Abstract
P16INK4a immunohistochemistry (IHC) is widely used to facilitate the diagnosis of human papillomavirus (HPV)-associated cervical precancerous lesions. While most p16 results are distinctly positive or negative, certain ones are ambiguous: they meet some but not all requirements for the “block-positive” pattern. It is unclear whether ambiguous p16 immunoreactivity indicates oncogenic HPV infection or risk of progression. Herein, we compared HPV genotypes and subsequent High-grade Squamous Intraepithelial Lesion (HSIL) outcomes among 220 cervical biopsies with a differential diagnosis of Cervical Intraepithelial Neoplasia 2 (CIN 2) based on hematoxylin and eosin morphology and varying degrees of p16 immunoreactivity. P16 results were classified as block-positive (n=40, 18%), negative (n=130, 59%), or ambiguous (n=50, 23%), a category we further grouped into three patterns: strong/basal (n=18), strong/focal (n=15), and weak/diffuse (n=17). 70% of ambiguous p16 lesions were negative for the most common low- and high-risk HPV types; the remaining 30% were positive for HPV 16, 18, 45, 58, 59, or 66. Three patterns revealed comparably low HPV detection rates (28%, 27% and 35%). During 12-month surveillance, HSILs were detected in 35% of the p16 block-positive group, 1.5% of negative group, and 16% of the ambiguous group. The accuracy of ambiguous p16 immunoreactivity in predicting oncogenic HPV and HSIL outcome is significantly lower than that of the block-positive pattern but greater than negative staining. Specific guidelines for this intermediate category should prevent diagnostic errors and help implement p16 IHC in general practice.
Keywords: P16 immunohistochemistry, CIN 2, Human papillomavirus
Introduction
Pathological diagnosis and classification of human papillomavirus (HPV)-associated cervical precancerous lesions have advanced from heavy reliance on morphologic interpretation to a new era that integrates biomarkers and HPV genotypes [1, 2]. In 2012, the Lower Anogenital Squamous Terminology (LAST) project recommended a two-tiered classification: Low-grade and High-grade Squamous Intraepithelial Lesion (LSIL, HSIL) [3]. The new classification eliminated Cervical Intraepithelial Neoplasia 2 (CIN 2), an intermediate category comprised of transient HPV infection (LSIL) along with transforming infection that carries true cancer risk (HSIL) [4]. The latter must be discerned in order to initiate an active intervention. Making this distinction solely based on hematoxylin and eosin (H&E) morphology can be challenging and subjective. Biomarker p16INK4a immunohistochemistry (p16 IHC), shown to improve diagnostic accuracy for HSIL, has been recommended by LAST as a supplement to aid this task [5, 6]. It is critical for pathologists to update their knowledge on how judiciously to use p16 as well as interpret its results [7, 8].
LAST defines “block-positive” p16 as supporting a diagnosis of HSIL, provided the staining meets certain criteria: (1) demonstrates strong nuclear with or without cytoplasmic signal, (2) extends from the basal layers upward at least one third of the epithelium, and (3) extends laterally over a significant distance. Following these criteria, most cases are straightforward to interpret: strong/diffuse (positive) or no stain (negative). When the signal is weak/focal, pathologists generally interpret it as negative as well. Apart from these uncomplicated cases, certain p16 results are ambiguous: they meet some but not all requirements for the “block-positive” pattern. One such example would be a strong nuclear and cytoplasmic staining that is nevertheless limited to a portion of the dysplastic epithelium. Whether equivocal lesions should be upgraded to HSIL based on such strong but limited staining has become an ongoing debate in daily practice.
It is unclear whether ambiguous p16 immunoreactivity indicates risk of progression or even the presence of oncogenic HPV. To answer this question, we compared HPV genotypes as well as subsequent HSIL outcomes among 220 cervical biopsies with differential diagnosis of CIN 2 on H&E morphology and varying degrees of p16 immunoreactivity. Our goal was to assess the ramifications of ambiguous p16 immunoreactivity and thereby to provide diagnostic guidance on this practical and important issue.
Materials and methods
Morphological diagnostic criteria and case selection
Institutional Review Board approval was obtained from the Mount Sinai School of Medicine. The pathology database was searched between January 2012 and June 2015 for cervical biopsies that had p16 IHC performed at the time of diagnosis. Blinded from p16 IHC result, authors (Y.L and M.A) reviewed all H&E slides and categorized lesions into unequivocal HSIL, unequivocal LSIL, and “morphologic CIN 2”. In cases where no agreement was reached between the two reviewers, one additional pathologist (T.K.) was consulted, and final consensus diagnosis was reached among the three at a multi-headed microscope. Only “morphologic CIN 2” cases were included in the study. Unequivocal HSIL and LSIL cases were excluded.
Our practice follows the morphological diagnostic criteria as recommended by LAST [3]. LSILs are lesions demonstrating koilocytosis and/or abnormal nuclear features and mitotic activity within the lower one third of the epithelium. The abnormal nuclear features refer to increased nuclear size, irregular nuclear membranes, and increased nuclear to cytoplasmic ratio. HSILs are lesions demonstrating abnormal nuclear features and mitotic activity involving the middle third or the full thickness of the epithelium.
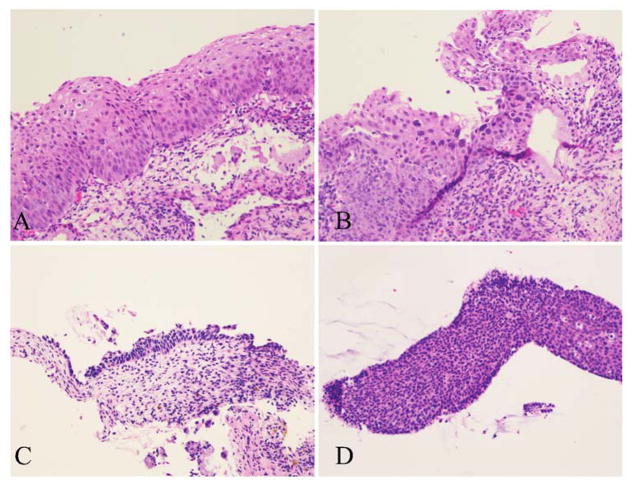
“Morphologic CIN 2” is a heterogeneous group consisting of lesions with certain histological features suspicious but not definitive for HSIL. Four of the most common histological features are illustrated in Fig 2: (A) mitoses located in the middle level of epithelium and/or the presence of atypical mitoses; (B) marked koilocytic atypia, defined as nuclear enlargement ≥ 5 times the size of an intermediate cell nucleus or multinucleation with ≥ 5 nuclei [9]; (C) eroded or thin epithelium displaying abnormal nuclear features; and (D) tangentially sectioned epithelium resulting in substantial expansion of the basal and parabasal layers.
Figure 2.
Four of the most common histological features trigger the differential diagnosis of CIN 2 and deployment of p16 IHC. A, Mitoses located in the middle level and/or atypical mitoses. B, Marked koilocytic atypia. C, Eroded or thin epithelium displaying abnormal nuclear features. D, Tangentially sectioned epithelium with substantial expansion of the basal and parabasal layers (H&E, original magnification ×200).
Medical charts were reviewed for patients’ subsequent clinical management and outcome within 12 months of the initial biopsy, including cervical cytology, biopsy and excisional procedure. We excluded patients with less than 12 months of follow-up, prior excisional procedures, or cervical glandular lesions.
P16 immunohistochemistry and interpretation
IHC was performed using a mouse monoclonal antibody to p16 (Roche E6H4™, catalog #725-4713) on a Ventana Benchmark LT automated immunostainer (Tucson AZ, USA) according to standard protocol. Positive and negative controls were included routinely. Positive signal was defined as nuclear ± cytoplasmic staining. Cytoplasmic staining alone was considered as negative. Without knowledge of the HPV results and follow-up outcome, authors (Y.L and M.A) reviewed all IHC slides and recorded a detailed description of p16 immunoreactivity for each lesion based on four parameters: (1) intensity: strong (dark brown color similar to the positive control) versus weak (yellow color significantly lighter than the positive control); (2) extent: diffuse (signal involves > 50% of the epithelium) versus focal (< 50% of the epithelium); (3) continuity: continuous (staining extends laterally over a significant distance) versus discontinuous (alternating clusters of either positively or negatively stained cells); and (4) location: positive cells reside in the lower third, two thirds, or full thickness of epithelium.
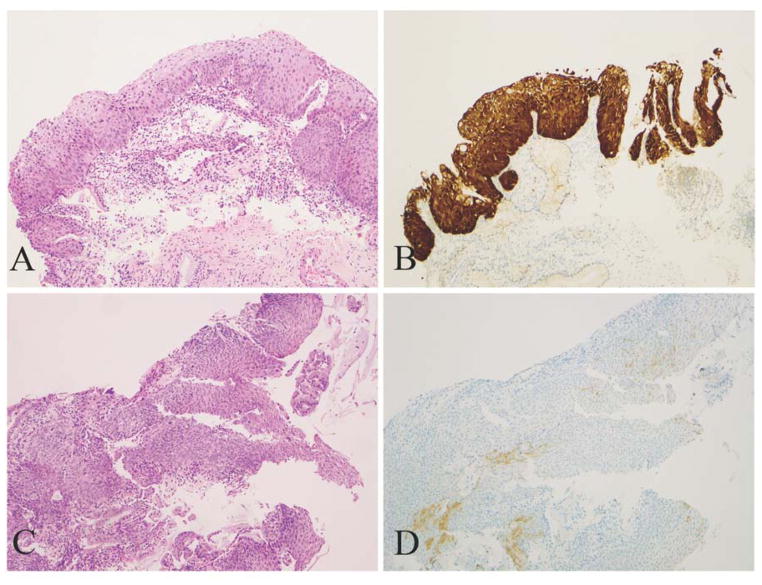
Based on these four parameters, lesions were categorized as block-positive, negative, and ambiguous pattern. Block-positive patterns (Fig. 1A and B) fulfilled all requirements described in LAST: strong and diffuse immunoreactivity extending from the basal layers upward more than one third of the epithelium and laterally over a significant distance. Negative results (Fig. 1C and D) were defined as total absence of staining or else weak, focal, and discontinuous staining.
Figure 1.
Morphologic CIN 2 lesions display distinctly positive and negative p16 results. A and B, A lesion displays strong/diffuse staining (block-positive in LAST terms) and tests positive for HPV 16. C and D, A lesion displays weak, discontinuous and focal staining (negative) and tests negative for HPV (A, C: H&E, original magnification ×100; B, D: corresponding p16 IHC, original magnification ×100).
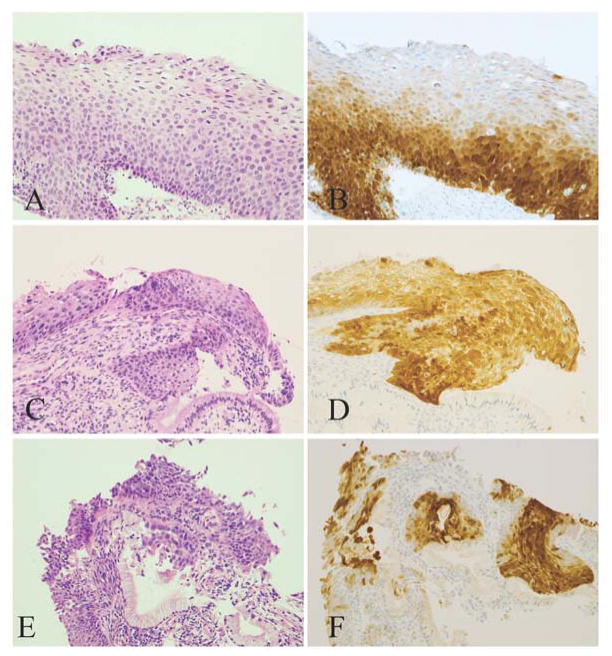
Certain p16 results that met some but not all requirements for the “block-positive” pattern were classified as ambiguous. Based on the four parameters, ambiguous results were further grouped into 3 patterns: (1) strong/basal (strong, diffuse, continuous staining of the lower third of epithelium without upward extension, Fig. 3A and B), (2) weak/diffuse (weak, diffuse, discontinuous staining reaching at least two thirds, Fig. 3C and D), and (3) strong/focal (strong, focal, and discontinuous staining located at any level of the epithelium, Fig. 3E and F). Consensus for each case was reached among the authors at a double-headed microscope.
Figure 3.
Morphologic CIN 2 lesions with ambiguous p16 immunoreactivity. A and B, A lesion displays strong/basal pattern and tests positive for HPV 16. Patient’s subsequent LEEP reveals HSIL. C and D, A lesion displays weak/diffuse pattern and tests negative for HPV. Subsequent biopsy reveals LSIL. E and F, A lesion displays strong/focal pattern and tests positive for HPV 58. Subsequent biopsy reveals LSIL. (A, C, and E: H&E, original magnification ×200; B, D, and F: corresponding p16 IHC, original magnification ×200).
HPV genotyping
The Maxwell 16 FFPE Tissue LEV DNA Kit (Promega, Madison, Wisconsin) was used for DNA extraction from tissue sections according to the manufacturer’s instructions. The concentrations of extracted DNAs were measured by a NanoDrop ND-2000 spectrophotometer. Real time PCR was performed with Roche Light cycler 480 and a High Resolution Melting Master kit (Roche, Indianapolis, IN) using PCR primers (GP5+ and GP6+) specifically targeted to L1 region covering the 13 most common high-risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66) and 10 low-risk types (6, 11, 13, 30, 32, 34, 40, 42, 53, 61) [10]. Beta-actin gene was used as a quality control for nucleic acid extraction and PCR reaction. Once the initial screening for HPV DNA was confirmed, type-specific primers and probes for HPV 16 or HPV 18 (targeting E6 region) were used to detect HPV 16 or HPV 18 by real time PCR. The probe pairs were labeled with fluorescein at the 3′ end and LightCycler-Red 640 for HPV 16 or LightCycler-Red 670 for HPV 18 at the 5′ end. When HPV 16/18 specific PCR was negative, Sanger Sequencing was performed to detect other HPV genotypes after amplification with GP5+ and GP6+ primer pairs. HPV subtypes other than HPV 16/18 were determined by aligning sequences in GeneBank.
Statistical analyses
Statistical analyses were performed in STATA 13 (Stata Corporation, College Station TX). Fisher’s test was used to determine the significance of associations between parameters. A p-value <0.001 was considered statistically significant.
RESULTS
Varying degrees of p16 immunoreactivity among morphologic CIN 2
A total of 220 cervical lesions (one per patient) with a differential diagnosis of CIN 2 based on H&E morphology were retrospectively studied. Patient ages ranged from 18 to 73 years old (mean 39). Five pathologists (two senior gynecological pathologists and three junior surgical pathologists) originally reported these cases as benign (n=88), LSIL/CIN 1 (n=90), HSIL/CIN 2 (n=35), and HSIL/CIN 3 (n=7).
40 cases (18%) revealed strong and diffuse staining corresponding to a block-positive result as described in LAST. 130 cases (59%) revealed negative results, including total absence of staining (n=71) or weak staining extending < 50% of the epithelium (n=59). The remaining 50 cases (23%) revealed ambiguous staining patterns: strong/basal (n=18, 8%), strong/focal (n=15, 7%), and weak/diffuse (n=17, 8%).
Oncogenic HPV subtypes
The 50 biopsies with ambiguous p16 were tested for oncogenic HPV subtypes. As shown in Table 1, 35 (70%) were negative for 13 high-risk and 10 low-risk strains, whereas 15 (30%) were positive, including HPV 16 (n=5), HPV 18 (n=4), HPV 66 (n=3), and one for each of HPV 45, 58, and 59. 13 of 18 (72%) strong/basal p16 lesions were negative for HPV; the remaining 5 (28%) were positive for HPV 16 (n=2), 45, 59, and 66. Among 15 strong/focal p16 lesions, 11 (73%) were negative, whereas 4 (27%) were positive for HPV 16, 18 (n=2), and 58. Among 17 weak/diffuse p16 lesions, 11 (65%) were negative, while 6 (35%) were positive for HPV 16 (n=2), 18 (n=2), and 66 (n=2). Upon reviewing H&E slides, lesions testing positive or negative for HPV DNA revealed significantly overlapping morphological features. Five cases with negative p16 staining were tested as negative controls, none revealing HPV. Another 5 cases with block-positive p16 staining were tested as positive controls, and all of them were positive for high-risk HPV, including 2 for HPV 16 and 3 for HPV 18.
Table 1.
The presence or absence of oncogenic HPV in morphologic CIN 2 lesions with ambiguous p16 immunoreactivity
| HPV DNA | Ambiguous P16 Immunoreactivity (n=50)
|
||
|---|---|---|---|
| Strong/basal (n=18) | Strong/focal (n=15) | Weak/diffuse (n=17) | |
|
| |||
| Absent (n=35) | 13 (72%) | 11 (73%) | 11 (65%) |
|
| |||
| Present (n=15) | 5 (28%) | 4 (27%) | 6 (35%) |
|
| |||
| Types | 16 (x2) 45 59 66 |
16 18 (x2) 58 |
16 (x2) 18 (x2) 66 (x2) |
HSIL outcome
Concurrent HSIL was present in 4 (10%) patients from the block-positive p16 group, 6 (12%) from the ambiguous group, and 1 (0.8%) from the negative group. All patients underwent surveillance at our facility for 12 months, including cytology, biopsy or excisional procedures. As shown in Table 2, 14 of 40 (35%) block-positive p16 patients revealed HSIL on subsequent procedures while the remaining 26 (65%) revealed LSIL or less. In the ambiguous p16 group, 8 out of 50 (16%) revealed HSIL, including 2 with strong/basal pattern (HPV 16, 45), 5 with strong/focal (HPV 16, 18×2, and negative ×2), and 1 with weak/diffuse pattern (HPV 16). Both HPV negative cases had concurrent HSIL lesions on biopsy. The remaining 42 (84%) revealed LSIL or less. In the negative p16 group, only 2 of 130 patients (1.5%) revealed HSIL; one of them had concurrent HSIL on biopsy. The remaining 128 (98.5%) revealed LSIL or less. The block-positive p16 group revealed a significantly higher percentage of HSIL outcomes than the ambiguous group during follow-up (p<0.001). Both block-positive and ambiguous p16 groups revealed a significantly higher percentage of HSIL outcomes than the negative group during follow-up (p<0.001).
Table 2.
Clinical outcomes for morphologic CIN 2 lesions with varying degrees of p16 immunoreactivity during 12-month follow-up
| P16 IHC | N | Concurrent HSIL | Follow-up within 12 months | |
|---|---|---|---|---|
|
| ||||
| HSIL | LSIL or less | |||
| Block-positive | 40 | 4 (10%) | 14 (35%)*# | 26 (65%) |
| Ambiguous | 50 | 6 (12%) | 8 (16%)* | 42 (84%) |
| Negative | 130 | 1 (0.8%) | 2 (1.5%) | 128 (98.5%) |
| Total | 220 | 11 (5%) | 24 (11%) | 196 (89%) |
Both block-positive and ambiguous p16 groups revealed significantly higher percentages of HSIL outcome than the negative group during 12-month follow-up (p<0.001).
The block-positive p16 group revealed a significantly higher percentage of HSIL outcome than the ambiguous group during 12-month follow-up (p<0.001).
Discussion
The use of p16 IHC has significantly increased in most practices since the publication of LAST recommendations [11–13]. Our group employs p16 primarily to triage lesions with the morphologic differential diagnosis of CIN 2 into LSIL vs. HSIL categories. Additionally, LAST recommends p16 to differentiate HSILs from their mimics, solve disagreement, and to search for subtle HSILs in cases with high-risk cytology. Regardless of application, the dramatic increase of p16 IHC has opened the door to a wide spectrum of staining results. We previously reported two notable p16-related diagnostic errors that led to over-diagnosis of HSIL in routine practice: (1) overusing p16 IHC on unequivocal LSIL and (2) upgrading questionable lesions to HSIL based on p16 non-block staining patterns [14]. In the present study, we focus on the latter, aiming to assess the ramifications of non-block p16 staining as well as to provide interpretative guidance on this practical and important issue.
Upon retrospective review, the majority of CIN 2 lesions (77%) in our series revealed p16 that was distinctly positive (18%) or negative (59%). In such instances, p16 IHC is a convenient adjunctive tool that promptly offers straightforward answers, especially for morphologically challenging cases. Multiple studies have established that among pathologists and even non-pathologists, p16 improves diagnostic accuracy for precancerous lesions [15, 16]. A pertinent study by Liao et al. showed that after receiving video-based training, 12 non-pathologist physicians were able to diagnose HSIL solely based upon p16 IHC with 88% sensitivity and 87% specificity, rates that were only slightly inferior to pathologists involved in the study (96% and 92%) [17].
Apart from these clear-cut cases, we found that difficulties arose when a p16 result met some but not all requirements for the “block-positive” pattern, instances we refer to as ambiguous pattern. 23% of morphologic CIN 2 lesions in our series followed this scenario. The ambiguous patterns we frequently encountered were strong/basal (8%), strong/focal (7%) and weak/diffuse (8%). For example, despite strong and continuous nuclear/cytoplasmic staining, the strong/basal pattern did not meet the height requirement for “block-positive” as specifically defined in LAST. Can we disregard strong/basal staining as negative, and if not, should we upgrade such lesions to HSIL?
To answer this question, we tested ambiguous p16 lesions for the presence/absence of HPV DNA. Surprisingly, 70% were in fact negative for the most common 13 high-risk and 10 low-risk HPV strains. Only 30% harbored HPV, including HPV 16/18 (60%) and the less common types 45, 58, 59, and 66 (40%). The 3 patterns revealed comparably low HPV detection rates: strong/basal (28%), strong/focal (27%), and weak/diffuse pattern (35%). By way of comparison, we tested 5 block-positive p16 lesions, all of which harbored HPV 16/18. Of the 5 negative p16 lesions tested, none were positive for HPV.
In regards to HPV detection rates, our ambiguous patterns are akin to the focal strong staining pattern described in several studies that employed semiquantitative p16 scoring methods. In a widely used scoring system, Klaes et al. categorized p16 results into focal (with a cutoff defined as < 25% of epithelium stained positive) vs. diffuse (>25% of epithelium) [18]. They detected HR-HPV in 27% of focal and 76% of diffuse cases. When Keating et al. increased the cutoff to 80%, HR-HPV remained low in the focal strong cases (30%) and high in the diffuse ones (70%) [19]. In contrast, block-positive p16 is strongly associated with the presence of HR-HPV as demonstrated by high positive predictive value (97%) as well as negative predictive value (86%) [20, 21].
We further compared surveillance results among CIN 2 lesions with varying degrees of p16 immunoreactivity. HSIL was subsequently detected in 16% of patients with ambiguous p16 lesions, including 2 strong/basal, 5 strong/focal, and 1 weak/diffuse pattern. The remaining patients, even those harboring HR-HPV, revealed LSIL or less for 12 months. Conversely, HSIL outcome was significantly higher in the block-positive p16 group (35%) and lower in the negative p16 group (1.5%). Our results are consistent with a large prospective study recently published by Miralpeix et al. that assessed p16’s status as a predictor of CIN 2 behavior [22]. They report that all “p16 negative CIN 2” regressed while 43% of “p16 positive CIN 2” persisted or progressed to CIN 3 at 12 months. We recommend “p16 ambiguous CIN 2” as a distinct category of cervical lesions that appear to pose an intermediate risk of persistence and progression.
P16 is a commonly used IHC marker; however, its interpretation is unique in the context of HPV-related lower anogenital lesions. Both quality and quantity of p16 immunoreactivity affect its specificity in predicting high-risk HPV and HSIL outcomes. One cannot simply designate it as positive or negative; rather, pathologists must consider multiple parameters such as staining intensity, extent, continuity, and location. When using p16 to triage CIN 2 lesions, pathologists need to be aware of the ramifications of an ambiguous p16 result. Specific guidelines for this intermediate category should prevent diagnostic errors while helping to implement LAST in general practice.
Acknowledgments
We thank John Stone for assistance in editing this article.
Footnotes
Disclosures: The authors declare that there are no conflicts of interest to disclose. No funding was received for this study.
References
- 1.Pinto AP, Crum CP, Hirsch MS. Molecular markers of early cervical neoplasia. Diagn Histopathol. 2010;16:445–54. doi: 10.1016/j.mpdhp.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuschieri K, Wentzensen N. HPV mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2008;17:2536–45. doi: 10.1158/1055-9965.EPI-08-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darragh TM, Colgan T, Cox JT, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-Associated Lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205–42. doi: 10.1097/LGT.0b013e31825c31dd. [DOI] [PubMed] [Google Scholar]
- 4.Castle PE, Stoler MH, Solomon D, et al. The relationship of community biopsy-diagnosed cervical intraepithelial neoplasia grade 2 to the quality control pathology-reviewed diagnoses: an ALTS report. Am J Clin Pathol. 2007;127:805–15. doi: 10.1309/PT3PNC1QL2F4D2VL. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron C, Ordi J, Schmidt D, et al. Conjunctive p16INK4a testing significantly increases accuracy in diagnosing high-grade cervical intraepithelial neoplasia. Am J Clin Pathol. 2010;133:395–406. doi: 10.1309/AJCPXSVCDZ3D5MZM. [DOI] [PubMed] [Google Scholar]
- 6.Galgano MT, Castle PE, Atkins KA, et al. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Pathol. 2010;34:1077–1087. doi: 10.1097/PAS.0b013e3181e8b2c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Bogaert LJ. P16INK4a immunocytochemistry/immunohistochemistry: need for scoring uniformization to be clinically useful in gynecological pathology. Ann Diagn Pathol. 2012;16:422–6. doi: 10.1016/j.anndiagpath.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Van Bogaert LJ. Cervical preneoplasia biomarkers: a conundrum for the community based gynecologic surgical pathologist. J Gynecol Oncol. 2014;25:3–5. doi: 10.3802/jgo.2014.25.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park K, Ellenson LH, Pirog EC. Low-grade squamous intraepithelial lesions of the cervix with marked cytological atypia-clinical follow-up and human papillomavirus genotyping. Int J Gynecol Pathol. 2007;26:457–62. doi: 10.1097/pgp.0b013e31802f64ab. [DOI] [PubMed] [Google Scholar]
- 10.Husman AM, Walboomers JM, Van den Brule AJ, et al. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76:1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 11.Castle PE. A LASTing impression: incorporating p16 immunohistochemistry into routine diagnosis of cervical neoplasia. Pathology Case Reviews. 2013;18:154–157. [Google Scholar]
- 12.Clinton LK, Miyazaki K, Ayabe A, et al. The LAST Guidelines in Clinical Practice: Implementing Recommendations for p16 Use. Am J Clin Pathol. 2015;144:844–9. doi: 10.1309/AJCPUXLP7XD8OQYY. [DOI] [PubMed] [Google Scholar]
- 13.Thrall MJ. Effect of lower anogenital squamous terminology recommendations on the use of p16 immunohistochemistry and the proportion of high-grade diagnoses in cervical biopsy specimens. Am J Clin Pathol. 2016;145:524–30. doi: 10.1093/ajcp/aqw032. [DOI] [PubMed] [Google Scholar]
- 14.Clark JL, Lu D, Kalir T, et al. Overdiagnosis of HSIL on cervical biopsy: errors in p16 immunohistochemistry implementation. Hum Pathol. 2016;55:51–6. doi: 10.1016/j.humpath.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Reuschenbach M, Wentzensen N, Dijkstra MG, et al. p16INK4a immunohistochemistry in cervical biopsy specimens: A systematic review and meta-analysis of the interobservor agreement. Am J Clin Pathol. 2014;142:767–72. doi: 10.1309/AJCP3TPHV4TRIZEK. [DOI] [PubMed] [Google Scholar]
- 16.Omori M, Hashi A, Nakazawa K, et al. Estimation of prognoses for cervical intraepithelial neoplasia 2 by p16INK4a immunoexpression and high-risk HPV in situ hybridization signal types. Am J Clin Pathol. 2007;128:208–217. doi: 10.1309/0UP5PJK9RYF7BPHM. [DOI] [PubMed] [Google Scholar]
- 17.Liao GD, Kang LN, Chen W, et al. P16 immunohistochemistry interpretation by nonpathologists as an accurate method for diagnosing cervical precancer and cancer. J Lower Gen Tract Dis. 2015;19:207–11. doi: 10.1097/LGT.0000000000000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaes R, Benner A, Friedrich T, et al. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol. 2002;26:1389–99. doi: 10.1097/00000478-200211000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Keating JT, Cviko A, Riethdorf S, et al. Ki-67, Cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol. 2001;25:884–91. doi: 10.1097/00000478-200107000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Sano T, Oyama T, Kashiwabara K, et al. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153:1741–8. doi: 10.1016/S0002-9440(10)65689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benevolo M, Mottolese M, Marandino F, et al. Immunohistochemical expression of p16INK4a is predictive of HR-HPV infection in cervical low-grade lesions. Mod Pathol. 2006;19:384–91. doi: 10.1038/modpathol.3800551. [DOI] [PubMed] [Google Scholar]
- 22.Miralpeix E, Genovés J, Solé-Sedeño JM, et al. Usefulness of p16INK4a staining for managing histological high-grade squamous intraepithelial cervical lesions. Mod Pathol. 2017;30:304–10. doi: 10.1038/modpathol.2016.168. [DOI] [PubMed] [Google Scholar]