ABSTRACT
The biogenesis of autophagosome, the double membrane bound organelle related to macro-autophagy, is a complex event requiring numerous key-proteins and membrane remodeling events. Our recent findings identify the extended synaptotagmins, crucial tethers of Endoplasmic Reticulum-plasma membrane contact sites, as key-regulators of this molecular sequence.
Autophagy (or macro-autophagy) is a conserved and highly regulated intracellular process necessary for homeostasis maintenance and characterized by the formation of a double membrane-bound organelle called autophagosome.1 Autophagosome biogenesis is initiated by the de novo formation of a cup-shaped structure, the phagophore, which emanates, totally or partially, from a specialized domain of the Endoplasmic Reticulum (ER), enriched in Phosphatidyl-inositol-3-phosphate (PI3P), called omegasome.2 It is clearly established that the ER membrane contributes to phagophore growing, but other membrane compartments participate as well to the process, such as Golgi apparatus, ERGIC (ER-to Golgi intermediate compartment) vesicles, endosomes and plasma membrane.3 ER forms a huge and dynamic membranous network and extends throughout the cell, allowing local signaling and membrane dynamics events. ER has been also demonstrated to establish contact sites between virtually all other endomembranes, via the specific mobilization of tethering complexes.4 Interestingly, such contact site domains involved at the ER-mitochondria interface, have been shown to participate directly in autophagosome biogenesis regulation.5 A comprehensive understanding of molecular mechanisms that regulate ER subdomains implication in autophagy is thus required to better draw the global picture of how and where autophagosome(s) can be formed. Accordingly, the plethora of specific membranous junctions made by the ER in eukaryotes point out other ER-driven contacts as potential sites of autophagosome biogenesis.
Based on this hypothesis, we investigated the putative involvement of ER-plasma membrane contact sites in autophagy. We first observed that nutrients removal, which is known to induce autophagy, was as well responsible for a considerable increase in the ER to plasma membrane tethering, as observed and quantified by electron microscopy. Interestingly, key-proteins of ER-plasma membrane contacts, called Extended Synaptotagmins (E-Syt1, E-Syt2 and E-Syt3),6 were shown to be upregulated in the same experimental conditions, suggesting that nutritional stress stabilizes these contacts. Accordingly, we detected the presence of autophagosomal structures at the immediate vicinity of plasma membrane: these vesicles, shown to be positive for autophagic markers such as LC3 and ATG16L1, were often associated with the ER facing the plasma membrane, suggesting that a subset of autophagosomes is formed at ER-plasma membrane contact sites.
This was further confirmed by STED microscopy, showing presence of LC3 at ER subdomains positive for E-Syt2 and E-Syt3. Moreover, the detection of the omegasome marker DFCP1 (also known as ZFYVE1) at these E-Syts-positive domains indicates that autophagosome biogenesis occurs at ER-plasma membrane contact sites. Therefore, we next analyzed the effects of modulation of these tethering factors on autophagy activity. Overexpression of E-Syt2 (or E-Syt3) was previously demonstrated to artificially increase the ER-plasma membrane contact site density:6 in this situation we noticed as well an increased number of newly formed autophagosomes, while the triple knock down of E-Syts (by silencing each of these proteins), leads to a partial abolition of the autophagy process. Interestingly, as we analyzed the levels of key proteins involved in autophagosome biogenesis, we noticed that the E-Syts triple knock down had no effect on proteins of the ULK1 complex7 such as ATG13 or ULK1 itself, while several regulators of the class III PI3Kinase (PI3KCIII), responsible for local PI3P synthesis and autophagosome biogenesis initiation, were affected. We indeed observed a decrease in the levels of some of its components, such as Beclin1, ATG14L and – to a lesser extent – of ATG16L1 and ATG5-ATG12 complex, both required for local lipidation of LC3 protein.
These findings led us to investigate the relationship that may exist between E-Syts platforms at the ER membrane and the synthesis of PI3P, which is considered as a molecular signature for nascent autophagosomal membrane identification.8,9 While the destabilization of ER-plasma membrane contact sites did not affect the lipidic enzymatic activity of the PI3KCIII member VPS34, we noticed, by a specific fluorescence-based technique, a decrease of the autophagy-associated PI3P only, while the endosomal PI3P pool was not affected. This suggests a role for E-Syts in the stabilization and/or targeting of the autophagy-specific PI3KCIII complex. Among the autophagy-related proteins which were found to be diminished by E-Syts triple knock-down there was VMP1 (vacuolar membrane protein 1),10 an ER-associated protein known to interact with Beclin1 and suggested to participate in PI3KCIII complex stabilization at the ER membrane during autophagosome biogenesis initiation. Interestingly, a recent study identifies VMP1 as a protein that could localize also to ER-driven contact sites in mammalian cells. We found indeed that VMP1 was highly co-localized with Beclin1 and E-Syts at the vicinity of the plasma membrane during the first steps of autophagy response, suggesting a dedicated role of VMP1 in the specific autophagosome assembly at ER-plasma membrane contact sites. This was finally demonstrated by co-immunoprecipitation of E-Syt2 with VMP1 and Beclin1.
Altogether, our data show that VMP1 works as a molecular link between the autophagy machinery (i.e. the Beclin1/ATG14L/VPS34-containing PI3KCIII complex) and the tethering proteins of ER-plasma membrane contact sites (i.e. the E-Syts), to ensure a local PI3P synthesis and, as a consequence, the biogenesis of a subset of autophagosomes at this particular zone of the cell (Fig. 1).
Figure 1.
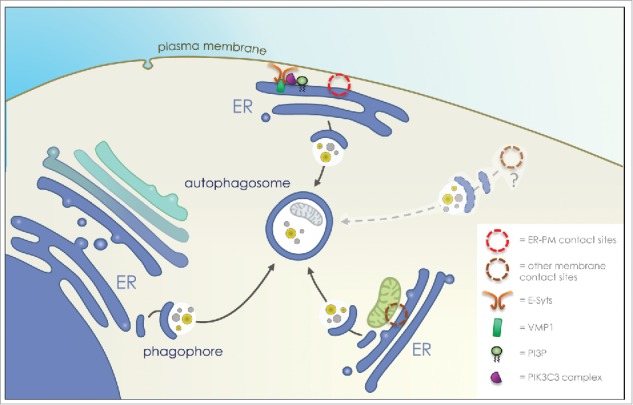
Autophagosome formation at ER-plasma membrane contact sites. Proposed scheme of action for E-Syts (extended synaptotagmins, regulators of Endoplasmic Reticulum (ER) – plasma membrane contact sites) and VMP1 (vacuolar membrane protein 1) in the recruitment of the autophagy machinery at ER-plasma membrane contact sites. The tethering protein E‐Syt2 forms a complex with VMP1 and Beclin1, two regulators of PI3KCIII (class III PI3kinase) complex activity, to initiate local PI3P (phosphatidylinositol-3-phosphate) synthesis and autophagosome biogenesis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Molino D., Zemirli N., Codogno P., and Morel E. (2017) The Journey of the Autophagosome through Mammalian Cell Organelles and Membranes. J. Mol. Biol. 429, 497–514 [DOI] [PubMed] [Google Scholar]
- 2.Nascimbeni A. C., Codogno P., and Morel E. (2017) Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J., [DOI] [PubMed] [Google Scholar]
- 3.Tooze S. A. (2013) Current views on the source of the autophagosome membrane. Essays Biochem 55, 29–38 [DOI] [PubMed] [Google Scholar]
- 4.Stefan C. J., Manford A. G., and Emr S. D. (2013) ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol 25, 434–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., and Yoshimori T. (2013) Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 [DOI] [PubMed] [Google Scholar]
- 6.Giordano F., Saheki Y., Idevall-Hagren O., Colombo S. F., Pirruccello M., Milosevic I., Gracheva E. O., Bagriantsev S. N., Borgese N., and De Camilli P. (2013) PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R., and Ktistakis N. T. (2013) Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem 55, 17–27 [DOI] [PubMed] [Google Scholar]
- 9.Lamb C. a, Yoshimori T., and Tooze S. A. (2013) The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14, 759–774 [DOI] [PubMed] [Google Scholar]
- 10.Molejon M. I., Ropolo A., Re A. Lo, Boggio V., and Vaccaro M. I. (2013) The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 3, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]


