Introduction
Plants are in constant association with microbial communities, and have evolved a multi-layered molecular defense strategy to protect themselves against pathogens. In the absence of specialized immune cells, plants rely on rapid alterations of signaling pathways within individual cells to achieve an appropriate defense response while balancing the trade-off with normal growth and metabolism. Much of our knowledge about the regulation of plant immune responses has been obtained using genetic and genomic approaches for identification of genes and gene expression patterns underlying defense pathways. However, genetic approaches are limited to examining the effects of losing or gaining gene functions, rather than dynamic regulation of cellular processes. Consequently, our current understanding of plant immune networks is far from complete. In recent years, there has been a significant shift from studying the underlying transcriptional networks to studying protein post-translational modification (PTM), which is a versatile regulatory process that rapidly alters the functional diversity of the proteome [1]. Current evidence shows that PTMs are critical for the rapid reprogramming of cells for defense signaling and for attenuating the response to achieve cellular homeostasis in all layers of the plant immune responses.
In this review, we seek to highlight discoveries that have demonstrated the power and versatility of PTMs as regulatory mechanisms for host cells to rapidly respond to pathogens and for pathogens to deploy effectors as a part of their virulence strategy.
Phosphorylation dynamics during pathogen perception and pattern-triggered immunity
At the front line of molecular defense strategies, plants deploy a molecular surveillance system through the cell surface-anchored pattern-recognition receptors (PRRs) to detect microbe-associated and damage-associated molecular patterns (MAMPs and DAMPs) and initiate pattern-triggered immunity (PTI) (Fig. 1) [2]. PTI is effective against a broad range of microbes and is characterized by rapid calcium influx, production of reactive oxygen species (ROS), mitogen activated protein kinase (MAPK) phosphorylation cascades, callose deposition, and defense gene expression [3]. The activation of PTI also leads to plant growth inhibition, highlighting the trade-off between growth and defense physiology. Therefore, understanding how this immune response is dynamically regulated by PTM to achieve rapid and transient induction has been a major focus in deciphering the mechanisms of PTI.
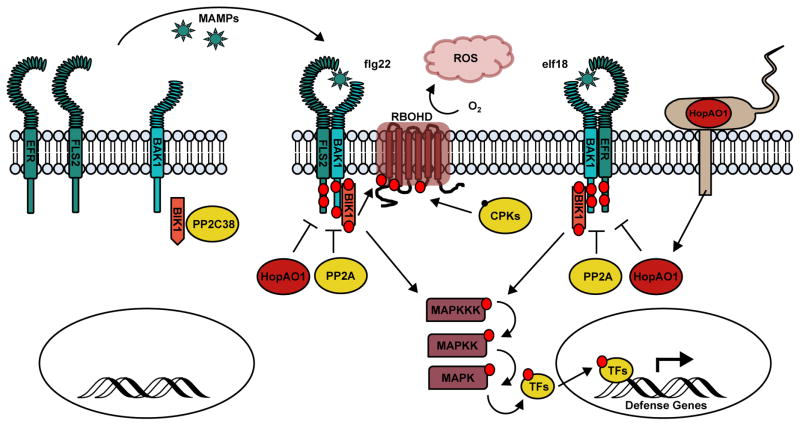
Figure 1. Post-translational signaling mechanisms for pattern-triggered immunity.
Cell surface-anchored pattern-recognition receptors (PRRs: FLS2, EFR) detect microbe-associated molecular patterns (MAMPs) and initiate pattern-triggered immunity (PTI). Activation of FLS2 and EFR by bacterial-derived peptides flagellin (flg22) and elongation factor Tu (elf18), stimulates the recruitment of the leucine-rich receptor-like kinase (LRR-RLK) BAK1 and the receptor-like cytoplasmic kinase (RLCK) BIK1, and induces auto- and trans-phosphorylation (red circles) of their cytoplasmic kinase domains. Phosphorylation of PRR-RLK-RLCK complex is attenuated by protein phosphatases (PP2A, PP2C38). MAMP perception triggers the production of reactive oxygen species (ROS) by the NADPH oxidase RBOHD at the plasma membrane, which is activated through phosphorylation by calcium-dependent protein kinases (CPKs) and BIK1. PTI signaling is propagated through mitogen activated protein kinase cascades (MAPKKK, MAPKK, MAPK) resulting in phosphorylation and activation of transcription factors (TFs) that induce transcriptional reprogramming for defense. Pathogens deliver effector proteins into the host cell to suppress PTI. For example, the Pseudomonas syringae effector HopAO1 is a protein tyrosine phosphatase that dephosphorylates PRRs to and inhibits the host immune response.
Activation of the PRRs, FLAGELLIN-SENSITIVE 2 (FLS2) and EF-TU RECEPTOR (EFR), by peptides derived from bacterial flagellin (flg22) and elongation factor Tu (elf18), stimulates the recruitment of the leucine-rich receptor-like kinase (LRR-RLK) BRI1-ASSOCIATED RECEPTOR KINASE (BAK1) and the receptor-like cytoplasmic kinase (RLCK) BOTRYTIS-INDUCED KINASE1 (BIK1). These plasma membrane (PM)-associated kinases are in turn activated by phosphorylation to initiate defense signal transduction [3,4]. BAK1 and BIK1 are shared by many PRR signaling pathways, including the growth hormone brassinosteroid (BR) signal mediated by the receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) [5]. Recent studies using targeted phospho-proteomics have revealed differential phosphorylation patterns for BAK1 in complex with distinct RLKs (i.e., FLS2, EFR, BIK1 and BRI1). For example, phosphorylation of BAK1 by BIK1 at four amino acids in the kinase domain and two in the activation loop is inhibited through phosphorylation mediated by FLS2 at Ser-286 [6••]. A similar inhibitory effect was observed for EFR through phosphorylation of Thr-455 in the BAK1 activation domain [6••]. The opposing activities of FLS2 and EFR against BIK1-mediated phosphorylation suggest that once BAK1 and BIK1 have become phosphorylated in response to MAMP perception, the PRRs can attenuate this immune response through inhibition of BAK1 phosphorylation mediated by BIK1.
Besides the opposing activities between BIK1 and FLS2/EFR on BAK1, phosphatases, such as SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PP2A), can also counteract the kinase activity to suppress PTI [7]. Upon flg22 treatment, PP2A activity has to be reduced to allow induction of BAK1 activity and PTI signaling [7]. Additionally, the PM-localized PP2C38 interacts with FLS2-BIK1 as well as EFR-BIK1 and dephosphorylates BIK1 to inhibit PAMP-induced ROS production and stomatal closure [8••]. Co-immunoprecipitation (Co-IP) experiments demonstrated that PP2C38 and BIK1 directly interact with each other. A combined approach using genetics and phospho-proteomic analysis indicated that MAMP treatment induces BIK1-dependent phosphorylation of PP2C38 at Ser-77, which significantly diminishes the PP2C38 activity and its interaction with the receptor complexes [8••]. This interplay between immune regulatory kinases and phosphatases is a major mechanism by which plant cells achieve homeostasis during infection.
Phosphorylation dynamics are essential for regulation of immune responses; therefore, they are also prime targets for the activity of bacterial type III effector proteins (TTEs). Pseudomonas syringae pv tomato DC3000 (Pst DC3000) delivers a protein tyrosine phosphatase (PTP), HopAO1, into host cells as part of its virulence strategy. When HopAO1 is ectopically expressed in Arabidopsis, it inhibits elf18- and flg22-induced ROS burst, MAPK activation, and resistance to Pst DC3000 [9]. HopAO1 interacts with the kinase domains of FLS2 and EFR in yeast and plant cells, and in vitro kinases assays demonstrated that the presence of HopAO1 results in a significant decrease in elf18-induced Tyr phosphorylation on EFR [9]. The ability of Pst DC3000 to colonize host tissue is compromised in strains harboring a HopAO1 deletion, and this defect is absent when inoculated into Arabidopsis fls2/efr double mutants, confirming that a major virulence function of HopAO1 is the targeted dephosphorylation of PRRs to inhibit MAMP-induced immune responses [9].
Ubiquitination and N-glycosylation contribute to PTI through degradation and cellular targeting of PRRs
PRR signaling dynamics are fine-tuned by ubiquitination, which facilitates their endocytosis and degradation by the 26S proteasome [10,11]. Yeast two-hybrid (Y2H) and Co-IP experiments using the BAK1 kinase domain as a bait demonstrated that flg22 treatment stimulates interaction between BAK1 and the plant U box (PUB) E3 ubiquitin ligases PUB12 and PUB13, and that the BAK1-PUB12/13 complex associates with FLS2 within 30 seconds of flg22 treatment [12]. Kinase assays using protoplasts expressing BAK1 or a kinase-inactive mutant indicated that BAK1 phosphorylates the C-terminal Armadillo (ARM) repeat domain of PUB13 in response to flg22 treatment, and chemical inhibition of kinase activity confirmed that BAK1-dependent phosphorylation is required for flg22-induced FLS2-PUB12/PUB13 interaction [12,13]. In vitro ubiquitination assays confirmed that PUB12 and PUB13 polyubiquitinate the cytosolic domain of FLS2, but not BAK1 or BIK1 [12]. Three additional, functionally redundant U-box E3 ubiquitin ligases, PUB22/23/24 have also been shown to negatively regulate PTI responses [14]. Upon flg22 perception, PUB22 ubiquitinates Exo70B2, a member of a vesicle tethering complex, and targets it for degradation; Exo70B2 is required for both early and late PTI responses [15,16]. Taken together, studies of ubiquitin-mediated regulation of PRRs indicate that their internalization and degradation likely serves to desensitize cells to continual MAMP stimulus, as well as to reset cells for new rounds of signaling [17].
Asparagine-linked protein glycosylation (N-glycosylation) contributes to protein folding and stability, secretion, and interactions with ligands and other proteins [18]. Treatment of Arabidopsis protoplasts with tunicamycin, which blocks N-glycosylation, and subsequent biochemical analyses showed that the extracellular LRR domains of both FLS2 and EFR are extensively N-glycosylated and that these modifications are required for their maturation and transport to the PM [19]. Site-directed mutagenesis has also been used to generate point mutations in EFR that disrupt individual N-X-(S/T) glycosylation motifs and affect discrete N-glycosylation events. When transiently expressed in Nicotiana benthamiana, which is deficient in elf18 perception, the point mutant of EFR, Asn-143-Gln, fails to elicit MAMP-induced ROS production [19], indicating that individual N-glycosylation sites also play a role in PRR stability and ligand recognition.
Downstream of the PRRs – MAPK cascades and production of reactive oxygen species
Upon pattern recognition and activation of PRR-RLK complexes, immune signaling is propagated by MAPK phosphorylation cascades leading to transcriptional reprogramming for defense [2]. Recent studies of the FLS2 signaling complexes have revealed a connection between PRR/RLK signals and the MAPK phosphorylation cascade, which has been a missing link in PTI signaling against bacterial pathogens. MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7 (MKKK7) was first identified in a large-scale proteomics screen for differential protein phosphorylation upon flg22 treatment [20]. MKKK7 interacts directly with FLS2, and quantitative phospho-proteomic analyses indicated that upon flg22 treatment, MKKK7 is phosphorylated at two different serine residues in a temporally distinct manner [21••]. MKKK7 appears to be a negative regulator of PTI because mkkk7 mutants enhanced MPK6 phosphorylation, increased expression of flg22-induced WRKY29 and FRK1, and enhanced resistance to Pst DC3000 [21••]. The upstream kinases that act on MKKK7 have yet to be firmly established.
Phosphatases that regulate the MAPK cascade include PROTEIN TYROSINE PHOSPHATASE 1 (PTP1) and the dual-specificity phosphatase (DSP) MKP1, which target MPK3 and MPK6, and down-regulate defense responses against Pst DC3000 [22–24]. Members of the PROTEIN PHOSPHATASE 2C (PP2C) family, AP2C1 and AP2C2, were recently shown to be negative regulators of PTI against P. syringae. The ap2c1 mutants exhibited enhanced activation of MPK3, 4, and 6 in response to flg22 and elf18 treatment, as well as to Pst DC3000 infection, resulting in upregulation of genes encoding SA biosynthetic enzymes, WRKY TFs, MAPKK/MAPK kinases, and ultimately enhanced callose deposition and bacterial resistance [25].
Another hallmark of PTI is the production of reactive oxygen species (ROS), which have antimicrobial activities and contribute to defense signaling, cell-wall reinforcement, and stomatal closure to limit pathogen entry into host tissues [26,27]. The respiratory burst oxidase homolog (RBOH) proteins, which are PM-localized NADPH oxidases, are required for PTI-triggered ROS production and are activated through phosphorylation by calcium-dependent protein kinases (CDPKs) and the RLCK BIK1 (Fig. 1) [26,28–30]. ROS signaling is linked to metabolic pathways important for defense against pathogens. GLUCOSE-6-PHOSPHATE DEHYDROGENASE (G6PD), a key enzyme of the oxidative pentose phosphate pathway, is regulated by cellular redox status and phosphorylation. G6PD generates NADPH and metabolic intermediates for numerous biosynthetic pathways and exhibits pathogen-inducible activity that is required for the ROS burst [31,32]. The Arabidopsis GLYCOGEN SYNTHASE KINASE3 (GSK3)/Shaggy-like kinase ASKα, well-known for its role in regulating abiotic stress and brassinosteroid signaling responses, also promotes PTI [31,33••]. Treatment of seedlings with multiple MAMPs including flg22, chitin, and the DAMP PEPTIDE 1 (PEP1), rapidly induces ASKα protein levels and enzymatic activity. Plants over-expressing ASKα display increased ROS production and transcription of PTI marker genes, while these responses are attenuated in askα mutants [33••]. In response to abiotic stress, ASKα phosphorylates G6PD6 at Thr-467 [31], and expression of phospho-mimetic (T467E)/phospho-null (T467A) G6PD6 variants in Arabidopsis revealed that phosphorylation at this residue is also critical for enhanced ROS production. Furthermore, infection by Pst DC3000 stimulates ASKα expression levels and kinase activity, and both askα and g6pd6 result in increased susceptibility to the bacterial infection while expression of wild type G6PD6 or T467E restores resistance [33••]. The results from this work suggest that upon MAMP treatment, ASKα phosphorylates G6PD6 at Thr-467 leading to increased ROS production and enhanced PTI [31,33••].
ROS can also influence growth-defense balance through crosstalk among hormones such as indole-3-acetic acid (IAA) for growth and salicylic acid (SA) and jasmonic acid (JA) for defense against biotrophic and necrotrophic pathogens, respectively. The H2O2 scavenging enzyme CATALASE 2 (CAT2) was recently shown to participate in SA-mediated repression of IAA biosynthesis during defense responses [34,35••]. SA binds to CAT2 and inhibits its activity resulting in H2O2 accumulation [35••]. The increase in H2O2 promotes sulfenylation of Tryptophan (Trp) Synthetase β subunit 1 (TSB1) to inhibit its activity leading to reduction of Trp levels and ultimately decreased auxin production [35•]. This PTM of TSB1 likely occurs on Cys-308 as indicated by inhibited enzymatic activity and the insensitivity of Cys-308-Ser to H2O2-mediated sulfenylation [35•]. These conclusions were supported by the finding that over-expression of TSB1 could rescue the reduced Trp and IAA concentrations in the cat2–1 mutant [35•]. CAT2 also interacts with and promotes the activity of ACYL CO-ENZYME A OXIDASE (ACX2 and ACX3), peroxisomal enzymes in the JA biosynthetic pathway. Treatment with SA, or induction of SA biosynthesis in response to biotrophic pathogens significantly inhibits the interaction among these enzymes leading to reduction in JA biosynthesis [35••]. Since some pathogens have the ability to stimulate IAA biosynthesis in plant tissues to facilitate infection [34] and/or take advantage of inhibitory SA/JA crosstalk in their favor, this work has revealed a mechanism by which host cells can counter these virulence strategies through SA-mediated inhibition of IAA and JA biosynthesis during defense signaling.
Post-translational dynamics during effector-triggered immunity and systemic acquired resistance
PTI can be overcome by pathogens through delivery of TTEs into host cells. The virulence function of effectors is often achieved through PTM of host proteins to manipulate immune signaling cascades [36]. In response, plants have evolved resistance (R) proteins, which are intracellular nucleotide-binding leucine-rich repeat (NB-LRR) receptors that either directly or indirectly detect the activity of effector proteins to initiate effector-triggered immunity (ETI). ETI commonly leads to calcium and ROS accumulation, massive transcriptional reprogramming, and induction of programmed cell death (PCD) at the site of infection that limits the spread of the pathogen [37–40]. ETI also triggers the biosynthesis of the defense hormone SA and expression of antimicrobial pathogenesis related (PR) proteins in infected (local) and systemic tissue leading to establishment of systemic acquired resistance (SAR), which protects against additional infection [41].
(1) PTMs at the host-pathogen interface
The Arabidopsis RPM-1 INTERACTING PROTEIN 4 (RIN4) is a multi-functional immune response regulator at the host-pathogen interface that is targeted by at least five P. syringae effectors and guarded by the host NB-LRRs RPM1 and RPS2 [42–45]. RPM1 could detect phosphorylation of RIN4-Thr166 by the effector (AvrB and AvrRpm1)-mediated expression and activation of a host RLCK, RIPK [44,46]. In this case, the perturbation caused by the effectors are detected by the cognate host NB-LRRs to trigger ETI. However, effectors normally evolved to help the pathogens. For example, a bacterial cysteine protease, AvrPphB, contributes to the dynamics by cleaving RIPK, thereby suppressing RPM1-mediated ETI, resulting in increased susceptibility to Pst DC3000 expressing AvrB [47•].
The host RIN4 also serves as a regulatory link between ETI and PTI. Upon flg22 perception, RIN4 is rapidly phosphorylated at Ser-141 leading to enhanced PTI [48]. This phosphorylation event is upstream of and independent from phosphorylation of RIN4 at Thr-166 that activates ETI in response to AvrB/AvrRpm1 and does not affect the cleavage of RIN4 by AvrRpt2 [48]. Phosphorylation of RIN4-Thr-166, which represses PTI signaling, is reduced in response to flg22 treatment but enhanced in the presence of AvrB offering insight into the mechanism by which P. syringae subverts the host’s initial defense responses [49••]. Additionally, triple phospho-mimic RIN4 mutants (T21D, S160D, or T166D) exhibit enhanced interaction with, and activation of the plasma membrane H+-ATPase 1 (AHA1), and consequently wider stomatal apertures and enhanced colonization of spray inoculated P. syringae [49••]. Although the evidence demonstrates that RIN4 pSer-141 requires interaction with FLS2, the kinase responsible for this modification remains elusive.
In addition to targeting upstream signaling components, such as RIN4, some effectors have evolved to help pathogens evade immune responses through PTM of downstream components, such as TFs. Co-IP experiments demonstrated that PopP2 from Ralstonia solanacearum acetylates WRKY41,70, and 33, which are positive regulators of defense gene expression, and reduces their ability to bind to gene promoters resulting in suppression of host immune responses [50••,51••]. To counteract this pathogen virulence strategy the NB-LRR RESISTANCE TO RALSTONIA SOLANACEARUM 1 (RRS1) is activated directly through effector-mediated PTMs of its extra C-terminal WRKY DNA binding domain [50–52]. Biochemical and proteomics analyses revealed that PopP2 also acetylates RRS1 on four lysine residues in its WRKY domain with Lys-1221 being a key residue for this modification [50••,51••]. The WRKY domain of RRS1-R from Arabidopsis accessions Ws-2 and Nd-1 has an extended C-terminus that is hypothesized to interact with negative regulatory cis-elements in the promoters of genes that initiate ETI [51•]. Therefore, PopP2-dependent acetylation of RRS1 would attenuate its ability to bind to W-box motifs present in defense gene promoters and consequently trigger ETI [50•,51•]. Taken together, the results of these two studies reveal that acetylation of WRKY TFs is a virulence function of PopP2, and that the integrated WRKY domain of RRS1 has evolved as a decoy for recognition of the effector activity and induction of ETI.
(2) NPRs are regulators of two opposing SA-mediated immune responses
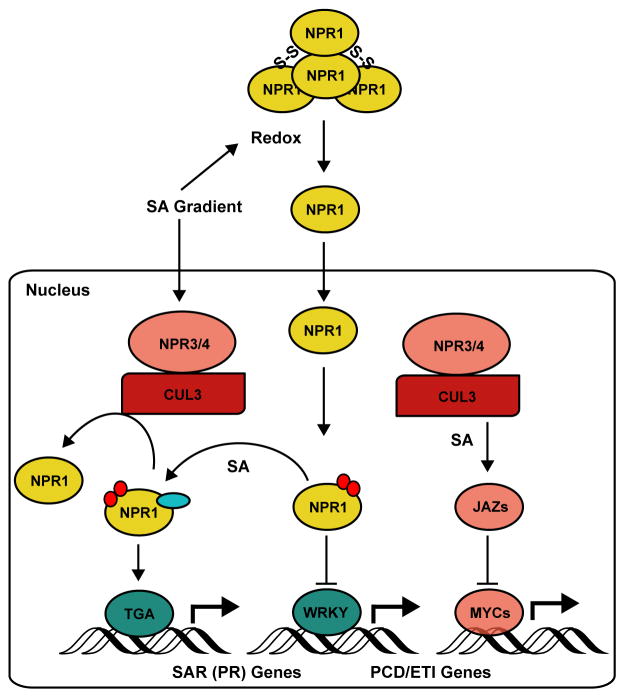
During ETI, the plant defense hormones against biotrophic and necrotrophic pathogens, SA and JA respectively, both accumulate in the infected tissue [53]. Although many studies support an antagonistic SA-JA relationship that depends on the transcription cofactor NPR1 [54,55], there is also considerable evidence indicating that these hormones have a cooperative role during ETI [56,57]. A recent study has now shown that JA plays a positive role in establishment ETI through a mechanism that does not require NPR1 or the canonical JA receptor, CORONATINE INSENSITIVE 1 (COI1), but requires the SA receptors NPR3 and NPR4. NPR3 and NPR4 interact with JAZ transcriptional repressors in yeast and in planta, and promote JAZ1 degradation during early stages of ETI to activate expression of JA biosynthesis and signaling genes (Fig. 2) [58••]. Given that NPR3/NPR4 are substrate adapters for an E3 ubiquitin ligase complex, the evidence suggests that ubiquitination is the PTM leading to JAZ degradation by the proteasome during early ETI responses.
Figure 2. NPRs are regulators of two opposing hormone-mediated immune responses.
NPR1 normally exists as a high molecular weight oligomer in the cytoplasm. In response to salicylic acid (SA) - induced changes in cellular redox, NPR1 monomers are released into the nucleus. At resting state, NPR1, which is phosphorylated (red circles) at Ser55/Ser59, interacts with WRKY, a transcriptional repressor of PR genes. Sumoylated (light blue oval) NPR1, which is phosphorylated at Ser11/Ser15 but dephosphorylated at Ser55/Ser59, interacts with the transcriptional activator TGA3 to promote PR gene expression and establishment of systemic acquired resistance (SAR). NPR3 and NPR4 mediate SA-dependent degradation of NPR1 by the proteasome. NPR3 and NPR4 also promote the SA-enhanced degradation of JAZ transcriptional repressors to induce JA gene expression, resulting in activation of JA synthesis and signaling during early effector-triggered immune (ETI) responses.
In contrast to ETI, which is a signal-specific local response, SAR confers broad-spectrum resistance in uninfected systemic tissues. NPR1 is both a positive regulator of SAR and a negative regulator of ETI and associated PCD (Fig. 2) [41]. In addition to SA-induced, redox-sensitive localization dynamics, the activity of NPR1 in the nucleus is tightly regulated by PTMs [59]. The discoveries of two opposing phosphorylation events (Ser55/Ser59 inhibits and Ser11/Ser15 activates NPR1) and their interplay with sumoylation, a PTM affecting NPR1 differential associations with TFs, TGAs and WRKYs, have revealed complex posttranslational regulation of NPR1-dependent transcription during immune responses [60••]. Sumoylation of NPR1, which is phosphorylated at Ser11/Ser15 but dephosphorylated at Ser55/Ser59, is also required for interaction with NPR3 and NPR4 leading to its degradation by the proteasome [60••,61]. Differences in the SA binding affinities for NPR3 (low) and NPR4 (high) and the opposing effects of SA binding to their interactions with NPR1 (i.e., SA facilitates NPR3-NPR1 interaction but disrupts NPR4-NPR1 interaction) [61], allows NPR1 degradation to occur in cells with high SA concentration (infection site) to remove inhibition on ETI and PCD. NPR1 is stabilized in systemic tissue where SA concentration is too low for NPR3-NPR1 interaction but high enough to disrupt NPR4-NPR1 interaction [61]. Thus, post-translational control of the NPR1 protein level allows PCD in local tissue and establishment of SAR in systemic tissues [61].
Conclusions
Several PTMs stand out as major players of immune regulation; notably, phosphorylation, ubiquitination, and sumoylation. Our understanding of other modifications such as S-nitrosylation, acetylation, and sulfenylation, and their importance to plant immune responses and pathogen virulence mechanisms is also expanding. Although phosphorylation is a prominent posttranslational mechanism for propagating rapid and transient immune responses, identifying the kinases upstream of a known substrate remains a major challenge. The use of targeted, quantitative proteomics and the application of proximity-dependent affinity labeling of protein complexes in vivo show promise for the discovery of unknown kinases [62,63••,64••]. Additionally, advances in the sensitivity of proteomics instrumentation is rapidly increasing our ability to detect PTMs that occur in low stoichiometry, and therefore will open the door to understanding how these modifications contribute to immune responses. Another exciting avenue for future studies, which will be paramount to understanding the dynamic immune system of plants, will be to connect protein kinases to their counteracting protein phosphatases and understand both the quantitative and the physiological significance of these opposing modifications. Gaining an in-depth understanding of how PTMs facilitate efficient immune responses and link PTI, ETI, and SAR will provide a dynamic and holistic view of plant immunity, and lead to new strategies for improving plant performance in the face of pathogen challenge.
Highlights.
Phosphorylation is a rapid and transient switch for PTI.
PTM links metabolic pathways important for defense.
PTM of TSB1 counters pathogen virulence strategies.
PTM at the host-pathogen interface during ETI.
NPRs are regulators of both ETI and SAR.
Acknowledgments
This work was supported by grants from National Institutes of Health (2R01-GM069594-09); the Howard Hughes Medical Institute-Gordon and Betty Moore Foundation (through Grant GBMF3032) to X.D.; and the Duke University Hargitt Fellowship to J.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Humphrey SJ, James DE, Mann M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol Metab. 2015;26:676–687. doi: 10.1016/j.tem.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol Plant. 2015;8:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 4.Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci U S A. 2013;110:12114–12119. doi: 10.1073/pnas.1302154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Wang Y, Li Z, Liu D, Xu J, Wei X, Yan L, Yang C, Lou Z, Shui W. Assessment of BAK1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J Proteomics. 2014;108:484–493. doi: 10.1016/j.jprot.2014.06.009. Using quantitative phospho-proteomics, this study revealed that BAK1 is differentially phosphorylated when in complex with different PRR complexes indicating that differential phosphorylation may be the molecular mechanisms for discrete regulation of BAK1-dependent pathways. [DOI] [PubMed] [Google Scholar]
- 7.Segonzac C, Macho AP, Sanmartin M, Ntoukakis V, Sanchez-Serrano JJ, Zipfel C. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014;33:2069–2079. doi: 10.15252/embj.201488698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Couto D, Niebergall R, Liang X, Bucherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. The Arabidopsis Protein Phosphatase PP2C38 Negatively Regulates the Central Immune Kinase BIK1. PLoS Pathog. 2016;12:e1005811. doi: 10.1371/journal.ppat.1005811. This study focuses on the negative regulation of PTI through the activity of the protein phosphatase PP2C38 on the cytoplasmic kinase BIK1, a component of several PRR complexes. PP2C38 was demonstrated to be a negative regulator of BIK1 activity and BIK1-mediated immunity through attentuation of PAMP-induced BIK1 phosphorylation, which decreases phosphorylation of the NADPH oxidase RBOHD by BIK1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh MH, Sklenar J, Derbyshire P, et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science. 2014;343:1509–1512. doi: 10.1126/science.1248849. [DOI] [PubMed] [Google Scholar]
- 10.Furlan G, Klinkenberg J, Trujillo M. Regulation of plant immune receptors by ubiquitination. Front Plant Sci. 2012;3:238. doi: 10.3389/fpls.2012.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trujillo M, Shirasu K. Ubiquitination in plant immunity. Curr Opin Plant Biol. 2010;13:402–408. doi: 10.1016/j.pbi.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Lu D, Xu G, Finlayson SA, He P, Shan L. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J Exp Bot. 2015;66:3353–3366. doi: 10.1093/jxb/erv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trujillo M, Ichimura K, Casais C, Shirasu K. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol. 2008;18:1396–1401. doi: 10.1016/j.cub.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 15.Kulich I, Pecenkova T, Sekeres J, Smetana O, Fendrych M, Foissner I, Hoftberger M, Zarsky V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14:1155–1165. doi: 10.1111/tra.12101. [DOI] [PubMed] [Google Scholar]
- 16.Pecenkova T, Hala M, Kulich I, Kocourkova D, Drdova E, Fendrych M, Toupalova H, Zarsky V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J Exp Bot. 2011;62:2107–2116. doi: 10.1093/jxb/erq402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A. Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014;164:440–454. doi: 10.1104/pp.113.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saijo Y. ER quality control of immune receptors and regulators in plants. Cell Microbiol. 2010;12:716–724. doi: 10.1111/j.1462-5822.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- 19.Haweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem. 2010;285:4629–4636. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 21••.Mithoe SC, Ludwig C, Pel MJ, Cucinotta M, Casartelli A, Mbengue M, Sklenar J, Derbyshire P, Robatzek S, Pieterse CM, et al. Attenuation of pattern recognition receptor signaling is mediated by a MAP kinase kinase kinase. EMBO Rep. 2016;17:441–454. doi: 10.15252/embr.201540806. This work identifies MAPKKK7 as an FLS2-interacting kinase, providing a link between PRR complexes and the MAPK phosphorylation cascade. Targeted proteomics indicated that MKKK7 is differentially phosphorylated at specific serine residues, and functional analyses demonstrated that this kinase negatively regulates flagellin-triggered immune responses through supression of MPK6 activity, ROS production, and defense gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson JC, Bartels S, Gonzalez Besteiro MA, Shahollari B, Ulm R, Peck SC. Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011;67:258–268. doi: 10.1111/j.1365-313X.2011.04588.x. [DOI] [PubMed] [Google Scholar]
- 23.Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci U S A. 2014;111:6846–6851. doi: 10.1073/pnas.1403248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, Buchala A, Metraux JP, Peck SC, Ulm R. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–2897. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shubchynskyy V, Boniecka J, Schweighofer A, Simulis J, Kvederaviciute K, Stumpe M, Mauch F, Balazadeh S, Mueller-Roeber B, Boutrot F, et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J Exp Bot. 2017 doi: 10.1093/jxb/erw485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Sewelam N, Kazan K, Schenk PM. Global Plant Stress Signaling: Reactive Oxygen Species at the Cross-Road. Front Plant Sci. 2016;7:187. doi: 10.3389/fpls.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015;56:1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- 29.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 30.Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci U S A. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, Rozhon W, Heuck A, Clausen T, Jonak C. Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2012;24:3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer T, Holscher C, Schwoppe C, von Schaewen A. Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 2011;66:745–758. doi: 10.1111/j.1365-313X.2011.04535.x. [DOI] [PubMed] [Google Scholar]
- 33••.Stampfl H, Fritz M, Dal Santo S, Jonak C. The GSK3/Shaggy-Like Kinase ASKalpha Contributes to Pattern-Triggered Immunity. Plant Physiol. 2016;171:1366–1377. doi: 10.1104/pp.15.01741. This study identifies GSK3/ASKα kinase as a positive regulator of plant immunity. GSK/ASKα directly phosphorylat and stimulat the activity of G6PD, a key enzyme of the oxidative pentose phosphate pathway, providing a link between protein phosphorylation cascades during PTI to primary metabolic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazan K, Manners JM. Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci. 2009;14:373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 35••.Yuan HM, Liu WC, Lu YT. CATALASE2 Coordinates SA-Mediated Repression of Both Auxin Accumulation and JA Biosynthesis in Plant Defenses. Cell Host Microbe. 2017;21:143–155. doi: 10.1016/j.chom.2017.01.007. This study demonstrates that the redox scavenging enzyme CAT2 mediates the inhibitory effect of the defense hormone SA on auxin and JA biosynthesis. The authors demonstrated that SA suppresses CAT2 causing increases in H2O2 and sulfenylation of the tryptophan synthesis enzyme TSB1. Inactivation of TSB1 by sulfenylation reduces tryptophan levels and limits this chemical precursor of auxin synthesis. CAT2 was also shown to have a positive effect on JA biosynthesis by promoting interaction between the JA biosynthetic enzymes, ACX2 and ACX3. Therefore, SA repression of CAT2 inhibits JA accumulation. [DOI] [PubMed] [Google Scholar]
- 36.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 37.Senthil-Kumar M, Mysore KS. Nonhost Resistance Against Bacterial Pathogens: Retrospectives and Prospects. Annu Rev Phytopathol. 2013 doi: 10.1146/annurev-phyto-082712-102319. [DOI] [PubMed] [Google Scholar]
- 38.Romeis T, Herde M. From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr Opin Plant Biol. 2014;20:1–10. doi: 10.1016/j.pbi.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Gao X, Cox KL, Jr, He P. Functions of Calcium-Dependent Protein Kinases in Plant Innate Immunity. Plants (Basel) 2014;3:160–176. doi: 10.3390/plants3010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zebell SG, Dong X. Cell-Cycle Regulators and Cell Death in Immunity. Cell Host Microbe. 2015;18:402–407. doi: 10.1016/j.chom.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu ZQ, Dong X. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu Rev Plant Biol. 2013 doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Wu S, Chen X, Liu C, Sheen J, Shan L, He P. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014;77:235–245. doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci U S A. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci U S A. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Elmore JM, Lin ZJ, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Russell AR, Ashfield T, Innes RW. Pseudomonas syringae Effector AvrPphB Suppresses AvrB-Induced Activation of RPM1 but Not AvrRpm1-Induced Activation. Mol Plant Microbe Interact. 2015;28:727–735. doi: 10.1094/MPMI-08-14-0248-R. This study reveals that the bacterial effector protein AvrPphB suppresses AvrB-mediated activation of RPM1, but not AvrRpm1-mediated activation, through cleavage of RIPK by AvrPphB. This indicates the presence of distinct mechanisms for recognition of AvrB and AvrRpm1 by the single R protein RPM1. [DOI] [PubMed] [Google Scholar]
- 48.Chung EH, El-Kasmi F, He Y, Loehr A, Dangl JL. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe. 2014;16:484–494. doi: 10.1016/j.chom.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 49••.Lee D, Bourdais G, Yu G, Robatzek S, Coaker G. Phosphorylation of the Plant Immune Regulator RPM1-INTERACTING PROTEIN4 Enhances Plant Plasma Membrane H(+)-ATPase Activity and Inhibits Flagellin-Triggered Immune Responses in Arabidopsis. Plant Cell. 2015;27:2042–2056. doi: 10.1105/tpc.114.132308. The P. syringae effector AvrB targets the host proteins RIN4 and RIPK. RIPK phosphorylates RIN4 at Thr-166, which is sensed by the NB-LRR RPM1 leading to activation of ETI. This work demonstrated that phosphomimetic RIN4 mutants result in reduced flagellin-triggered responses, and enhanced susceptibility to P. syringae due to inhibition of stomatal defense through enhanced plasma membrane H(+)-ATPase activity. The authors demonstrate that basal phosphorylation of RIN4 Thr-166 decreases upon flagellin treatment, but not in the presence of AvrB, suggesting that AvrB targets RIN4 to enhance pathogen entry through stomata. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. This study, in combination with the work from Le Roux et al. (2015), offers significant insight into how NB-LRRs act as a surveillance system for the activity of a pathogen effectors. Using two RRS1 alleles, RRS1-S (Arabidopsis Col-0) and RRS1-R (Arabidopsis Ws-2 and Nd-1), the authors demonstrate that the integrated WRKY domain present at the C terminus acts as a decoy for the biochemical activity of the bacterial effector PopP2, which normally acetylates WRKY TFs that are positive regulators of defense. PopP2 acetylates the RRS1-S WRKY domain and inhibits AvrRps4 Recognition. PopP2-dependent RRS1 acetylation reduces its ability to bind defense gene promoters. The authors suggest that the extended C terminus of RRS1-R is essential for PopP2-dependent ETI. [DOI] [PubMed] [Google Scholar]
- 51••.Le Roux C, Huet G, Jauneau A, Camborde L, Tremousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. This study, in combination with the work from Sarris et al. (2015), offers significant insight into how NB-LRRs act as a surveillance system for the activity of a pathogen effectors. Using an RRS1 allele that is present in Arabidopsis Ws-2 and Nd-1, RRS1-R, the authors demonstrate that the integrated WRKY domain present at the C terminus acts as a decoy for the biochemical activity of the bacterial effector PopP2, which normally acetylates WRKY TFs that are positive regulators of defense. PopP2 acetylates conserved lysines, specifically Lys-1221 in the RRS1-R WRKY domain, which reduces its ability to bind defense gene promoters and triggers ETI. [DOI] [PubMed] [Google Scholar]
- 52.Sohn KH, Segonzac C, Rallapalli G, Sarris PF, Woo JY, Williams SJ, Newman TE, Paek KH, Kobe B, Jones JD. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 2014;10:e1004655. doi: 10.1371/journal.pgen.1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caarls L, Pieterse CM, Van Wees SC. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015;6:170. doi: 10.3389/fpls.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 56.Beckers GJ, Spoel SH. Fine-Tuning Plant Defence Signalling: Salicylate versus Jasmonate. Plant Biol (Stuttg) 2006;8:1–10. doi: 10.1055/s-2005-872705. [DOI] [PubMed] [Google Scholar]
- 57.Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Liu L, Sonbol F-M, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, Dong X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nature Communications. 2016;7:13099. doi: 10.1038/ncomms13099. In this study, the authors provide strong evidence that a plant defense hormone, JA, is a positive regulator of RPS2-mediated ETI. They demonstrate that during ETI, early induction of JA synthesis following SA accumulation is activated through the SA receptors NPR3 and NPR4, instead of the JA receptor COI1. NPR3 and NPR4 promote degradation of JAZ proteins, transcriptional repressors of JA signaling. This discovery offers a mechanistic understanding of how plants can defend against a biotrophic pathogen without increasing susceptibility to necrotrophic pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Withers J, Dong X. Posttranslational Modifications of NPR1: A Single Protein Playing Multiple Roles in Plant Immunity and Physiology. PLoS Pathog. 2016;12:e1005707. doi: 10.1371/journal.ppat.1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60••.Saleh A, Withers J, Mohan R, Marques J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X. Posttranslational Modifications of the Master Transcriptional Regulator NPR1 Enable Dynamic but Tight Control of Plant Immune Responses. Cell Host Microbe. 2015;18:169–182. doi: 10.1016/j.chom.2015.07.005. This work demonstrates that NPR1 is sumoylated by SUMO3, and that this PTM serves to activate defense responses by switching NPR1’s association with WRKY70 (transcriptional repressor) to TGA3 (transcriptional activator). This study also refines the model of NPR1 phosphorylation dynamics by showing that NPR1 is phosphorylated at Ser55/Ser59 during resting state, and that SA induces dephosphorylation of Ser55/Ser59, which is a requirement for NPR1 activation by sumoylation and phosphorylation at Ser11/Ser15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature. 2012;486:228–232. doi: 10.1038/nature11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang T, Schneider JD, Zhu N, Chen S. Identification of MAPK Substrates Using Quantitative Phosphoproteomics. Methods Mol Biol. 2017;1578:133–142. doi: 10.1007/978-1-4939-6859-6_10. [DOI] [PubMed] [Google Scholar]
- 63••.Kanshin E, Giguere S, Cheng J, Tyers MD, Thibault P. Machine learning of global phosphoproteomic profiles enables discrimination of direct versus indirect kinase substrates. Mol Cell Proteomics. 2017 doi: 10.1074/mcp.M116.066233. Proteomics approaches to quantitative identification of PTMs have rapidly advanced in recent years. This work describes methodology for determining biologically relevant kinase substrates using a combination of techniques including selective chemical inhibition, quantitative phosphoproteomics, and machine learning. This platform was applied to identify direct targets of Cdc28 and Snf1 kinases in S. cerevisiae; however, the methods are broadly applicable and can be applied in other species to facilitate global phosphorylation network profiling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Li P, Li J, Wang L, Di LJ. Proximity labeling of interacting proteins: Application of BioID as a discovery tool. Proteomics. 2017 doi: 10.1002/pmic.201700002. Proximity-dependent labeling of interacting proteins using a promiscuous biotin ligase fused to a bait protein is a powerful technique that has the capability of identifying members of protein complexes, as well as rapid transient interactors such as kinases or phosphatases. This review describes the widely used BioID platform and its applications to different target protein types, and compares/contrasts this method to other proximity-dependent labeling approaches. [DOI] [PubMed] [Google Scholar]