Abstract
In this study, we suggest a designer vaccine adjuvant that can mimic the drainage of pathogens into lymph nodes and activate innate immune response in lymph nodes. By the amination of multivalent carboxyl groups in poly-(γ-glutamic acid) (γ-PGA) nanomicelles, the size was reduced for rapid entry into lymphatic vessels, and the immunologically inert nanomicelles were turned into potential activators of inflammasomes. Aminated γ-PGA nanomicelles (aPNMs) induced NLRP3 inflammasome activation and the subsequent release of proinflammatory IL-1β. The NLRP3-dependent inflammasome induction mechanism was confirmed through enzyme (cathepsin B and caspase-1) inhibitors and NLRP3 knockout mice model. After the aPNMs were combined with a clinically evaluated TLR3 agonist, polyinosinic–polycytidylic acid sodium salt (aPNM-IC), they triggered multiple arms of the innate immune response, including the secretion of pro-inflammatory cytokines by both inflammasomes and an inflammasome-independent pathway and the included type I interferons.
Keywords: vaccine adjuvant, nanoparticles, innate immunity, antigen-presenting cells, lymphatic vessel, type I interferon
Introduction
The rational design and synthesis of pathogen-mimicking nanomaterials are required for the development of efficient immunomodulatory agents because the interaction of nanomaterials with immune cells is highly dependent on their size, shape, and surface characteristics.1–7 Lymphatic vessels have evolved to drain pathogens into lymph nodes, which provide a first line of defense against invading viruses and the initiation/subsequent direction of adaptive immune responses for the removal of pathogens.8–10 Nodal antigen-presenting cells (APCs) detect danger signals through pattern recognition receptors and initiate multiple arms of innate immune response such as the activation of inflammasomes and the secretion of proinflammatory cytokines and type I interferons (IFNs).11,12 The activation of inflammasomes as an innate immune system is a critical step in the initiation of an effective adaptive immune response and a major goal of many nanoadjuvant systems.13–18 Inflammasomes are cytosolic multiprotein complexes that can detect intracellular pathogens/danger signals and initiate inflammatory processes.13 The most well-characterized inflammasomes consist of three major components of the Nod-like receptor (NLR)-family protein: NLRP3, procaspase-1, and the ASC adapter.19 Materials that trigger lysosomal destabilization, release cathepsin B, and detect danger signals may be useful as inflammasome inducers.20 It has been shown that the NLRP3 inflammasome is activated by classical adjuvants, including aluminum hydroxide and saponin, which requires both Toll-like receptor (TLR) and NLRP3 signaling in APCs.21–24 Following the recruitment and activation of caspase-1 in the NLRP3 inflammasome pathway, the biologically inactive precursor pro-IL-1β is processed into IL-1β. In fact, chitosan or SiO2 nanoparticles are known to trigger caspase-1 cleavage and IL-1β secretion in macrophages and keratinocytes, which are mediated by the NLRP3 inflammasome.25,26 However, a high concentration (>100 µg mL−1) is required to activate inflammasomes. Thus, the design and synthesis of a nanoadjuvant that can traffic through lymphatic vessels, migrate into lymph nodes efficiently, and trigger innate immune responses are essential for the development of synthetic vaccines.
In this study, we designed and synthesized a nanoadjuvant that mimicked the drainage of pathogens into lymph nodes and activated the innate immune system.
Materials and methods
Materials
Poly-(γ-glutamic acid) (γ-PGA; molecular weight =50 kDa) was provided by BioLeaders Corporation (Daejeon, Republic of Korea). Cholesteryl chloroformate (97%), 1,1′-carbonylbis-1H-imidazole, 1,2-ethanediamine, and polyinosinic–polycytidylic acid sodium salt, poly-(I:C), were purchased from Sigma-Aldrich (St Louis, MO, USA). IRDye800 NHS ester was purchased from Li-COR Biosciences (Lincoln, NE, USA).
Methods
Preparation of aminated γ-PGA nanomicelles (aPNMs)
Cholesteryl chloroformate (2.25 g, 5 mmol) was slowly added to a solution of 1,2-ethanediamine (16.7 mL, 250 mmol) in toluene (anhydrous, 250 mL) in an ice bath. The reaction mixture was reacted at 25°C for 16 hours, washed with deionized water, dried over anhydrous magnesium sulfate, and evaporated in a rotary evaporator. The residue was dissolved in dichloromethane (20 mL) and then added to methanol (20 mL). The resulting suspension was filtered to remove biscarbamate using syringe filtration (1 µm, polytetrafluoroethylene). The filtrate was evaporated in a rotary evaporator to obtain a white solid. To synthesize γ-PGA-CH, 500 mg of γ-PGA (3.876 mmol) in DMSO (10 mL) and 46 mg of cholesterol amine (0.036 mmol) in tetrahydrofuran (10 mL) were reacted in the presence of 1,1′-carbonyldiimidazole (63 mg, 0.3876 mmol) at 40°C for 18 hours. The reaction mixture was evaporated in a rotary evaporator to remove the tetrahydrofuran and then was poured into acetone. After centrifugation, the precipitate was collected, dried, and mixed with NaHCO3 solution and stirred for 4 hours. Amberlite IR-120H beads were treated by ion exchange. After filtration with beads, the reaction mixture was dialyzed through a 10- to 12-kDa molecular weight cutoff membrane against deionized water for 2 days, and the solution was freeze-dried. Next, the γ-PGA-CH conjugate was aminated in the presence of ethylenediamine in DMSO. For amination, γ-PGA-CH (100 mg, 0.775 mmol) and ethylenediamine (20.6 mL, 310 mmol) were slowly mixed in DMSO (20 mL) containing 1,1′-carbonyldiimidazole (752.8 mg, 4.65 mmol) under constant stirring for 12 hours. In order to generate the complexes of γ-PGA-CH-NH2-poly-(I:C) (aPNM-IC), a γ-PGA-CH-NH2 nanoparticle solution was added to a solution of poly-(I:C) at a specified mass ratio of 1:2 (γ-PGA-CH-NH2/poly-(I:C)) and reacted for 2 hours to form a stable complex.
Characterization of aPNMs
The size distribution and zeta potential of the multivalent polymer nanocomplex were analyzed by dynamic light scattering (DLS) using an electrophoretic light scattering photometer (ELS-Z, Osaka, Japan) and a field-emission scanning electron microscope (JEOL Ltd., Tokyo, Japan). The size change of aPNMs in phosphate-buffered saline (PBS) and serum at body temperature (37°C) was also measured by using DLS for 6 days. The critical micelle concentration (CMC) was determined via the fluorescence spectroscopic method, using pyrene as a hydrophobic fluorescence probe. Aliquots of pyrene solutions (1.5×10−5 M in acetone) were added to the containers, and the solvent was removed under reduced pressure. Aqueous phthalocyanine conjugate solutions of different concentrations, from 5×10−3 to 5 mg mL−1, were added to the containers containing pyrene. The mixtures were incubated for 24 hours at 20°C, and the excitation spectra were recorded on an F-2500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan), from 300 to 360 nm with an emission wavelength of 390 nm. The ratio (I337.5/I323) of the fluorescent intensities, at 337.5 and 323 nm, was calculated to evaluate the hydrophobicity around the pyrene molecules, and the rising point of I337.5/I323 was defined as the CMC. The CMC of aPNM was ~0.5 mg mL−1. The surface morphology and size of the aPNMs were analyzed by high-resolution transmission electron microscopy (JEOL Ltd.). The samples were stained with 2% uranyl acetate (Sigma-Aldrich).
Animals
Female C57BL/6 (H-2b) mice that were 6 weeks of age were purchased from ORIENT (Sungnam, Republic of Korea). The mice were kept under pathogen-free conditions. NLRP3 knockout mice were kindly provided by Professor Gabriel Nuňez (University of Michigan, Ann Arbor, MI, USA) and have been previously described.27 All experiments involving mice were performed in accordance with the Korean Food and Drug Administration guidelines for the care and use of laboratory research animals. This study was reviewed and approved by the Institutional Animal Care and Use Committee of Sungkyunkwan University School of Medicine (SUSM; Permission 16–56). SUSM is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility and abide by the Institute of Laboratory Animal Resources guide.
Generation of bone marrow-derived dendritic cells (BMDCs) and bone marrow-derived macrophages (BMDMs) from mice
Both the femurs and tibiae were collected, and the muscle attachments were carefully removed. Intact bones were soaked in 70% (v/v) ethanol for 1 minute for disinfection and then washed with PBS. Next, both ends of the bones were cut with scissors, and the marrow was flushed with Roswell Park Memorial Institute (RPMI) 1640 medium using a 1-mL syringe with a 26-G needle. Clusters within the marrow suspension were disintegrated by vigorous pipetting. After one wash (490× g, 5 minutes) in RPMI 1640 medium, the red blood cells were depleted with 0.83 M NH4Cl buffer (Sigma-Aldrich). Bone marrow cells (2×106 cells) were collected and cultured in 100-mm Petri dishes containing 10 mL of RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 50 IU mL−1 penicillin, 50 mg mL−1 streptomycin, and 20 ng mL−1 mouse recombinant granulocyte macrophage colony-stimulating factor (R&D Systems, Minneapolis, MN, USA). Bone marrow cells (5×105 cells) were collected and cultured in 100-mm Petri dishes containing 10 mL DMEM supplemented with 20% heat-inactivated fetal bovine serum, 50 IU mL−1 penicillin, 50 mg mL−1 streptomycin, and 20 ng mL−1 recombinant murine macrophage colony-stimulating factor (PeproTech, Rocky Hill, NJ, USA). After 7 days, nonadherent and loosely adherent cells (BMDCs) or adherent cells (BMDMs) were harvested, washed, and used for in vitro experiments.
Immunofluorescence staining and imaging
For intracellular staining, BMDMs and BMDCs were fixed with 4% (w/v) paraformaldehyde solution on ice for 30 minutes, permeabilized with 0.1% saponin/5% bovine serum albumin/PBS for 20 minutes, and then incubated with primary antibodies against ASC (1:200; Adipogen, San Diego, CA, USA) and NLRP3 (1:200; Abcam, London, UK) overnight at 4°C. After washing three times with PBS, the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat (1:1,000; Abcam) and tetramethylrhodamine (TRITC)-conjugated donkey anti-rabbit (1:1,000; Abcam) for 1 hour at room temperature. After washing three times with PBS, the cells were stained with Hoechst 33342 (trihydrochloride, trihydrate; Invitrogen, Carlsbad, CA, USA). Florescence images were obtained by using a DeltaVision™ PD instrument (Applied Precision Technologies, Issaquah, WA, USA) with a filter set (DAPI: excitation 360/40, emission 455/50; FITC: excitation 490/20, emission 525/36; TRITC: excitation 555/25, emission 605/52; Omega Optical, Brattleboro, VT, USA). In order to analyze the intracellular localization of aPNMs in BMDMs and BMDCs, the cells were treated with aPNM-FITC overnight at 37°C and were washed twice with PBS, and lysosomes were stained with 50 nM LysoTracker® red DND-99 (Thermo Fisher Scientific, Waltham, MA, USA) for 30 minutes at 37°C. The cells were then washed, fixed, and examined by using the DeltaVision PD instrument.
In vitro cytokine assay
BMDMs or BMDCs were cultured in 6-well plates at a density of 1×106 cells/well and cultured with 400 ng mL−1 lipopolysaccharide (LPS) for 3 hours at 37°C. The aPNMs were added to the wells in a 1 mL total volume at a concentration of 1, 2, 5, or 10 µg mL−1. In some experiments, BMDMs or BMDCs were incubated with a caspase-1 inhibitor (Ac-YVAD-cmk; Sigma-Aldrich) or cathepsin B inhibitor (CA-074; Sigma-Aldrich). After a 4-hour treatment, the culture supernatants were collected and analyzed for IL-1β levels by using cytokine-specific enzyme-linked immunosorbent assay (ELISA; BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions.
Ex vivo cytokine assay
To assay the induction of inflammasomes in lymph nodes, aPNMs (50 µg) and carboxyl-terminated γ-PGA nanomicelles were inoculated into the footpad of 6-week-old C57BL/6 mice and NLRP3 knockout mice after priming with LPS (5 µg) for 24 hours. Next, 3 and 6 hours after aPNM injection, the lymph nodes of the mice were removed. The dissected lymph nodes were transferred to round-bottomed microfuge tubes and snap-frozen in liquid nitrogen. Next, 300 µL of ice-cold lysis buffer (R0278; Sigma-Aldrich) was added, and the lymph nodes were homogenized by using an electric homogenizer (Z359971; Sigma-Aldrich). Another 600 µL of lysis buffer was added during homogenization. After homogenization was completed, the contents were stirred for 2 hours at 4°C. The supernatants were collected following centrifugation at 16,000× g for 20 minutes at 4°C. IL-1β was analyzed by using cytokine-specific ELISA (BD Biosciences) according to the manufacturer’s instructions.
Quantitative PCR for cytokines
Total RNA was extracted by using an RNeasy mini kit (Qiagen, Hilden, Germany), and 1 µg of total RNA was used for first-strand cDNA synthesis with the GoScript™ Reverse Transcription System (Promega, Madison, WI, USA) with random primers according to the manufacturer’s instructions. Quantitative PCR was performed by using the StepOnePlus™ Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA). Quantitative PCR amplification was conducted in a volume of 20 µL containing 10 µL of SYBR Green PCR Master Mix (Applied Biosystems), 7 µL of distilled water, 5 pmol each of forward and reverse oligonucleotide primers, and 1 µL of cDNA template. The following primers were specific to conserved regions: mouse tumor necrosis factor-alpha (TNF-α) 5′-TCCCAGGTTCTCTTCAAGGGA-3′ (forward) and 5′-GGTGAGGAGCACGTAGTCGG-3′ (reverse), mouse IL-6 5-ACAACCACGGCCTTCCCTACTT-3′ (forward) and 5′-CACGATTTCCCAGAGAACATGTG-3′ (reverse), and mouse IFN-β 5′-TTCAAGTGGAGAGCAGTTGAG-3′ (forward) and 5′-CATCAACTATAAGCAGCTCCA-3′ (reverse; Bioneer, Daejeon, Republic of Korea). GAPDH served as a reference gene to normalize target mRNA levels. The samples were run in triplicate, and melting curve analysis was performed to confirm the amplification specificity of the PCR products.
In vivo fluorescence imaging
For in vivo imaging, C57BL/6 mice were anesthetized with 300 µL of 2.5% avertin solution (2,2,2-tribromoethanol-tertamyl alcohol; Sigma-Aldrich), and the imaging areas were treated with a depilatory cream. aPNM-IRDye800 (50 µg in 50 µL of water) was intradermally injected into the forepaw pad. aPNM-IRDye800 was tracked by using a custom-made whole body optical imaging system at various experimental time points. Near-infrared spectroscopy images (0.5-second exposure) of the axillary lymph nodes were acquired using a 785-nm, 500-mW diode laser as an excitation light source and 835/45-nm band-pass emission filter. All images were processed by using Simple PCI software (Compix Inc., Cranberry Township, PA, USA).
In situ histofluorescence
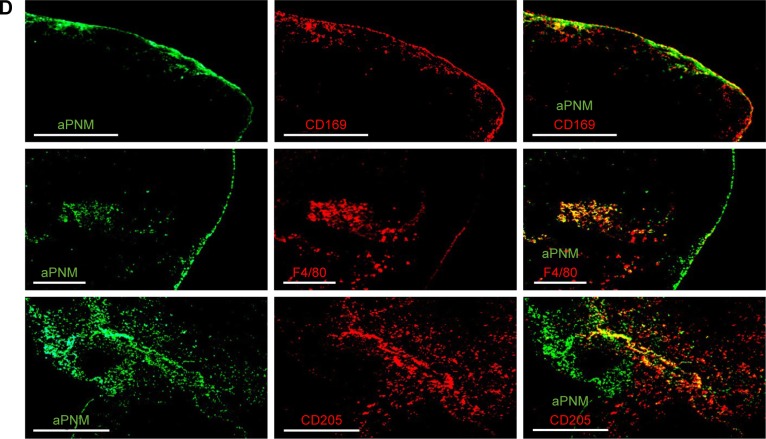
In order to analyze the in situ distribution of aPNMs, the axillary lymph node was dissected 24 hours after the injection of 50 µg of aPNM-FITC and embedded in Tissue-Tek OCT compound (SAKURA, Tokyo, Japan) followed by freezing in liquid nitrogen. Cryosections (10 µm) were prepared by using a Leica cryostat CM1850 (Leica Micro-systems, Wetzlar, Germany) and transferred to glass slides. The sections were fixed with cold acetone for 5 minutes, dried, and frozen at −20°C until use. The slides were washed with PBS and blocked with PBS containing 1% bovine serum albumin for 1 hour at room temperature. After additional washing, the slides were stained with rat anti-mouse F4/80 (Serotec, Oxford, UK), CD169 (Siglec-1; Serotec), and CD205 (DEC-205; Serotec) overnight at 4°C to label the macrophages and dendritic cells (DCs), respectively. The slides were then stained with TRITC-conjugated anti-rat IgG secondary antibodies (BD Biosciences) for 1 hour at room temperature. The slides were washed twice with PBS and then treated with 2 µg mL−1 Hoechst 33342 in PBS for 10 minutes. After the final wash, the slides were mounted in 50% glycerol (in PBS) and examined by using a fluorescence microscope (Olympus IX71; Olympus Optical, Tokyo, Japan) and DeltaVision PD instrument.
Statistical analysis
All results are expressed as mean differences and were tested for significance using Student’s t-test, wherein significance is indicated by p-values <0.05 (*), <0.01 (**), and <0.001 (***). p<0.05 was considered to indicate a significant difference. All values are expressed as the mean ± SD. GraphPad Prism software was used for all statistical analyses (GraphPad Software, San Diego, CA, USA).
Results and discussion
Characterization of lymph node targeting aPNMs
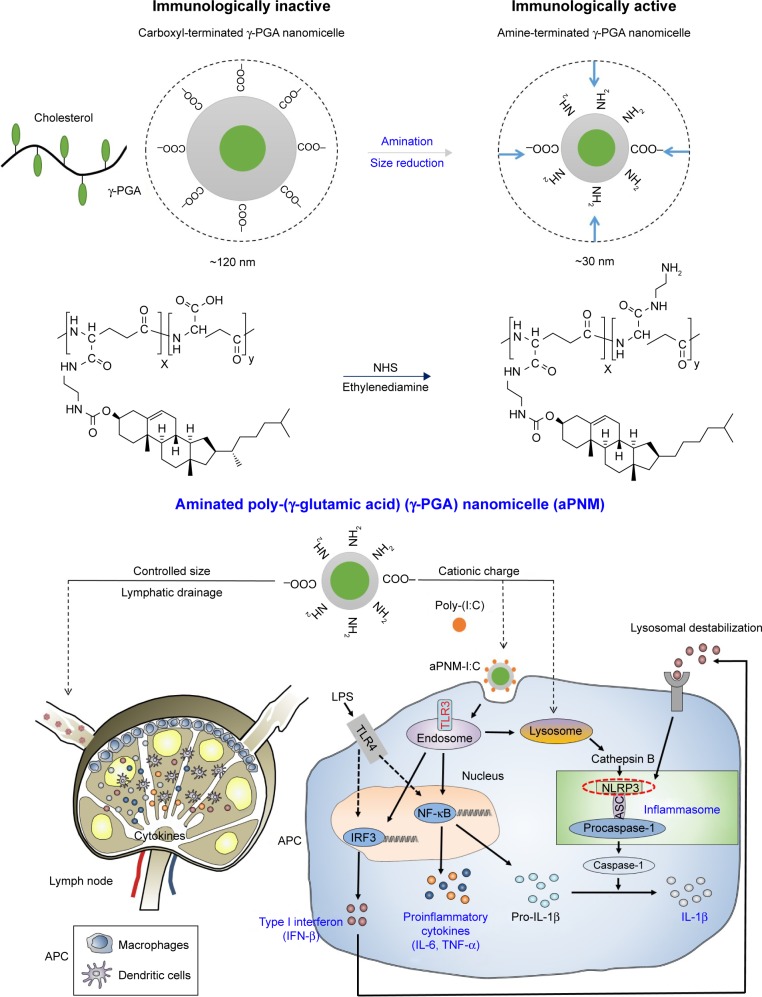
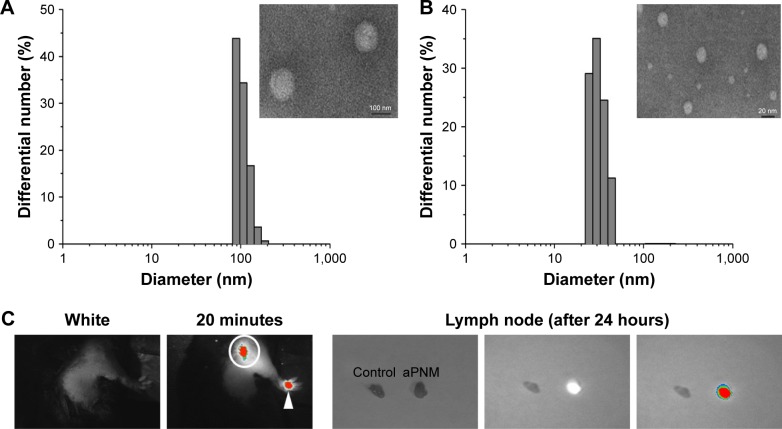
We synthesized size-controlled aPNMs as a novel inflammasome inducer that could function even at low concentrations (<10 µg mL−1). Furthermore, the inflammasome inducer was combined with poly-(I:C), a TLR3 agonist, to activate multiple arms of the innate immune system (Scheme 1).28 We synthesized γ-PGA nanomicelles by conjugating the main chain of γ-PGA with hydrophobic cholesterol groups.29 However, the size of γ-PGA nanomicelles should be tuned to ensure efficient migration through lymphatic vessels.1 In order to improve the targeting efficiency of nanoparticles into lymph nodes, three main research approaches have been suggested: size-tuning, hitch-hiking on albumin, and PEGylation.30–34 Regarding size-tuning, it has been reported that 15–70 nm nanoparticles are optimal for rapid entry into lymphatic vessels and migration into lymph nodes. The amination of γ-PGA nanomicelles with carboxylate groups on their surfaces reduced the size of nanomicelles tô30 nm (Scheme 1; Figure 1A and B). When the size change of aPNM was measured in PBS and serum at body temperature, the size was kept during a span of 6 days (Figure S1). When aPNMs were injected into the footpad, they migrated into the lymph nodes easily because of their modulated size and were taken up by APCs such as macrophages (stained with anti-CD169 and anti-F4/80) and DCs (stained with anti-CD205; Figure 1C and D). Because aPNMs have a positive charge, they can interact with the negatively charged cell membrane of APCs, enter them, and induce lysosomal destabilization, resulting in the activation of the NLRP3 inflammasome signaling pathway (Figure S2; Scheme 1). The amine groups also facilitated loading of anionic charged immunostimulatory materials such as poly-(I:C), which can induce the innate immune response through different signaling pathways.35 Scheme 1 presents a schematic illustration of the nanoadjuvants that target lymph nodes and trigger multiple arms of the innate immune response (IL-1β secretion by inflammasomes, release of inflammasome-independent proinflammatory cytokines [TNF-α and IL-6], and type I IFN [IFN-β]).36
Scheme 1.
Schematic illustrations of γ-PGA nanomicelles and inflammasome pathway.
Notes: Aminated γ-PGA nanomicelles facilitate efficient migration through lymphatic vessels, targeting of APCs in the lymph nodes, and induction of inflammasomes after lysosomal destabilization. When aPNMs combine with anionic poly-(I:C), a TLR3 agonist, they trigger multiple arms of the innate immune response (IL-1β secretion by inflammasomes, inflammasome-independent secretion of proinflammatory [TNF-α and IL-6], and type I interferon [IFN-β]).
Abbreviations: APCs, antigen-presenting cells; aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); IFN-β, interferon-β; LPS, lipopolysaccharide; poly-(I:C), polyinosinic–polycytidylic acid; TLR, Toll-like receptor; TNF-α, tumor necrosis factor-alpha.
Figure 1.
Lymph node targeting aPNMs.
Notes: Size and size distribution of (A) carboxyl-terminated and (B) amine-terminated γ-PGA nanomicelles measured by dynamic light scattering and transmission electron microscopy. In vivo trafficking of aPNMs to lymph nodes. (C) In vivo NIR fluorescence image of IRDye800-labeled aPNMs 20 minutes after injection into the footpad (triangle: footpad, circle: lymph node). (D) Immunohistofluorescence analysis of the dissected lymph nodes of a mouse injected with FITC-labeled aPNMs. The slides were stained with anti-CD169, anti-F4/80, or anti-CD205. Scale bar is 250 µm. Magnification ×4 (Olympus IX 71, Olympus, Tokyo, Japan).
Abbreviations: aPNMs, amine-terminated γ-PGA nanomicelles; FITC, fluorescein isothiocyanate; γ-PGA, poly-(γ-glutamic acid); NIR, near-infrared.
Induction of inflammasome-related immune response by aPNMs
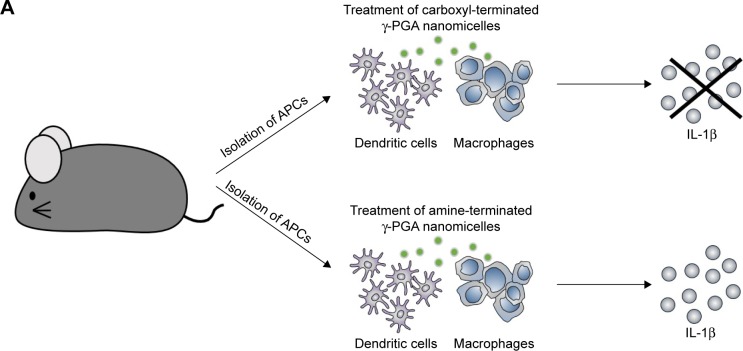
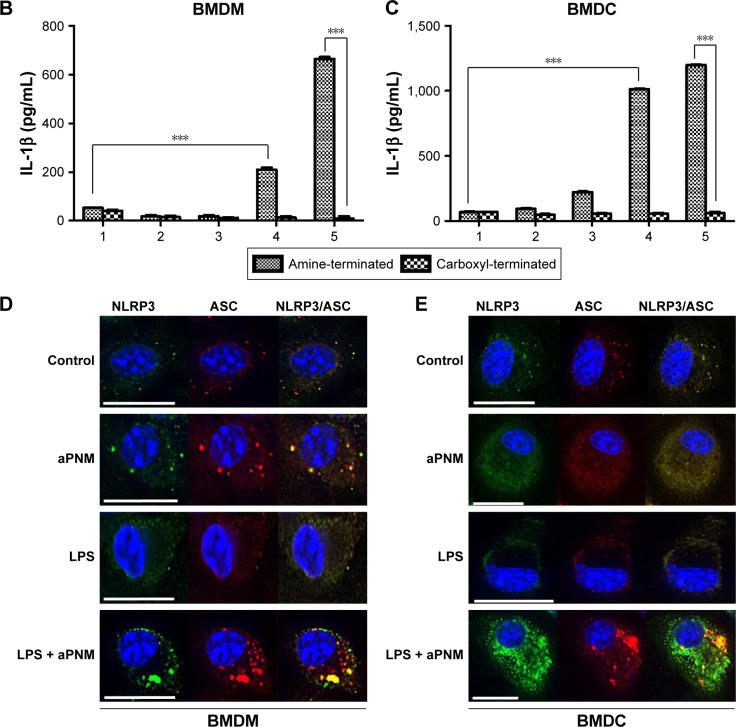
We first investigated whether the designed aPNMs induced inflammasomes. In order to investigate the role of the amine moiety in aPNMs as a stimulus of inflammasomes, we evaluated the concentration of IL-1β secreted by APCs treated with aPNMs and carboxyl-terminated γ-PGA nanomicelles (Figure 2). As shown in Figure 2B and C, the secretion of IL-1β was enhanced after the incubation of BMDCs and BMDMs with aPNMs for 4 hours and after priming with LPS (400 ng mL−1) for 3 hours. In contrast, the activation of BMDCs and BMDMs with carboxyl-terminated γ-PGA nanomicelles at the same concentration did not result in an increased secretion of IL-1β. Although the control group was also primed with LPS, there was no increase in the secretion of IL-1β. Thus, the enhanced secretion of IL-1β after incubation of BMDCs and BMDMs with aPNMs may have been related to the amine moiety of aPNMs. In order to investigate whether LPS and aPNMs induce the formation of NLRP3 inflammasomes in BMDCs and BMDMs, the interaction between NLRP3 and ASC was assessed by immunofluorescence. We pretreated APCs with LPS (400 ng mL−1) or medium only for 3 hours and then incubated them with medium alone or aPNMs for 4 hours. Many colocalized intracellular complexes of NLRP3 (green) and ASC (red) in both BMDCs and BMDMs were detected after the stimulation of LPS with aPNMs (Figure 2D and E). However, few complexes of NLRP3 and ASC were found in the control, LPS-only, and aPNM-only groups. The experimental results were consistent with the assembly of the inflammasome mechanism shown in Scheme 1. Therefore, the experimental data demonstrate that simultaneous treatment of aPNMs with LPS induced the activation and assembly of NLRP3 inflammasomes efficiently in both BMDCs and BMDMs.
Figure 2.
Induction of inflammasome response.
Notes: (A) Scheme of ex vivo experiment. (B, C) Secretion of IL-1β after incubation of BMDMs and BMDCs with various concentrations of aPNMs or carboxyl-terminated γ-PGA nanomicelles for 4 hours and after priming with LPS (400 ng mL−1) for 3 hours. The concentration unit of the X-axis is µg mL−1. ***p<0.001. Scale bar is 15 µm. (1: control, 2: 1 µg mL−1, 3: 2 µg mL−1, 4: 5 µg mL−1, 5: 10 µg mL−1). Immunofluorescent images (100×) of the inflammasome complex (NLRP3/ASC) of BMDM (D) and BMDC (E).
Abbreviations: APC, antigen-presenting cells; aPNMs, amine-terminated γ-PGA nanomicelles; BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages; γ-PGA, poly-(γ-glutamic acid); LPS, lipopolysaccharide.
Mechanism study of inflammasome activation
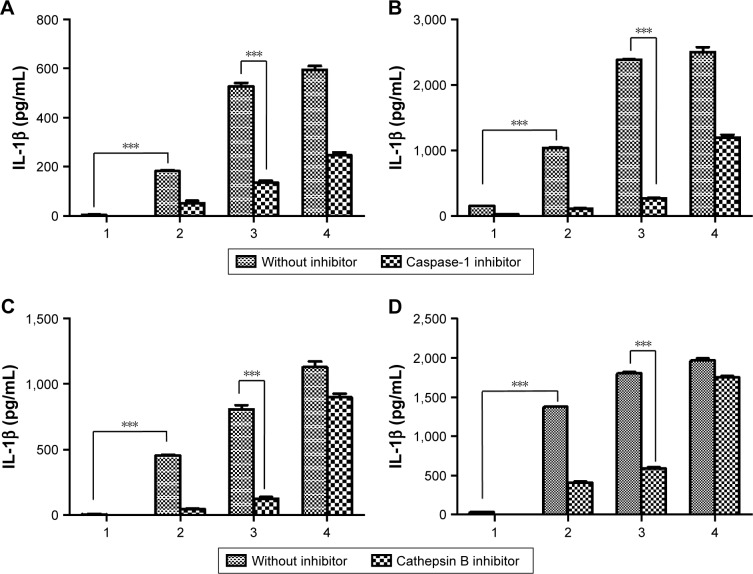
We also investigated the mechanism of inflammasome activation by inhibitors (cathepsin B inhibitor [CA-074] and caspase-1 inhibitor [Ac-YVAD-cmk]) of the inflammasome signaling pathway as shown in Schemes 1 and S1.37 The activation of caspase-1 is essential for inducing the active form of pro-IL-1β into IL-1β in the whole inflammasome pathway.11 Thus, the inhibition of caspase-1 activity can attenuate the effect of inflammasome activators. Mature IL-1β has been implicated in many immune responses. In order to evaluate the effect of inhibitors, we incubated BMDCs or BMDMs with aPNMs and inhibitors simultaneously after priming with LPS for 3 hours. As shown in Figures 3 and S3, IL-1β was decreased after treatment with a caspase-1 inhibitor (Ac-YVAD-cmk) and cathepsin B inhibitor (CA-074) in both BMDCs and BMDMs. As a control, we used another inflammasome inducer, poly(deoxyade nylic–deoxythymidylic) (poly-(dA:dT)), which is known to stimulate the AIM2 inflammasome (Scheme S2).38 Although the AIM2 inflammasome signaling pathway differs from that of the NLRP3 inflammasome, the final pathway for the activation of caspase-1 to produce IL-1β is the same. After incubation with poly-(dA:dT) and the caspase-1 inhibitor, we observed a dramatic decrease in IL-1β (Figure 3A and B). However, the cathepsin B inhibitor did not result in a large difference in the concentration of secreted IL-1β because the cathepsin B inhibitor affects only the NLRP3 inflammasome, not the AIM2 inflammasome (Figure 3C and D).11
Figure 3.
Effect of inflammasome inhibitors.
Notes: Secretion of IL-1β after incubation of BMDMs (A) and BMDCs (B) with a caspase-1 inhibitor. Secretion of IL-1β after incubation of BMDMs (C) and BMDCs (D) with a cathepsin B inhibitor. All data were obtained in triplicate and are presented as the mean ± SD. ***p<0.001. The concentration unit of the X-axis is µg mL−1, and poly-(dA:dT) is 2 µg mL−1 (1: control, 2: 5 µg mL−1, 3: 10 µg mL−1, 4: poly-(dA:dT)).
Abbreviations: BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages; poly-(dA:dT), poly(deoxyadenylic–deoxythymidylic).
In vivo induction of inflammasome
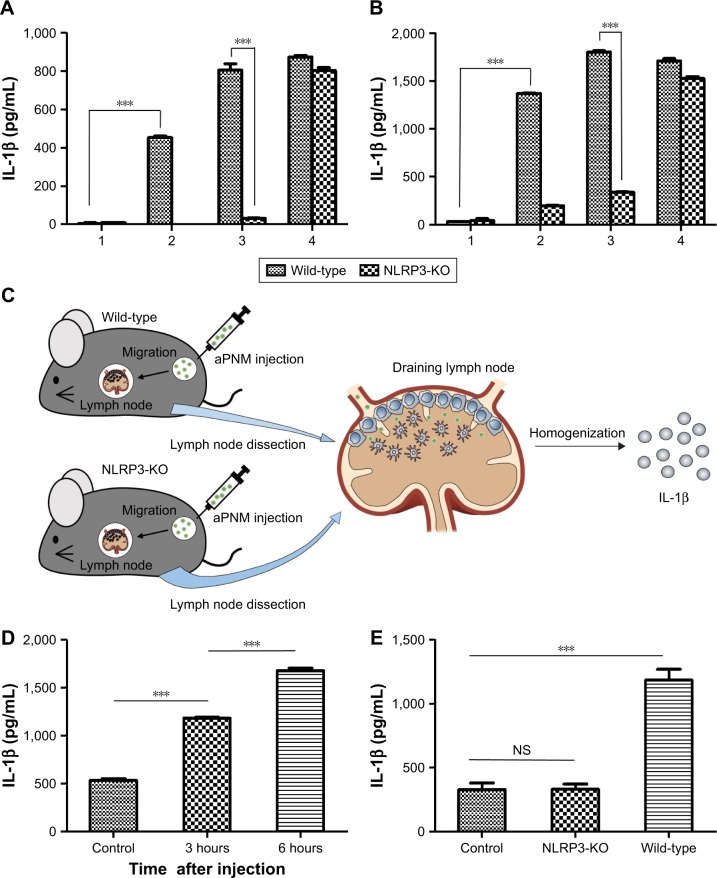
In order to investigate inflammasome induction in vivo, we used NLRP3 knockout mice (NLRP3−/−) to assess inflammasome induction by aPNMs.39 When both BMDMs and BMDCs isolated from NLRP3−/− mice were treated with aPNMs, the secretion of IL-1β was greatly inhibited (Figure 4A and B). This implies that the NLRP3 inflammasome-inducing pathway is closely related to the production of IL-1β by APCs treated with aPNMs. In contrast, the secretion of IL-1β by both BMDMs and BMDCs isolated from NLRP3−/− mice was not inhibited when they were treated with poly-(dA:dT). As we observed in the in vitro experiment (Figure 3), the induction of inflammasomes by poly-(dA:dT) is related to the AIM2 pathway, not the NLRP3 pathway.40
Figure 4.
In vitro and in vivo induction of inflammasomes in lymph nodes.
Notes: Secretion of IL-1β after incubation of BMDMs (A) and BMDCs (B) isolated from wild-type or NLRP3-KO mice (NLRP3−/−) with aPNMs. The concentration unit of the X-axis is µg mL−1, and poly-(dA:dT) is 2 µg mL−1. (1: control, 2: 5 µg mL−1, 3: 10 µg mL−1, 4: poly-(dA:dT)). (C) Scheme of in vivo and ex vivo experiment. (D) Secretion of IL-1β in lymph nodes at different time points after injection of aPNMs. (E) Different degrees of inflammasome induction between wild-type and NLRP3-KO mice. All data were obtained in triplicate and are presented as the mean ± SD. ***p<0.001. NS, not significant.
Abbreviations: aPNMs, amine-terminated γ-PGA nanomicelles; BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages; γ-PGA, poly-(γ-glutamic acid); KO, knock out; poly-(dA:dT), poly(deoxyadenylic–deoxythymidylic).
We also determined whether aPNMs could act as an efficient inflammasome inducer after their injection into mice. We injected aPNMs after priming with LPS (5 µg) for 24 hours. After aPNMs were subcutaneously injected, lymph nodes were collected from the immunized mice and used for ex vivo analysis of IL-1β. As shown in Figure 4C, the secreted concentration of IL-1β in wild-type mice immunized with aPNMs increased up to 6 hours after injection. Furthermore, there was a large difference between NLRP3−/− and wild-type mice at 6 hours after injection (Figure 4D). The experimental results are all consistent with those observed in the in vitro system. However, we found no differences in the secretion of IL-1β between mouse groups injected with carboxyl-terminated γ-PGA nanomicelles and the control group (Figure S4). The experimental results also suggest that aPNMs can move into lymph nodes efficiently and activate the inflammasome of APCs.
Triggering multiple arms of innate immune response by aPNMs/poly-(I:C)
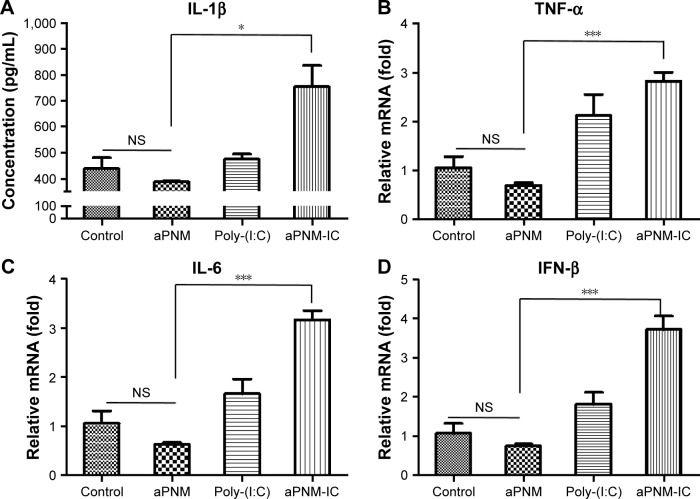
Although LPS was used as a TLR4 agonist, which was required for the efficient induction of inflammasomes, the toxicity of LPS limits its clinical application.41 Based on our experimental results, another TLR agonist that has been tested for human vaccines was combined with the novel inflammasome inducer aPNMs. We investigated whether aPNMs could activate multiple arms of the innate immune system after combining them with poly-(I:C), a TLR3 agonist that is a synthetic analog of double-stranded RNA.42,43 This agonist can lead to the induction of protective cellular genes, including type I IFN and proinflammatory cytokines, to directly limit viral replication and help direct subsequent adaptive immune responses.44 After aPNMs coated with poly-(I:C) were immunized into mice, the innate immune response in lymph nodes was analyzed. In order to investigate the effects of the combined nanoadjuvant aPNM-IC on multiple arms of the innate immune response, we determined whether other inflammasome-independent proinflammatory cytokines and type I IFN were secreted in addition to inflammasome induction.36 The secretion of IL-1β was higher in the mouse groups treated with aPNM-IC (Figure 5A). The secretion levels of TNF-α and IL-6 were also markedly increased in the mouse groups treated with aPNM-IC compared with those treated with aPNMs or poly-(I:C) only (Figure 5B and C). IFN-β, a representative type I IFN, was also produced at high levels in mouse groups treated with aPNM-IC (Figure 5D). The intrinsic immunological effects of poly-(I:C) on the production of proinflammatory cytokines (TNF-α and IL-6) and type I IFN (IFN-β) were synergistically enhanced by the combination of poly-(I:C) with aPNMs. This synergistic effect may be related to the enhanced delivery of poly-(I:C) into endosomes, where TLR3 is located, through the help of aPNMs. Coupling of the type I IFN pathway with inflammasome activation may also contribute to the synergistic effect (Scheme S2).44 The experimental results demonstrate that well-designed multiple arms of the innate immune response can be tailored by the careful selection and combination of immunomodulatory compounds with aPNMs, although aPNMs stimulate APCs only through the NLRP3 inflammasome pathway.
Figure 5.
Triggering of multiple arms of the innate immune response in lymph nodes by aPNM-IC.
Notes: Secretion levels of (A) IL-1β, (B) TNF-α, (C) IL-6, (D) IFN-β. All data were obtained in triplicate and are presented as the mean ± SD. *p<0.05, ***p<0.001. NS, not significant.
Abbreviations: aPNM-IC, amine-terminated γ-PGA nanomicelles + poly-(I:C); poly-(I:C), polyinosinic–polycytidylic acid; γ-PGA, poly-(γ-glutamic acid); IFN-β, interferon-β; TNF-α, tumor necrosis factor-alpha.
Conclusion
In this study, we found that size-controlled aPNMs can be used as a designer vaccine adjuvant to activate multiple arms of the innate immune response in lymph nodes. By aminating carboxyl groups in γ-PGA nanomicelles, the immunologically inert nanomicelles were converted into potential activators of inflammasomes, similar to conventional adjuvants such as alum. Both in vitro and in vivo experimental results revealed that aPNMs induced NLRP3 inflammasome activation and subsequent release of proinflammatory IL-1β. When aPNMs were combined with a clinically evaluated TLR3 agonist, poly-(I:C) (aPNM-IC), they triggered multiple arms of the innate immune response, including the secretion of proinflammatory cytokines by both inflammasomes and an inflammasome-independent pathway and the induction of type I IFNs. In the future studies, it is necessary to assess the reciprocal interaction between the aminated nanomicelles and surface of biological membranes when the aminated nanomicelles are used for biomedical applications because positively charged nanomaterials can induce unexpected side effects.45,46 However, their targeted application in the control of infectious diseases as well as cancer immunotherapy, which requires well-designed innate and adaptive immune responses, is expected to produce positive results. The aPNMs can be used as a multifunctional nanoadjuvant that can tailor the immune response in lymph nodes by the incorporation of various types of immunostimulatory compounds, eg, other TLR ligands, nucleotide-oligomerization domain ligands, and stimulator of IFN genes ligands. Furthermore, the amine group of aPNMs will also facilitate the loading of antigens (ie, recombinant proteins, peptides, and genes) when used to design a vaccine for infectious diseases and cancer immunotherapy.47–51
Supplementary materials
The stability of aPNM in PBS and serum at body temperature.
Note: The stability test results conducted at body temperature showed that the size of aminated micelles was kept during a time span of 6 days.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); PBS, phosphate-buffered saline.
Intracellular location of aPNM-FITC measured by fluorescence microscopy.
Notes: BMDMs and BMDCs were stained with 50 nM LysoTracker® red (Thermo Fisher Scientific, Waltham, MA, USA) and treated with aPNM-FITC overnight. Some of aPNM-FITC was colocalized with LysoTracker, while others were in the cytosol. Scale bar is 5 µm. Magnification ×60 (Delta vision PD instrument; GE Healthcare, Little Chalfont, UK).
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages; DIC, differential interference contrast; FITC, fluorescein isothiocyanate; γ-PGA, poly-(γ-glutamic acid).
Effect of inflammasome inhibitors.
Notes: Secretion of IL-1β after incubation of BMDMs (A) and BMDCs (B) with caspase-1 inhibitor. Secretion of IL-1β after incubation of BMDMs (C) and BMDCs (D) with cathepsin B inhibitor. All data were obtained in triplicate and are presented as a mean value ± SD. ***p<0.001. The concentration unit of X-axis is µg mL−1 (1: control, 2: 1 µg mL−1, 3: 2 µg mL−1, 4: 5 µg mL−1, 5: 10 µg mL−1).
Abbreviations: BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages.
In vivo induction of inflammasomes in lymph nodes.
Notes: Secretion of IL-1β in lymph nodes at different time points after injection of carboxyl-terminated γ-PGA nanomicelles. All data were obtained in triplicate and are presented as the mean ± SD. NS, not significant.
Abbreviation: γ-PGA, poly-(γ-glutamic acid).
Mechanism study of inflammasome activation by inhibitors of inflammasome signaling pathway.
Notes: Cathepsin B was inhibited by CA-074, and caspase-1 was inhibited by Ac-YVAD-cmk. NLRP3 was depleted in NLRP3-KO mice.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); IFN-β, interferon-β; KO, knock out; LPS, lipopolysaccharide; TNF-α, tumor necrosis factor-alpha.
Signaling pathway of different types of inflammasomes.
Notes: Poly-(dA:dT) stimulates the AIM2 inflammasome. The final pathway for the activation of caspase-1 to produce IL-1β is the same.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); poly-(dA:dT), poly(deoxyadenylic–deoxythymidylic); TLR4, toll-like receptor 4.
Acknowledgments
The authors acknowledge the financial support from the National Research Foundation of Korea grant funded by the Korean government (The Ministry of Science, ICT and Future Planning; Grant Numbers 2017R1A5A1014560 and 2016R1A2B4015056) and grant funded by the Ministry of Health and Welfare (Grant Number HI14C2680). Prof Sung Jae Shin is also one of the corresponding authors for this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Li WA, Choi Y, et al. Injectable, spontaneously assembling, inorganic scaffolds modulate immune cells in vivo and increase vaccine efficacy. Nat Biotechnol. 2015;33(1):64–72. doi: 10.1038/nbt.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvine DJ, Swartz MA, Szeto GL. Engineering synthetic vaccines using cues from natural immunity. Nat Mater. 2013;12(11):978–990. doi: 10.1038/nmat3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu M, Wang R, Nie G. Applications of nanomaterials as vaccine adjuvants. Hum Vaccin Immunother. 2014;10(9):2761–2774. doi: 10.4161/hv.29589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain S, Vanoirbeek JA, Hoet PH. Interactions of nanomaterials with the immune system. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(2):169–183. doi: 10.1002/wnan.166. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Noh YW, Heo MB, Cho MY, Lim YT. Multifunctional hybrid nanoconjugates for efficient in vivo delivery of immunomodulating oligonucleotides and enhanced antitumor immunity. Angew Chem Int Ed Engl. 2012;51(38):9670–9673. doi: 10.1002/anie.201204989. [DOI] [PubMed] [Google Scholar]
- 7.Niikura K, Matsunaga T, Suzuki T, et al. Gold nanoparticles as a vaccine platform: influence of size and shape on immunological responses in vitro and in vivo. ACS Nano. 2013;7(5):3926–3938. doi: 10.1021/nn3057005. [DOI] [PubMed] [Google Scholar]
- 8.Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PLoS One. 2012;7(6):e38258. doi: 10.1371/journal.pone.0038258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neeland MR, Shi W, Collignon C, et al. The Lymphatic immune response induced by the adjuvant AS01: a comparison of intramuscular and subcutaneous immunization routes. J Immunol. 2016;197(7):2704–2714. doi: 10.4049/jimmunol.1600817. [DOI] [PubMed] [Google Scholar]
- 10.Tozuka M, Oka T, Jounai N, et al. Efficient antigen delivery to the draining lymph nodes is a key component in the immunogenic pathway of the intradermal vaccine. J Dermatol Sci. 2016;82(1):38–45. doi: 10.1016/j.jdermsci.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21(7):677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neeland MR, Elhay MJ, Nathanielsz J, Meeusen ENT, de Veer MJ. Incorporation of CpG into a liposomal vaccine formulation increases the maturation of antigen-loaded dendritic cells and monocytes to improve local and systemic immunity. J Immunol. 2014;192(8):3666–3675. doi: 10.4049/jimmunol.1303014. [DOI] [PubMed] [Google Scholar]
- 13.Hickman HD. Illuminating inflammasome activity in vivo. Nat Med. 2016;22(1):22–23. doi: 10.1038/nm.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebre F, Hearnden CH, Lavelle EC. Modulation of immune responses by particulate materials. Adv Mater. 2016;28(27):5525–5541. doi: 10.1002/adma.201505395. [DOI] [PubMed] [Google Scholar]
- 15.Neumann S, Burkert K, Kemp R, Rades T, Rod Dunbar P, Hook S. Activation of the NLRP3 inflammasome is not a feature of all particulate vaccine adjuvants. Immunol Cell Biol. 2014;92(6):535–542. doi: 10.1038/icb.2014.21. [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Flavin K, Kopf I, et al. Functionalization of carbon nanoparticles modulates inflammatory cell recruitment and NLRP3 inflammasome activation. Small. 2013;9(24):4194–4206. doi: 10.1002/smll.201300481. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J Immunol. 2008;181(1):17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp FA, Ruane D, Claass B, et al. Uptake of particulate vaccine adjuvants by dendritic cells activates the NALP3 inflammasome. Proc Natl Acad Sci U S A. 2009;106(3):870–875. doi: 10.1073/pnas.0804897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halle A, Hornung V, Petzold GC, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9(8):857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornung V, Bauernfeind F, Halle A, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kono H, Orlowski GM, Patel Z, Rock KL. The IL-1-dependent sterile inflammatory response has a substantial caspase-1-independent component that requires cathepsin C. J Immunol. 2012;189(7):3734–3740. doi: 10.4049/jimmunol.1200136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruwona TB, Xu H, Li X, Taylor AN, Shi YC, Cui Z. Toward understanding the mechanism underlying the strong adjuvant activity of aluminum salt nanoparticles. Vaccine. 2016;34(27):3059–3067. doi: 10.1016/j.vaccine.2016.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun B, Ji Z, Liao YP, et al. Engineering an effective immune adjuvant by designed control of shape and crystallinity of aluminum oxyhydroxide nanoparticles. ACS Nano. 2013;7(12):10834–10849. doi: 10.1021/nn404211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.den Brok MH, Büll C, Wassink M, et al. Saponin-based adjuvants induce cross-presentation in dendritic cells by intracellular lipid body formation. Nat Commun. 2016;7:13324. doi: 10.1038/ncomms13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bueter CL, Lee CK, Rathinam VA, et al. Chitosan but not chitin activates the inflammasome by a mechanism dependent upon phagocytosis. J Biol Chem. 2011;286(41):35447–35455. doi: 10.1074/jbc.M111.274936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Dunphy DR, Jiang X, et al. Processing pathway dependence of amorphous silica nanoparticle toxicity: colloidal vs pyrolytic. J Am Chem Soc. 2012;134(38):15790–15804. doi: 10.1021/ja304907c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanneganti TD, Ozoren N, Body-Malapel M, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440(7081):233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 28.Ribes S, Ebert S, Regen T, et al. Toll-like receptor stimulation enhances phagocytosis and intracellular killing of nonencapsulated and encapsulated Streptococcus pneumoniae by murine microglia. Infect Immun. 2010;78(2):865–871. doi: 10.1128/IAI.01110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noh YW, Hong JH, Shim SM, et al. Polymer nanomicelles for efficient mucus delivery and antigen-specific high mucosal immunity. Angew Chem Int Ed Engl. 2013;52(30):7684–7689. doi: 10.1002/anie.201302881. [DOI] [PubMed] [Google Scholar]
- 30.Reddy ST, van der Vlies AJ, Simeoni E, et al. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007;25(10):1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 31.Reddy ST, Rehor A, Schmoekel HG, Hubbell JA, Swartz MA. In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. J Control Release. 2006;112(1):26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Noh YW, Kong SH, Choi DY, et al. Near-infrared emitting polymer nanogels for efficient sentinel lymph node mapping. ACS Nano. 2012;6(9):7820–7831. doi: 10.1021/nn301949y. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Moynihan KD, Zheng Y, et al. Structure-based programming of lymph-node targeting in molecular vaccines. Nature. 2014;507(7493):519–522. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Koker S, Cui J, Vanparijs N, et al. Engineering polymer hydrogel nanoparticles for lymph node-targeted delivery. Angew Chem Int Ed Engl. 2016;55(4):1334–1339. doi: 10.1002/anie.201508626. [DOI] [PubMed] [Google Scholar]
- 35.Starkhammar M, Kumlien Georen S, Swedin L, Dahlen SE, Adner M, Cardell LO. Intranasal administration of poly(I:C) and LPS in BALB/c mice induces airway hyperresponsiveness and inflammation via different pathways. PLoS One. 2012;7(2):e32110. doi: 10.1371/journal.pone.0032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson NS, Duewell P, Yang B, et al. Inflammasome-dependent and -independent IL-18 production mediates immunity to the ISCO-MATRIX adjuvant. J Immunol. 2014;192(7):3259–3268. doi: 10.4049/jimmunol.1302011. [DOI] [PubMed] [Google Scholar]
- 37.Man SM, Hopkins LJ, Nugent E, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A. 2014;111(20):7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalantari P, DeOliveira RB, Chan J, et al. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep. 2014;6(1):196–210. doi: 10.1016/j.celrep.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y, Jiang J, Said-Sadier N, et al. Activation of the NLRP3 inflammasome by vault nanoparticles expressing a chlamydial epitope. Vaccine. 2015;33(2):298–306. doi: 10.1016/j.vaccine.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10(3):241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bohannon JK, Hernandez A, Enkhbaatar P, Adams WL, Sherwood ER. The immunobiology of TLR4 agonists: from endotoxin tolerance to immunoadjuvants. Shock. 2013;40(6):451–462. doi: 10.1097/SHK.0000000000000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forte G, Rega A, Morello S, et al. Polyinosinic-polycytidylic acid limits tumor outgrowth in a mouse model of metastatic lung cancer. J Immunol. 2012;188(11):5357–5364. doi: 10.4049/jimmunol.1103811. [DOI] [PubMed] [Google Scholar]
- 43.Kadowaki N, Antonenko S, Liu YJ. Distinct CpG DNA and polyinosinic-polycytidylic acid double-stranded RNA, respectively, stimulate CD11c− type 2 dendritic cell precursors and CD11c+ dendritic cells to produce type I IFN. J Immunol. 2001;166(4):2291–2295. doi: 10.4049/jimmunol.166.4.2291. [DOI] [PubMed] [Google Scholar]
- 44.Malireddi RK, Kanneganti TD. Role of type I interferons in inflammasome activation, cell death, and disease during microbial infection. Front Cell Infect Microbiol. 2013;3:77. doi: 10.3389/fcimb.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaller T, Puntes V, Casals E, Duschl A, Oostingh GJ. In vitro investigation of immunomodulatory effects caused by engineered inorganic nanoparticles – the impact of experimental design and cell choice. Nanotoxicology. 2009;3(1):46–59. [Google Scholar]
- 46.Najafi-Hajivar S, Zakeri-Milani P, Mohammadi H, et al. Overview on experimental models of interactions between nanoparticles and the immune system. Biomed Pharmacother. 2016;83:1365–1378. doi: 10.1016/j.biopha.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 47.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2(8):469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 48.Hubbell JA, Thomas SN, Swartz MA. Materials engineering for immunomodulation. Nature. 2009;462(7272):449–460. doi: 10.1038/nature08604. [DOI] [PubMed] [Google Scholar]
- 49.Naahidi S, Jafari M, Edalat F, Raymond K, Khademhosseini A, Chen P. Biocompatibility of engineered nanoparticles for drug delivery. J Control Release. 2013;166(2):182–194. doi: 10.1016/j.jconrel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 50.Lee IH, Kwon HK, An S, et al. Imageable antigen-presenting gold nanoparticle vaccines for effective cancer immunotherapy in vivo. Angew Chem Int Ed Engl. 2012;51(35):8800–8805. doi: 10.1002/anie.201203193. [DOI] [PubMed] [Google Scholar]
- 51.Liu H, Kwong B, Irvine DJ. Membrane anchored immunostimulatory oligonucleotides for in vivo cell modification and localized immunotherapy. Angew Chem Int Ed Engl. 2011;50(31):7052–7055. doi: 10.1002/anie.201101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The stability of aPNM in PBS and serum at body temperature.
Note: The stability test results conducted at body temperature showed that the size of aminated micelles was kept during a time span of 6 days.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); PBS, phosphate-buffered saline.
Intracellular location of aPNM-FITC measured by fluorescence microscopy.
Notes: BMDMs and BMDCs were stained with 50 nM LysoTracker® red (Thermo Fisher Scientific, Waltham, MA, USA) and treated with aPNM-FITC overnight. Some of aPNM-FITC was colocalized with LysoTracker, while others were in the cytosol. Scale bar is 5 µm. Magnification ×60 (Delta vision PD instrument; GE Healthcare, Little Chalfont, UK).
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages; DIC, differential interference contrast; FITC, fluorescein isothiocyanate; γ-PGA, poly-(γ-glutamic acid).
Effect of inflammasome inhibitors.
Notes: Secretion of IL-1β after incubation of BMDMs (A) and BMDCs (B) with caspase-1 inhibitor. Secretion of IL-1β after incubation of BMDMs (C) and BMDCs (D) with cathepsin B inhibitor. All data were obtained in triplicate and are presented as a mean value ± SD. ***p<0.001. The concentration unit of X-axis is µg mL−1 (1: control, 2: 1 µg mL−1, 3: 2 µg mL−1, 4: 5 µg mL−1, 5: 10 µg mL−1).
Abbreviations: BMDCs, bone marrow-derived dendritic cells; BMDMs, bone marrow-derived macrophages.
In vivo induction of inflammasomes in lymph nodes.
Notes: Secretion of IL-1β in lymph nodes at different time points after injection of carboxyl-terminated γ-PGA nanomicelles. All data were obtained in triplicate and are presented as the mean ± SD. NS, not significant.
Abbreviation: γ-PGA, poly-(γ-glutamic acid).
Mechanism study of inflammasome activation by inhibitors of inflammasome signaling pathway.
Notes: Cathepsin B was inhibited by CA-074, and caspase-1 was inhibited by Ac-YVAD-cmk. NLRP3 was depleted in NLRP3-KO mice.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); IFN-β, interferon-β; KO, knock out; LPS, lipopolysaccharide; TNF-α, tumor necrosis factor-alpha.
Signaling pathway of different types of inflammasomes.
Notes: Poly-(dA:dT) stimulates the AIM2 inflammasome. The final pathway for the activation of caspase-1 to produce IL-1β is the same.
Abbreviations: aPNM, amine-terminated γ-PGA nanomicelles; γ-PGA, poly-(γ-glutamic acid); poly-(dA:dT), poly(deoxyadenylic–deoxythymidylic); TLR4, toll-like receptor 4.