 1).
Table I.
Results of MALDI-TOF MS analysis for the identification of 150 non-fermenting Gram-negative bacilli (NFGNB) showing the level of discrimination achieved using two different cut-offs and additional step of on-plate formic acid extraction
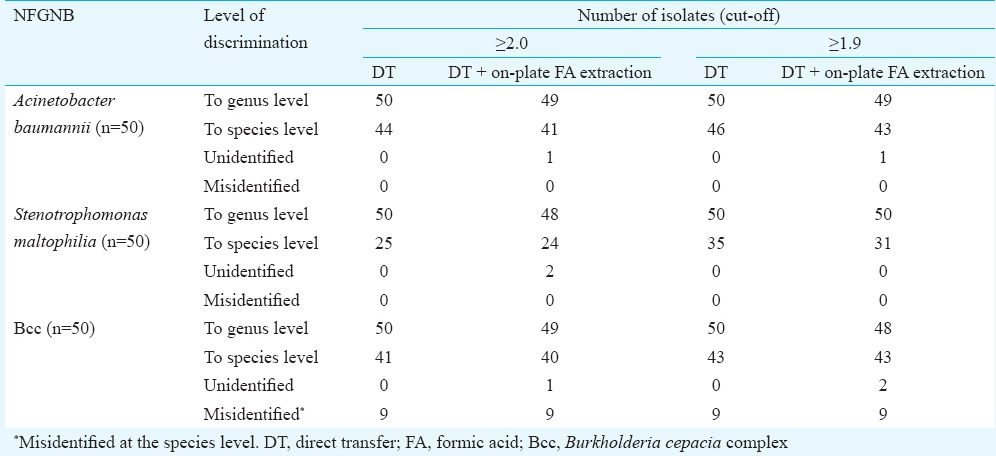
Fig. 1.
MALDI-TOF MS based principal component analysis dendrogram for 50 isolates of Acinetobacter baumannii. The isolates fell into two distinct groups based on their main spectra. Distance level represents diversity in percentage. Numbers indicate isolates in study.
Stenotrophomonas maltophilia
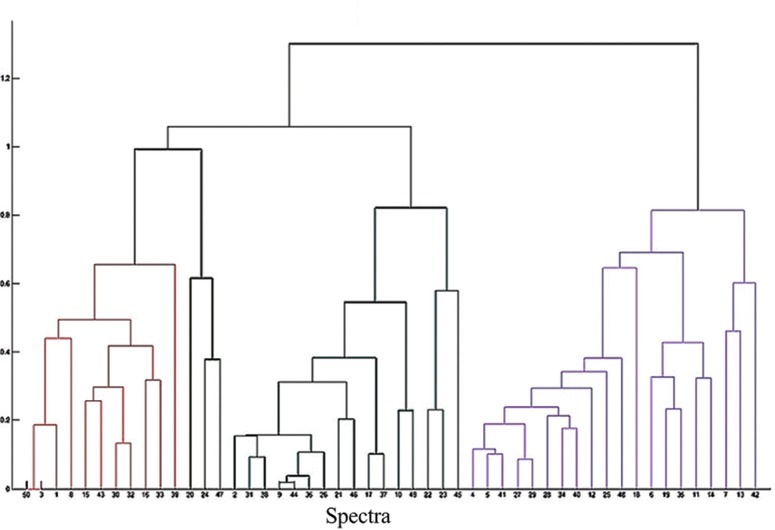
All 50 isolates presumptively identified on the basis of biochemical reactions were confirmed to be S. maltophilia by species-specific PCR. The concordance rate of MALDI-TOF MS was 100 per cent for the genus and 50 per cent (25/50) for the correct species identification. An increase in species identification to 70 per cent (35/50) was noted when altered cut-off of ≥1.9 was considered. Four per cent of isolates remained unidentified with FA on-plate extraction while there were none with direct transfer. No isolate was misidentified by MALDI-TOF (Table I). PCA dendrogram revealed two distinct groups with four clusters representing all the 50 isolates of S. maltophilia (Fig. 2).
Fig. 2.
MALDI-TOF MS based principal component analysis dendrogram for 50 isolates of Stenotrophomonas maltophilia. The isolates fell into four distinct groups based on their main spectra.
Burkholderia cepacia complex (Bcc)
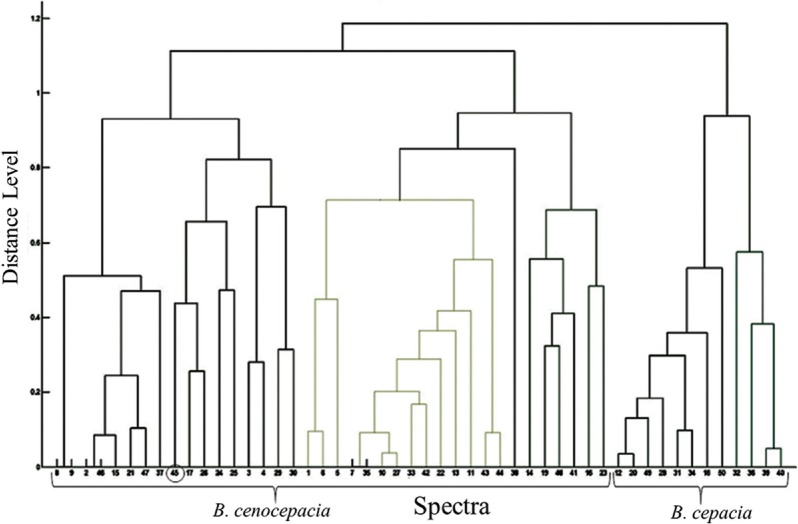
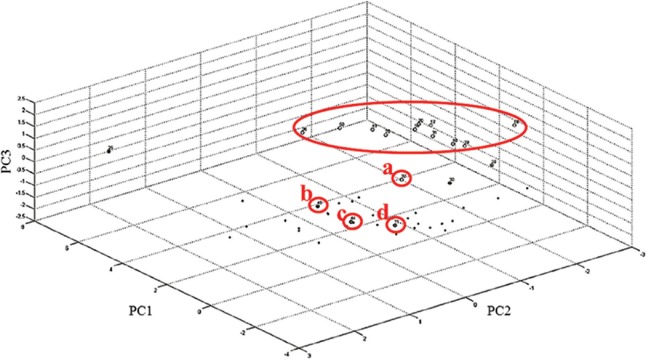
Of the 50 isolates labelled as Bcc by conventional methods, E-MLST identified 37 isolates as B. cenocepacia (35-B. cenocepacia IIIA, 2-B. cenocepacia IIIB) and 13 as B. cepacia. MALDI-TOF MS gave a concordance rate of 100 per cent for genus identification and 82 per cent (41/50) for correct species identification when a cut-off of ≥2 was considered. An increase of four per cent was noted with ≥1.9 cut-off. Direct transfer method could identify all the 50 isolates, while two and four per cent remained unidentified by FA on-plate extraction method using cut-off ≥2 and ≥1.9, respectively (Table I). There were no misidentifications with respect to 37 B. cenocepacia isolates; however, they were not resolved to lineage level of IIIA and IIIB. Of the 13 B. cepacia isolates, only four were correctly identified while the rest were misidentified as B. cenocepacia by MALDI-TOF MS. The overall concordance rate of MALDI-TOF MS for correct identification up to species level was 100 per cent for B. cenocepacia and 30.77 per cent for B. cepacia (Table II). The recA sequencing correctly identified 29 of 37 B. cenocepacia and 12 of 13 B. cepacia, giving a concordance of 78.37 and 92.30 per cent, respectively. PCA dendrogram constructed by MALDI-TOF MS placed all B. cenocepacia and 12 of 13 B. cepacia isolates into two distinct groups (Fig. 3). Cluster analysis also identified all B. cenocepacia and 12 of 13 B. cepacia correctly, 11 of which were closely placed (Fig. 4). Only one B. cepacia isolate remained ‘misidentified’ by both the methods.
Table II.
Comparison of MALDI-TOF MS and recA sequencing in identification of 50 Burkholderia cepacia complex isolates and their concordance with multilocus sequence typing (MLST)
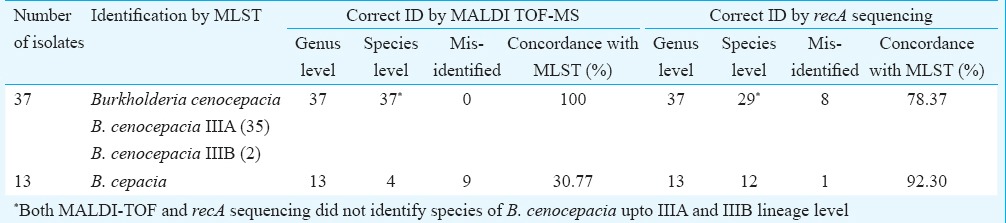
Fig. 3.
MALDI-TOF MS based principal component analysis dendrogram for 50 isolates of Burkholderia cepacia complex. All 37 B. cenocepacia isolates were grouped together; 12 of 13 B. cepacia isolates formed a distinct group. The 13th isolate (45), misidentified by MALDI-TOF MS as B. cenocepacia, is encircled.
Fig. 4.
Cluster diagram of 50 Burkholderia cepacia complex isolates based on three principal components. Eleven of 13 isolates of B. cepacia were grouped together (encircled). Isolate number 32 (a), though identified as B. cepacia, has been placed far apart from rest of the isolates. Isolate number 45 (b), which was misidentified by MALDI-TOF MS as B. cenocepacia, was placed well outside the B. cepacia cluster. Isolate numbers 11 (c) and 44 (d), which were B. cenocepacia IIIB by multilocus sequence typing, were grouped along with the B. cenocepacia IIIA isolates.
Discussion
In spite of continuous advancements in the field of diagnostics, the identification of NFGNB by conventional methods alone is neither satisfactory nor reliable. Keeping in view the inter-species differences in these NFGNB, the need for reliable identification beyond genus level cannot be understated. The importance of speciation of A. calcoaceticus- A. baumannii complex is well known. Not only is A. calcoaceticus an environmental non-pathogenic species, but the other species of the complex also vary in their clinical outcomes and show contrasting susceptibilities to antimicrobial agents14,15. Toh et al16 have highlighted the role of MALDI-TOF MS in Acinetobacter speciation. Within the genus Stenotrophomonas, much of the clinical significance is attached to S. maltophilia in being both environmental as well as opportunist pathogen17. The different species within Bcc not only differ in their geographical distribution, but also in their pathogenic potential, ranging from environmental contaminants to those posing an epidemic threat necessitating infection control measures2.
MALDI-TOF MS has not only reduced the average turnaround time but also the cost per isolate, with no loss in accuracy18,19. MALDI-TOF MS is a promising tool, but it needs optimization. This study was aimed to validate MALDI-TOF MS identification of NFGNB by comparing with the gold standard molecular techniques. An overall concordance of 100 per cent was obtained in correctly identifying the genus of NFGNB using MALDI-TOF MS, which corroborated well with earlier studies7,8. For the identification of isolates up to species level, a concordance of 73.33 per cent with the molecular methods was obtained. However, when the cut-off was optimized to ≥1.9, the identification rate increased to 82.6 per cent without any compromise in accuracy. Earlier studies have shown rates in the range of 72-77 per cent8,13,20 for species-level identification of NFGNB. It was worth noting that although the score was less for a few isolates of A. baumannii and S. maltophilia, yet there were no misidentifications. This was in contrast to the work of Espinal et al21 in which a few species of genus Acinetobacter were misidentified. One possible explanation could be the upgradation of the database in the Biotyper version 3 used in our study. Further, the isolates of A. baumannii and S. maltophilia which were not resolved up to species level due to scores <1.9 could also be correctly characterized by their close grouping in the PCA dendrogram. Spinali et al22 have also shown an upcoming role of MALDI-TOF MS in microbial typing. Though the concordance in species identification for B. cepacia isolates was only 30.77 per cent, similar to that reported by Fehlberg et al23, when they were subjected to PCA dendrogram, all but one isolate were placed correctly in their respective B. cenocepacia and B. cepacia groups. The cluster analysis method was also able to correctly group together 11 out of 13 B. cepacia isolates. In the current study, recA sequencing misidentified several isolates giving a concordance of 78.37 per cent for B. cenocepacia and 92.30 per cent for B. cepacia. Correct species identification in as low as 65.4 per cent of Bcc isolates using single gene target, i.e. recA sequencing has been noted earlier also24. A probable explanation could be that classification on the basis of a single gene will have lower discriminatory power than that based on a group of seven putative housekeeping genes, as done in E-MLST.
B. cenocepacia IIIA was found to be the most prevalent species in our setup25, and MALDI-TOF MS was able to identify all these isolates, although not to the lineage level. MALDI-TOF MS has shown promising results in correct identification of commonly occurring Bcc isolates elsewhere in the world as well26. Thus, harnessing the various applications of MALDI-TOF MS, beyond mere matching with the latest database, may not only improve the sensitivity of microbial identification but also give an insight into the predominant strains circulating in the patient population on the basis of different groups formed by PCA dendrogram.
The FA extraction step has aided in improving identification rates in case of Gram-positive cocci27, but such correlation has not been found in case of NFGNBs13. This pre-analytic step of extraction has not shown any significant improvement in our study also. Our findings support a cut-off of ≥1.9 for reliable species identification of these NFGNBs.
In conclusion, leaving aside the high setup cost, the prompt identification with a high discriminatory power at low running cost per isolate makes MALDI-TOF MS a suitable tool for the characterization of difficult to identify microorganisms in high-throughput laboratories.
Footnotes
Conflicts of Interest: None.
References
- 1.Slama TG. Gram-negative antibiotic resistance: There is a price to pay. Crit Care. 2008;12(Suppl 4):S4. doi: 10.1186/cc6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gautam V, Singhal L, Ray P. Burkholderia cepacia complex: Beyond Pseudomonas and Acinetobacter. Indian J Med Microbiol. 2011;29:4–12. doi: 10.4103/0255-0857.76516. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative Gram-negative bacteria. Clin Infect Dis. 1998;27(Suppl 1):S93–9. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 4.Barin J, Martins AF, Heineck BL, Barth AL, Zavascki AP. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann Clin Microbiol Antimicrob. 2013;12:15. doi: 10.1186/1476-0711-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vila J, Pachón J. Therapeutic options for Acinetobacter baumannii infections: An update. Expert Opin Pharmacother. 2012;13:2319–36. doi: 10.1517/14656566.2012.729820. [DOI] [PubMed] [Google Scholar]
- 6.Lee SY, Shin JH, Kim SH, Shin MG, Suh SP, Ryang DW. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based VITEK MS system for the identification of Acinetobacter species from blood cultures: Comparison with VITEK 2 and MicroScan systems. Ann Lab Med. 2015;35:62–8. doi: 10.3343/alm.2015.35.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnini S, Ghelardi E, Brucculeri V, Morici P, Lupetti A. Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol. 2015;15:124. doi: 10.1186/s12866-015-0459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, et al. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting Gram-negative bacilli isolated from cultures from cystic fibrosis patients. J Clin Microbiol. 2012;50:2034–9. doi: 10.1128/JCM.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautam V, Ray P, Vandamme P, Chatterjee SS, Das A, Sharma K, et al. Identification of lysine positive non-fermenting Gram negative bacilli (Stenotrophomonas maltophilia and Burkholderia cepacia complex) Indian J Med Microbiol. 2009;27:128–33. doi: 10.4103/0255-0857.49425. [DOI] [PubMed] [Google Scholar]
- 10.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13:807–15. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 11.Whitby PW, Carter KB, Burns JL, Royall JA, LiPuma JJ, Stull TL. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J Clin Microbiol. 2000;38:4305–9. doi: 10.1128/jcm.38.12.4305-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spilker T, Baldwin A, Bumford A, Dowson CG, Mahenthiralingam E, LiPuma JJ. Expanded multilocus sequence typing for Burkholderia species. J Clin Microbiol. 2009;47:2607–10. doi: 10.1128/JCM.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford BA, Burnham CA. Optimization of routine identification of clinically relevant Gram-negative bacteria by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and the Bruker Biotyper. J Clin Microbiol. 2013;51:1412–20. doi: 10.1128/JCM.01803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang YC, Sheng WH, Li SY, Lin YC, Wang JT, Chen YC, et al. Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with Acinetobacter bacteremia. Clin Infect Dis. 2011;52:352–60. doi: 10.1093/cid/ciq154. [DOI] [PubMed] [Google Scholar]
- 15.Lim YM, Shin KS, Kim J. Distinct antimicrobial resistance patterns and antimicrobial resistance-harboring genes according to genomic species of Acinetobacter isolates. J Clin Microbiol. 2007;45:902–5. doi: 10.1128/JCM.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toh BE, Paterson DL, Kamolvit W, Zowawi H, Kvaskoff D, Sidjabat H, et al. Species identification within Acinetobacter calcoaceticus-baumannii complex using MALDI-TOF MS. J Microbiol Methods. 2015;118:128–32. doi: 10.1016/j.mimet.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–25. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 18.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48:1169–75. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westblade LF, Garner OB, MacDonald K, Bradford C, Pincus DH, Mochon AB, et al. Assessment of reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry for bacterial and yeast identification. J Clin Microbiol. 2015;53:2349–52. doi: 10.1128/JCM.00187-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homem de Mello de Souza HA, Dalla-Costa LM, Vicenzi FJ, Camargo de Souza D, Riedi CA, Filho NA, et al. MALDI-TOF: A useful tool for laboratory identification of uncommon glucose non-fermenting Gram-negative bacteria associated with cystic fibrosis. J Med Microbiol. 2014;63(Pt 9):1148–53. doi: 10.1099/jmm.0.076869-0. [DOI] [PubMed] [Google Scholar]
- 21.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin Microbiol Infect. 2012;18:1097–103. doi: 10.1111/j.1469-0691.2011.03696.x. [DOI] [PubMed] [Google Scholar]
- 22.Spinali S, van Belkum A, Goering RV, Girard V, Welker M, Van Nuenen M, et al. Microbial typing by matrix-assisted laser desorption ionization-time of flight mass spectrometry: Do we need guidance for data interpretation? J Clin Microbiol. 2015;53:760–5. doi: 10.1128/JCM.01635-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehlberg LC, Andrade LH, Assis DM, Pereira RH, Gales AC, Marques EA. Performance of MALDI-ToF MS for species identification of Burkholderia cepacia complex clinical isolates. Diagn Microbiol Infect Dis. 2013;77:126–8. doi: 10.1016/j.diagmicrobio.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Cesarini S, Bevivino A, Tabacchioni S, Chiarini L, Dalmastri C. RecA gene sequence and multilocus sequence typing for species-level resolution of Burkholderia cepacia complex isolates. Lett Appl Microbiol. 2009;49:580–8. doi: 10.1111/j.1472-765X.2009.02709.x. [DOI] [PubMed] [Google Scholar]
- 25.Singhal L, Gautam V, Kaur M, Ray P. In vitro activity of doripenem against Burkholderia cepacia complex isolates from non-cystic fibrosis patients. Antimicrob Agents Chemother. 2011;55:1320–1. doi: 10.1128/AAC.01386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambiase A, Del Pezzo M, Cerbone D, Raia V, Rossano F, Catania MR. Rapid identification of Burkholderia cepacia complex species recovered from cystic fibrosis patients using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Microbiol Methods. 2013;92:145–9. doi: 10.1016/j.mimet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 27.McElvania Tekippe E, Shuey S, Winkler DW, Butler MA, Burnham CA. Optimizing identification of clinically relevant Gram-positive organisms by use of the Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J Clin Microbiol. 2013;51:1421–7. doi: 10.1128/JCM.02680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]