Abstract
RNA-Seq enables the generation of extensive transcriptome information providing the capability to characterize transcripts (including alternative isoforms and polymorphism), to quantify expression and to identify differential regulation in a single experiment. To reveal the capacity of new anti-HIV ABX464 candidate in modulating the expression of genes, datasets were generated and validated using RNA-seq approach. This comprehensive dataset will be useful to deepen the comprehensive understanding of the progression of human immunodeficiency virus (HIV) associated with mucosal damage in the gastrointestinal (GI) tract and subsequent inflammation, providing an opportunity to generate new therapies, diagnoses, and preventive strategies.
Subject terms: High-throughput screening, Data processing, Data acquisition
Background & Summary
RNA sequencing (RNA-seq) transcriptome analysis using massively parallel next generation sequencing technology provides the capability to understand global changes in gene expression throughout a range of tissue samples and have become the gold standard for in depth characterization of novel transcript isoforms. RNA-seq (Quantification) is used to analyse gene expression of certain biological objects under specific conditions. It is a cost-effective quantification method that produces high reproducibility, high accuracy and wide dynamic range. As described in our related manuscript submitted earlier in Scientific Reports1, our aim in this study is to consolidate the protective effect of ABX464 in DSS-induced colitis comparing transcript levels between bone marrow-derived macrophages (BMDMs) ABX464-treated stimulated or not by LPS. To perform this task we sequenced here 24 samples of C57BL6 strains of mice using BGI RNA sequencing (RNA-seq) technology2,3.
ABX464 is a first-in-class antiviral drug candidate used to treat patients infected with human immunodeficiency virus. ABX464 is an orally available, well-tolerated small molecule that blocks HIV replication through the modulation of RNA biogenesis. ABX464 demonstrates anti-viral activity in treatment-naïve patients and induces a long-lasting control of the viral load in HIV-infected humanized mice after treatment arrest. This treatment might improve current therapies by hampering the appearance of virus resistance and, more importantly, lead to a long-lasting control of HIV.
Despite the successful control of viremia, many HIV-infected individuals treated with antiretroviral therapy (ART) exhibit residual inflammation associated with non-AIDS-related morbidity and mortality. Several reports have shown that measures of inflammation and immune activation are the best independent predictors of disease progression in HIV-infected individuals. It is also known that elevated inflammation and immune activation predict non-AIDS-associated morbidities and mortality, even in well-suppressed treated HIV-infected individuals. Mucosal damage to the gastrointestinal (GI) tract with resulting microbial translocation significantly contributes to the heightened and persistent chronic inflammation and immune activation characteristic to HIV infection and might contribute to virus persistence during ART. Recently, a dextran sulfate sodium (DSS)-induced nonhuman primate (NHP) colitis model in SIV-infected rhesus macaques a natural host species for SIV that does not manifest GI tract damage or chronic immune activation during infection, was used to demonstrate that DSS-exposure results in colitis with elevated levels of plasma SIV RNA, sCD14, LPS, CRP and mucosal CD4 T cell loss4,5. The pathogenic potential of bacterial translocation has also been studied in humanized mice exposed to DSS to provide evidence that HIV chronic infection contributes to systemic inflammation under DSS-exposure6,7. Given the similarities in colitis induction between DSS-exposed macaques and mice5, the antiviral activity of ABX464 in humanized mice model8 and its pharmacodynamics and pharmacoketics profiles in rodent, NHP8 and humans9, the anti-inflammatory properties of this drug in DSS-treated mice were tested (Chebli et al.). The results presented in this manuscript based on the use of DSS-induced colitis in mice, show for the first time that an anti-HIV drug can specifically act on the immune system to resolve the inflammation.
The principal aim of this study was to provide a comprehensive resource to deposit the raw RNA-seq data sets underpinning the advanced high-resolution transcriptome analysis of ABX464-treated BMDMs stimulated with LPS. To this end the data from three distinct group of BMDM cultures were analyzed for different gene expression levels using RNA-seq. We performed a high-resolution transcriptome analysis of ABX464-treated BMDMs stimulated or unstimulated with LPS. In this work, the RNA-seq libraries were sequenced using the BGISEQ-500 platform which use the DNA nanoball technology. Approximately 1 billion 50-bp single-end RNA-seq reads were generated, with an average of over 45 million sequence reads per sample.
24 intestinal macrophages mRNA profiles of three distinct group of BMDM cultures from mice were generated by deep sequencing, using Illumina protocol. The sequence reads that passed quality filters were analyzed at the transcript isoform level with two methods: HISAT10 and Bowtie2 (ref. 11) followed by genes expression quantification using a software package called RSEM12.
Using an optimized data analysis workflow, about 45 million sequence reads per sample were mapped to the mouse genome (build mm9) and identified 1,383 expressed genes in the LPS-stimulated BMDMs among which 439 and 397 genes increased and decreased in expression >=1.5 log2-fold, respectively. Altered expression of some genes was confirmed with qPCR, demonstrating the high degree of sensitivity of the RNA-seq method. Hierarchical clustering of differentially expressed genes uncovered several as yet uncharacterized genes that may contribute to inflammatory function. Data analysis with HISAT10/Bowtie2 (ref. 11) and RSEM12 workflows revealed a significant overlap yet provided complementary insights in transcriptome profiling. A comprehensive RNA-seq data resource was provided on the anti-inflammatory properties of ABX464 that warrant exploration in both HIV and inflammatory ulcerative colitis (UC) disease. The data generated here will be valuable to the vision research community for characterizing global changes in gene expression and facilitate a better understanding of the complexities during anti-AIDS drugs treatment and then can help to develop better ways to combat HIV.
Methods
Macrophages RNA-seq preparation
Cell isolates from the femurs of C57BL6 mice (in Table 1 column sample; 58, 67, 69 stands for the number of the mouse from which cells were derived, the final number 1–8 stands for the different stimulation time points and treatments) were cultured for 9 days in the presence of GM-CSF (50 ng ml−1; Peprotech). Cell cultures were treated for the last 3 days with either ABX464 (5 μM) suspended in DMSO or DMSO alone. After additional stimulation for 6 h with LPS (4 μg ml−1; Invivogen) or unstimulation (NS), the supernatants were analyzed for cytokine production using a cytometric bead array technology (CBA; Becton Dickinson). The detection limits were 5 pg ml−1 for IL-6, 52.7 pg ml−1 for MCP-1, 17.5 pg ml−1 for IL-10, 7.3 pg ml−1 for TNF α, and 10.7 pg ml−1 for IL12p70 (Results presented in ref. 1). The cells were lysed in the Qiazol and the RNAs were purified with the miRNeasy kit from Qiagen. Dosage and analysis are performed using Nanodrop/Qubit and 2,100 Bioanalyzer (Agilent Technologies).
Table 1. Summary of sequencing data for each sample.
| Sample | Sequencing Strategy | Time Point | Stimulation | Treatment | Raw Data Size (bp) | Raw Reads Number | Clean Data Size (bp) | Clean Reads Number | Clean Data Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| Clean Data Rate (%)=Clean Reads Number/Raw Reads Number. | |||||||||
| 581 | SE50 | 12 h | NS | DMSO | 2177780600 | 43555612 | 2177704450 | 43554089 | 99.99 |
| 582 | SE50 | 12 h | + LPS | DMSO | 2286641850 | 45732837 | 2284394800 | 45687896 | 99.90 |
| 583 | SE50 | 12 h | NS | ABX464 | 2286729500 | 45734590 | 2286667900 | 45733358 | 99.99 |
| 584 | SE50 | 12 h | + LPS | ABX464 | 2286677050 | 45733541 | 2272614700 | 45452294 | 99.38 |
| 585 | SE50 | 48 h | NS | DMSO | 2177833800 | 43556676 | 2177769250 | 43555385 | 99.99 |
| 586 | SE50 | 48 h | + LPS | DMSO | 2286674400 | 45733488 | 2271449550 | 45428991 | 99.33 |
| 587 | SE50 | 48 h | NS | ABX464 | 2286672950 | 45733459 | 2278218550 | 45564371 | 99.63 |
| 588 | SE50 | 48 h | + LPS | ABX464 | 2286705150 | 45734103 | 2284648050 | 45692961 | 99.91 |
| 671 | SE50 | 12 h | NS | DMSO | 2286656900 | 45733138 | 2277603650 | 45552073 | 99.60 |
| 672 | SE50 | 12 h | + LPS | DMSO | 2177390000 | 43547800 | 2177332500 | 43546650 | 99.99 |
| 673 | SE50 | 12 h | NS | ABX464 | 2286644550 | 45732891 | 2286555900 | 45731118 | 99.99 |
| 674 | SE50 | 12 h | + LPS | ABX464 | 2286680700 | 45733614 | 2286621650 | 45732433 | 99.99 |
| 675 | SE50 | 48 h | NS | DMSO | 2286664900 | 45733298 | 2286609750 | 45732195 | 99.99 |
| 676 | SE50 | 48 h | + LPS | DMSO | 2286695750 | 45733915 | 2285342400 | 45706848 | 99.94 |
| 677 | SE50 | 48 h | NS | ABX464 | 2286657850 | 45733157 | 2285755750 | 45715115 | 99.96 |
| 678 | SE50 | 48 h | + LPS | ABX464 | 2177678700 | 43553574 | 2177622700 | 43552454 | 99.99 |
| 691 | SE50 | 12 h | NS | DMSO | 2286665700 | 45733314 | 2278600650 | 45572013 | 99.64 |
| 692 | SE50 | 12 h | + LPS | DMSO | 2177614000 | 43552280 | 2177556650 | 43551133 | 99.99 |
| 693 | SE50 | 12 h | NS | ABX464 | 2286655650 | 45733113 | 2284221650 | 45684433 | 99.89 |
| 694 | SE50 | 12 h | + LPS | ABX464 | 2286686550 | 45733731 | 2286455400 | 45729108 | 99.98 |
| 695 | SE50 | 48 h | NS | DMSO | 2177711450 | 43554229 | 2177646300 | 43552926 | 99.99 |
| 696 | SE50 | 48 h | + LPS | DMSO | 2286707650 | 45734153 | 2279763650 | 45595273 | 99.69 |
| 697 | SE50 | 48 h | NS | ABX464 | 2286548450 | 45730969 | 2286495900 | 45729918 | 99.99 |
| 698 | SE50 | 48 h | + LPS | ABX464 | 2286689250 | 45733785 | 2277338850 | 45546777 | 99.59 |
RNA-seq library preparation and sequencing
DNase I was used to digestion step, DNase I degrade double-stranded and single-stranded DNA contaminant in RNA samples. RNA molecules were purified using oligo (dT)-attached magnetic beads. The RNA intensity was checked using a 2,100 Bioanalyzer (Agilent Technologies) and only high quality samples with an RNA Integrity Number (RIN) value greater than or equal to 7 were used to construct the sequencing library. Then mRNA were fragmented into small pieces using fragmentation reagent. In cDNA synthesis first-strand cDNA was generated using random hexamer-primed reverse transcription, then was followed by a second-strand cDNA synthesis. Typically, 1 μg of total RNA was used with the TruSeq RNA library preparation kit (Illumina) in accordance with low-throughput protocol, except that SuperScript III reverse transcriptase (Invitrogen) was used to synthesize first strand cDNA. In the step of end repair the synthesized cDNA was subjected to end-repair and then was 3′ adenylated. Adaptors were ligated to the ends of these 3′ adenylated cDNA fragments. We used PCR to amplify the cDNA fragments with adaptors from previous step. PCR products are purified with the SPRI beads, and dissolved in EB solution. In the circularization step the double stranded PCR products were heat denatured and circularized by the splint oligo sequence. The single strand circle DNA (ssCir DNA) were formatted as the final library. After PCR enrichment and purification of adapter-ligated fragments, the concentration of DNA with adapters was determined with quantitative PCR (Applied Biosystems 7,500) using primers QP1 5′- AATGATACGGCGACCACCGA-3′ and QP2 5′- CAAGCAGAAGACGGCATACGAGA-3′. The length of the DNA fragment was measured using a 2,100 Bioanalyzer.
The library was amplified with phi29 to make DNA nanoball (DNB) which have more than 300 copies of one molecular. The DNBs were load into the patterned nanoarray and single end 50 bases reads were generated in the way of sequenced by synthesis. Then, RNA-seq libraries were sequenced using the BGISEQ-500 sequencer. Base-calling was performed by the BGISEQ-500 software version 0.3.8.1111.
Data filtering
‘Dirty’ raw reads were defined as reads which contain the sequence of adaptor, high content of unknown bases and low quality reads. These reads were removed before downstream analysis to decrease data noise. Filtering steps are as follows:
The quality of the RNA-seq libraries was first evaluated using the FastQC v0.11.5 software. Then, the reads were subjected to standard quality control (QC) criteria according to the following parameters: (1) trimming and cleaning reads that aligned to primers and/or adaptors, (2) reads with over 50% of low-quality bases (quality value ≤5) in one read, and (3) reads with over 10% unknown bases (N bases). After filtering, the remaining reads are called ‘clean reads’ and stored as FASTQ format13.
RNA-seq data analysis
24 samples of C57BL6 strains of mice were sequenced using RNA-Seq technology2,3, averagely generating 45,188,386 raw sequencing reads and then 45,120,825 clean reads after filtering low quality (see Data Filtering in method page). Table 1 briefly summarizes information of sequencing data for each sample.
After filtering, clean reads are mapped to reference using HISAT10/Bowtie2 (ref. 11) tools. The average mapping ratio with reference gene is 83.67% and Table 2 lists separate mapping rate for each sample. The average genome mapping ratio is 91.47%. Strict quality control was conducted for each sample to evaluate whether the sequencing data are qualified.
Table 2. Alignment statistics of reads align to reference gene.
| Sample | Total Reads | Total Mapped Reads (%) | Unique Match (%) | Multi-position Match (%) | Total Unmapped Reads (%) |
|---|---|---|---|---|---|
| Total Mapped Reads (%)=Unique Match (%)+Multi-position Match (%). | |||||
| 581 | 43554089 | 84.66 | 77.56 | 7.10 | 15.34 |
| 582 | 45687896 | 85.29 | 78.41 | 6.89 | 14.71 |
| 583 | 45733358 | 82.68 | 76.23 | 6.45 | 17.32 |
| 584 | 45452294 | 84.61 | 78.50 | 6.11 | 15.39 |
| 585 | 43555385 | 82.86 | 76.04 | 6.82 | 17.14 |
| 586 | 45428991 | 84.19 | 78.26 | 5.94 | 15.81 |
| 587 | 45564371 | 83.41 | 76.80 | 6.61 | 16.59 |
| 588 | 45692961 | 84.16 | 78.07 | 6.09 | 15.84 |
| 671 | 45552073 | 85.11 | 77.91 | 7.20 | 14.89 |
| 672 | 43546650 | 83.76 | 77.23 | 6.53 | 16.24 |
| 673 | 45731118 | 79.29 | 70.46 | 8.84 | 20.71 |
| 674 | 45732433 | 83.78 | 77.60 | 6.18 | 16.22 |
| 675 | 45732195 | 82.10 | 75.61 | 6.48 | 17.90 |
| 676 | 45706848 | 84.12 | 77.18 | 6.94 | 15.88 |
| 677 | 45715115 | 84.41 | 78.28 | 6.13 | 15.59 |
| 678 | 43552454 | 82.24 | 76.51 | 5.73 | 17.76 |
| 691 | 45572013 | 83.71 | 76.64 | 7.07 | 16.29 |
| 692 | 43551133 | 83.26 | 76.50 | 6.75 | 16.74 |
| 693 | 45684433 | 84.69 | 77.78 | 6.91 | 15.31 |
| 694 | 45729108 | 82.98 | 76.35 | 6.63 | 17.02 |
| 695 | 43552926 | 83.34 | 76.54 | 6.80 | 16.66 |
| 696 | 45595273 | 85.49 | 79.07 | 6.43 | 14.51 |
| 697 | 45729918 | 82.68 | 75.71 | 6.97 | 17.32 |
| 698 | 45546777 | 85.18 | 78.79 | 6.40 | 14.82 |
Sequencing data saturation analysis is used to measure whether the depth of sequencing data is sufficient for informatics analysis. With the number of sequenced reads increasing, the number of identified genes is also increased. However, when the number of sequenced reads reaches a certain amount, the growth curve of identified genes flattens, indicating that the number of identified genes tends to reach the saturation.
The distribution of the reads on reference gene reflects whether each part of the gene body is evenly sequenced. If the randomness is good, the reads in every position (from 5' terminal to 3' terminal) would be evenly distributed. If the randomness is poor, reads preference to specific gene region will directly affect subsequent bioinformatics analysis. We use the distribution of reads on the reference genes to evaluate the fragmentation randomness, as shown in Fig. 1.
Figure 1. Reads distribution on reference gene.
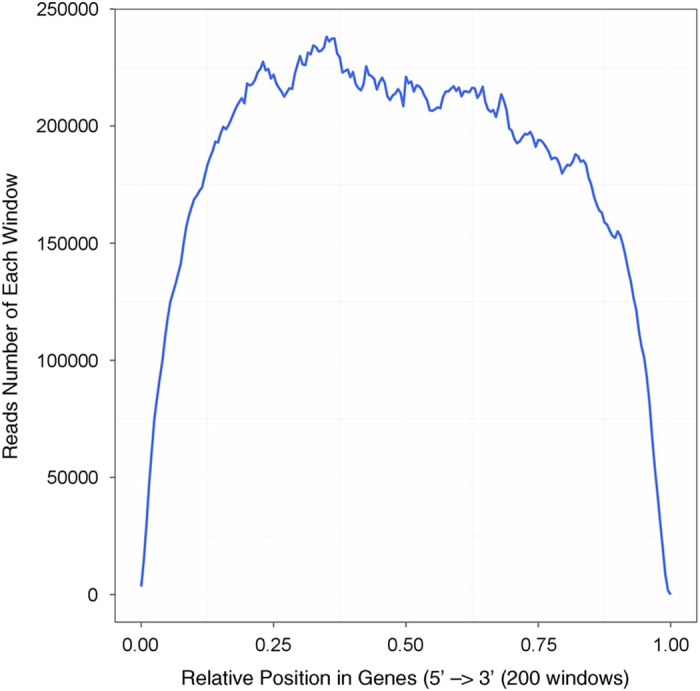
Because of variable lengths of reference genes, the average length of genes is divided into N equal parts. Each equal part is called a window. X-axis shows the relative position of genes, and Y-axis shows the number of reads in each window.
Gene’s expression level is quantified by a software package called RSEM12. The number of identified expressed genes were counted and its proportion to total gene number in database for each sample was calculated. Meanwhile, the distribution of gene number on different expression level for each sample are shown as Fig. 2, to get a general idea about the expression levels of each gene. The file all.gene.FPKM.xls available on Figshare repository is an expression table for all samples with brief gene description and annotation.
Figure 2. Histogram distribution of genes on expression level of each sample.
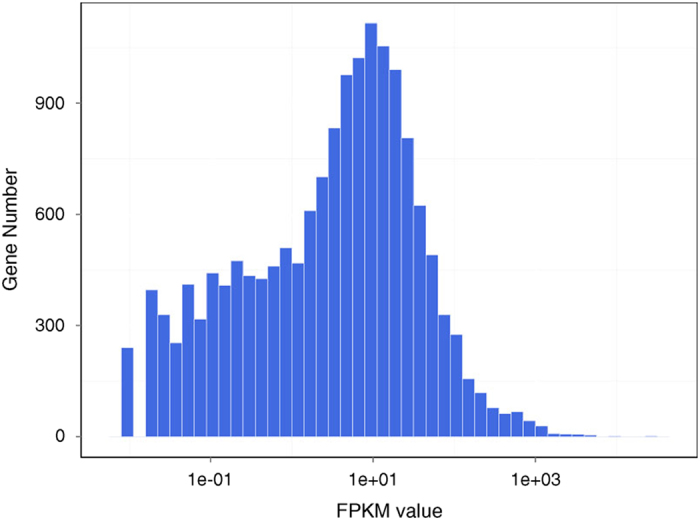
X-axis is FPKM value (the coordinate has been changed by logarithm for better view). Y-axis is gene number of corresponding FPKM.
DEGs (Differentially Expressed Genes) screening is aimed to find differential expressed genes14 between samples and perform further function analysis on them. Poisson distribution method was used to perform this analysis.
All expressed genes of each pairwise are stored in *.GeneDiffExp.xls and screened DEGs are stored in *.GeneDiffExpFilter.xls. They have the same file format and are listed in Figshare repository (Data Citation 1).
For result list of each control-treatment pair above (*.GeneDiffExp.xls), scatter plots were draw for all expressed genes (Fig. 3) to present the distribution of DEGs in screening threshold dimensions. Also, a histogram is shown to demonstrate the number of genes that are significantly up-down in each pairwise comparison (Fig. 4).
Figure 3. Scatter plots of all expressed genes in each pairwise.
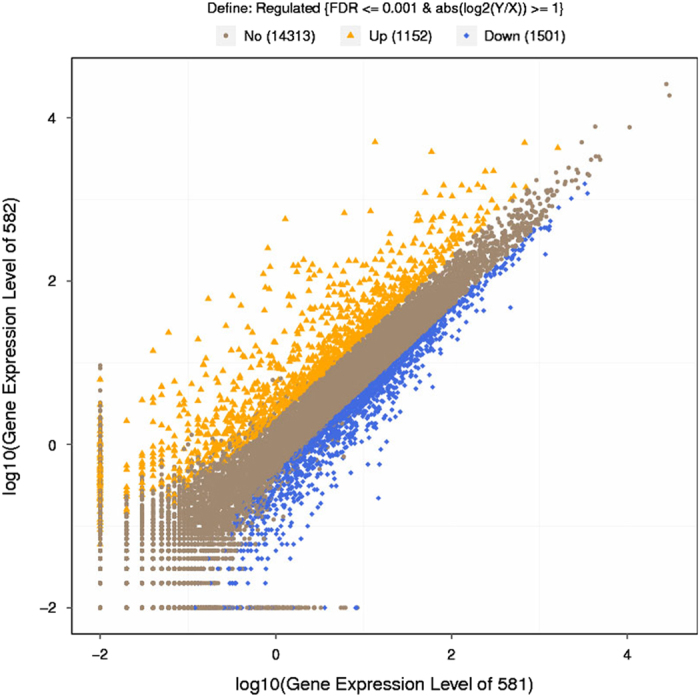
X-axis and Y-axis present log2 value of gene expression. Blue means down-regulation gene, orange means upregulation gene and brown means non-regulation gene. If a gene expressed just in one sample, its expression value in another sample will be replaced by the minimum value of all expressed genes in control and case samples. Screening threshold is on top legend.
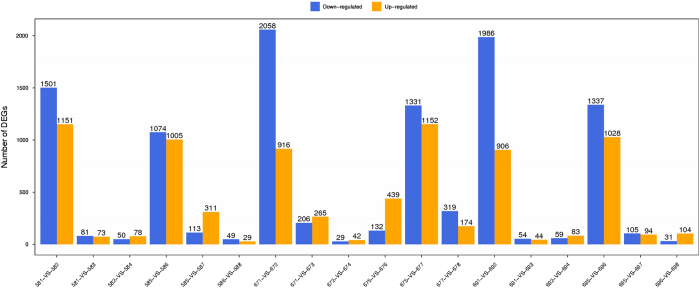
Figure 4. Statistic of differentially expressed genes.
X axis represents pairwise and Y axis means number of screened DEGs. Blue bar denotes down-regulated genes and orange bar for the up-regulated.
Genes with similar expression patterns usually have same functional correlation. We also provide clustering analysis of differentially expressed genes with cluster15,16 and javaTreeview software17 according to the provided cluster plans for DEGs. Complete intersection and union DEGs heatmap for each cluster plan were provided (Fig. 5).
Figure 5. Intersection heatmap of DEGs for each cluster plan.
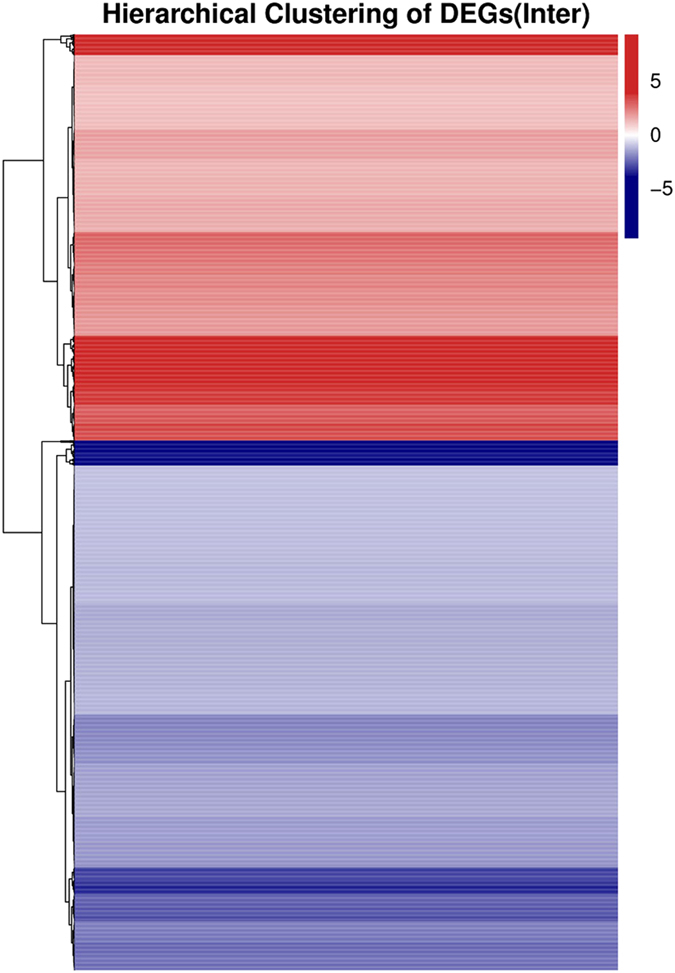
Only genes that differentially expressed in all pairwises of cluster plan are used to build this heatmap. Gradient color barcode at the right top indicates log2 (FC) value (FC, Fold-Change of expression in treatment case to expression in control case). Each row represents a DEG and each column represents a condition pairwise (for some reason in R method, if there is only one pairwise, the pairwise name doesn’t appear in bottom). DEGs with similar fold-change value are clustered both at row and column level.
Annotation analysis of Gene Ontology (GO) are performed for screened DEGs and then generate a report named GOView.html under the data folder of Figshare (Data Citation 1).
After getting GO annotation for DEGs, WEGO software18 was used to do GO functional classification to help understand the distribution of gene functions of the specie from the macro level.
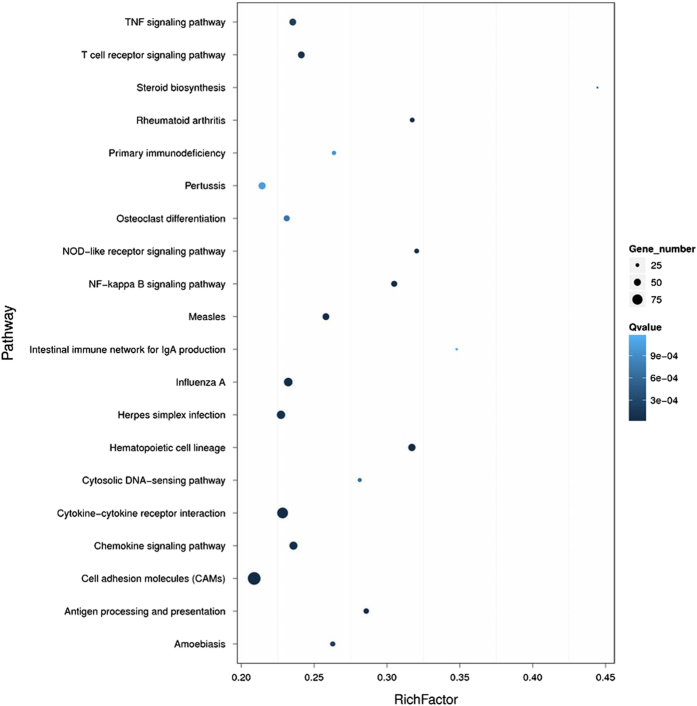
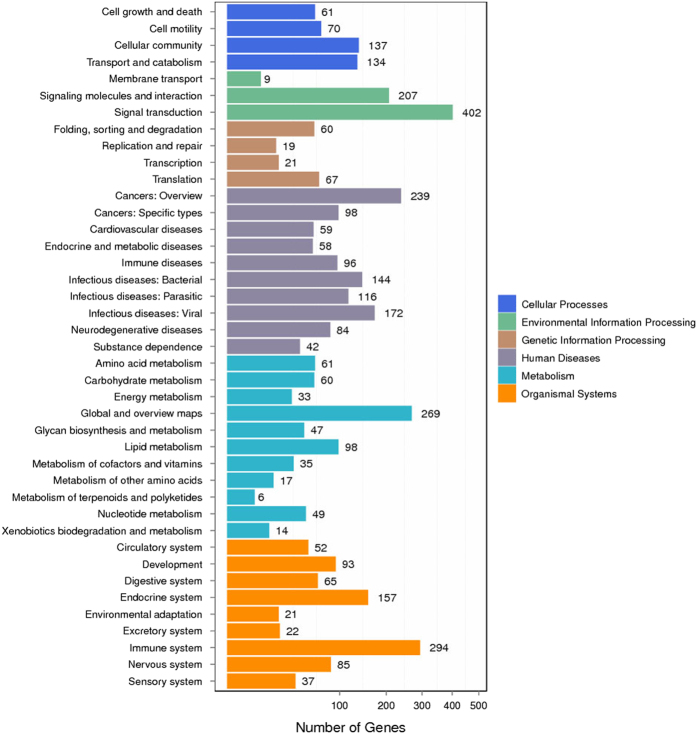
Genes usually interact with each other to play roles in certain biological functions. We perform pathway enrichment analysis of DEGs based on KEGG database19 and generate a report for DEGs in each pairwise respectively, stored in Figshare folder (Data Citation 1). In addition, we generate a scatter plot for the top 20 of KEGG enrichment results (Fig. 6) and a bar plot for the statistics of KEGG terms types (Fig. 7).
Figure 6. Statistics of pathway enrichment of DEGs in each pairwise.
RichFactor is the ratio of differentially expressed gene numbers annoted in this pathway term to all gene numbers annoted in this pathway term. Greater richFator means greater intensiveness. Qvalue is corrected pvalue ranging from 0~1, and less Qvalue means greater intensiveness. We just display the top 20 of enriched pathway terms.
Figure 7. KEGG classification on DEGs for each pairwise.
X axis means number of DEGs. Y axis represents second KEGG pathway terms. All second pathway terms are grouped in top pathway terms indicated in different color.
Different proteins often form complex proteins through complicated interactions to perform their biological functions. Protein-protein interaction network analysis integrates several famous interaction network databases such as BIND, BioGrid and HPRD and constructs interaction networks for DEGs' coding proteins. The analysis result documents are showed by Medusa software20 and webpage version is presented by the report network_en.html under Figshare folder (Data Citation 1).
Statistical data analysis
Referring to ‘The significance of digital gene expression profiles’14, we have developed a strict algorithm to identify differentially expressed genes between two samples. Denote the number of unambiguous clean tags (which means reads in RNA-seq) from gene A as x, given every gene's expression occupies only a small part of the library, x yields to the Poisson distribution:
The total clean tag number of the sample 1 is N1, and total clean tag number of sample 2 is N2; gene A holds x tags in sample 1 and y tags in sample 2. The probability of gene A expressed equally between two samples can be calculated with:
We do correction on P-value to differential gene expression test using Bonferroni method21. Since DEG analysis generate a large multiplicity problems in which thousands of hypothesis (is gene x differentially expressed between the two groups) are tested simultaneously, correction for false positive (type I errors) and false negative (type II) errors are performed using FDR method22. Assume that we have picked out R differentially expressed genes in which S genes really show differential expression and the other V genes are false positive. If we decide that the error ratio ‘Q=V/R’ must stay below a cutoff (e.g., 5%), the FDR was set to a number no larger than 0.05. We use 'FDR ≤0.001 and the absolute value of Log2Ratio ≥1' as the default threshold to judge the significance of gene expression difference. More stringent criteria with smaller FDR and bigger fold-change value can be used to identify DEGs.
Code availability
FastQC v0.11.5 software.
Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Data Records
The RNA-seq data from this study have been submitted and archived to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under sample accession number GSE97062 (Data Citation 2).
In addition, the complete datasets, including expression profiles (Data Citation 1), figures and report analysis files (Data Citation 3, 4, 5) are available on the Figshare data management platform.
Data Citation 1 describe quantitative analysis with all the figures and tables relative to each gene. Data Citation 3 provide supplementary material about statistical analysis such as tables and graphs to show in detail up and down regulated genes in LPS-stimulated BMDMs. Data Citation 4 show in Excel tables the results (quality, saturation) of mapping step for each library. Finally, Data Citation 5 is dedicated to functions analysis to highlight expression, distribution of gene and their potential role in tissue.
Technical Validation
Quality control—RNA integrity
To determine the quality of the RNA-seq libraries, RNA intensity was checked using a 2,100 Bioanalyzer (Agilent Technologies). The integrity of the total RNA was calculated by the RNA Integrity Number (RIN) algorithm, which can be used to estimate RNA quality and degradation level. Higher RIN values indicate better integrity of total RNA, with the highest RIN value being 10. In this study, only total RNAs with a RIN value of greater than or equal to 7 were used to construct the sequencing library, which shows RNA with high integrity was used.
Quality validation and analyses
We applied FastQC v0.11.5 software to determine data quality and analysed several measures, including the sequence quality per base, GC content per sequence, sequence duplication levels, and quality score distribution over all sequences of the fastq files13. A representative summary plots are depicted in Figshare repository (Data Citation 3). Here, the quality scores per base were high, with a median quality score above 30 suggesting high quality sequences across all bases. The GC distribution per base over all sequences was examined. The GC composition pattern was similar to theoretical distribution indicating that the samples were contaminant free. In addition, sequence complexity was examined. Approximately 15% of sequences were shown over 10 to 100 times, either from highly expressed transcripts duplications. The quality score distribution over all sequences was analysed to see if a subset of sequences had universally poor quality. The average quality for most sequences was high, with scores above 37, which indicated that a significant proportion of the sequences in a run had overall high. All other fastqc files were shown to have similar quality metrics.
Additional Information
How to cite this article: Manchon, L. et al. RNA sequencing analysis of activated macrophages treated with the anti-HIV ABX464 in intestinal inflammation. Sci. Data 4:170150 doi: 10.1038/sdata.2017.150 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help and support from many collaborators for their help in RNA-seq experiments and also BGI Genomics Company for sequencing services they provided us.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811035
- 2017. NCBI Gene Expression Omnibus. GSE97062
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4757974
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4810825
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811077
References
- Chebli K. et al. The anti-HIV candidate ABX464 dampens intestinal inflammation by triggering IL22 production in activated macrophages. Scientific Reports 7, 4860 https://dx.doi.org/doi:10.1038/s41598-017-04071-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. RNA-Seq: a revolutionary tool for Transcriptomics. Nature Reviews Genetics. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A. et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Estes J. D. et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 6, e1001052 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X. P. et al. Experimental colitis in SIV-uninfected rhesus macaques recapitulates important features of pathogenic SIV infection. Nat Commun 6, 8020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nischang M. et al. Humanized mice recapitulate key features of HIV-1 infection: a novel concept using long-acting anti-retroviral drugs for treating HIV-1. PLoS ONE 7, e38853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer U. et al. Inadequate clearance of translocated bacterial products in HIV-infected humanized mice. PLoS Pathog. 6, e1000867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos N. et al. Long lasting control of viral rebound with a new drug ABX464 targeting Rev—mediated viral RNA biogenesis. Retrovirology 12, 30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D. et al. Pharmacokinetics and tolerability of ABX464, a novel first in class compound to treat HIV infection, in healthy HIV-uninfected subjects. Journal of Antimicrobial Chemotherapy. 72, 820–828 (2017). [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B. & Salzberg S. L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 10, 25–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. & Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P. et al. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research. 38, 1767–1771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S. & Claverie J. M. The significance of digital gene expression profiles. Genome Res. 10, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- Eisen M. B. et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95, 1165–1188 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M. J. L. et al. Open Source Clustering Software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- Saldanha A. J. Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- Ye J. et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper S. D. & Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics 21, 4432–4433 (2005). [DOI] [PubMed] [Google Scholar]
- Abdi H. Bonferroni and Sidak corrections for multiple comparisons: Encyclopedia of Measurement and Statistics (Sage, Thousand Oaks, CA, 2007). [Google Scholar]
- Benjamini Y. & Yekutieli D. The control of the false discovery rate in multiple testing under dependency: The Annals of Statistics (Institute of Mathematical Statistics, 2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811035
- 2017. NCBI Gene Expression Omnibus. GSE97062
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4757974
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4810825
- Chebli K., Papon L., Hahne M., Manchon L., Tazi J. 2017. Figshare. https://dx.doi.org/10.6084/m9.figshare.4811077