Abstract
The soybean cyst nematode (SCN), Heterodera glycines, is a serious economic threat to soybean-producing regions worldwide. A new SCN population (called race X12) was detected in Shanxi province, China. Race X12 could reproduce on all the indicator lines of both race and Heterodera glycines (HG) type tests. The average number of females on Lee68 (susceptible control) was 171.40 with the lowest Female Index (FI) 61.31 on PI88788 and the highest FI 117.32 on Pickett in the race test. The average number of females on Lee68 was 323.17 with the lowest FI 44.18 on PI88788 and the highest FI 97.83 on PI548316 in the HG type test. ZDD2315 and ZDD24656 are elite resistant germplasms in China. ZDD2315 is highly resistant to race 4, the strongest infection race in the 16 races with FI 1.51 while being highly sensitive to race X12 with FI 64.32. ZDD24656, a variety derived from PI437654 and ZDD2315, is highly resistant to race 1 and race 2. ZDD24656 is highly sensitive to race X12 with FI 99.12. Morphological and molecular studies of J2 and cysts confirmed the population as the SCN H. glycines. This is a new SCN race with stronger virulence than that of race 4 and is a potential threat to soybean production in China.
Keywords: Heterodera glycines, race, soybean, soybean cyst nematode
Soybean cyst nematode (SCN), H. glycines, has been a major pest on soybeans (Glycine max Merr.) in China, the United States, the Russian Far East, and some other countries planting soybeans for many years (Koenning and Wrather, 2010; Woo et al., 2014; Kim et al., 2016). Planting SCN-resistant cultivars has formed the primary basis for controlling SCN and has been widely used in soybean-producing regions in the world, especially in the United States (Niblack et al., 2008; Koenning and Wrather, 2010).
ZDD2315 is an elite resistant germplasm in China with few SCN populations parasitizing on it. It has shown resistance to races 1 to 6 and 11 in our previous tests. Race X12 came from an SCN-infected field in Shanxi province, China. ZDD2315 is highly resistant to race 4, the strongest infection race while susceptible to race X12. This indicates that race X12 is a new race with some new virulent genes which makes it different from race 4.
The objective of this article is to show the virulence of race X12 in more detail with both race (Riggs and Schmitt, 1988) and HG type test (Niblack et al., 2002). Some other elite resistant lines, such as ZDD2315 and ZDD24656, were also used to test the virulence of race X12. The results of this article may be helpful for navigating the use of resistance in soybean breeding programs and the future control of the SCN population.
Materials and Methods
Plant materials:
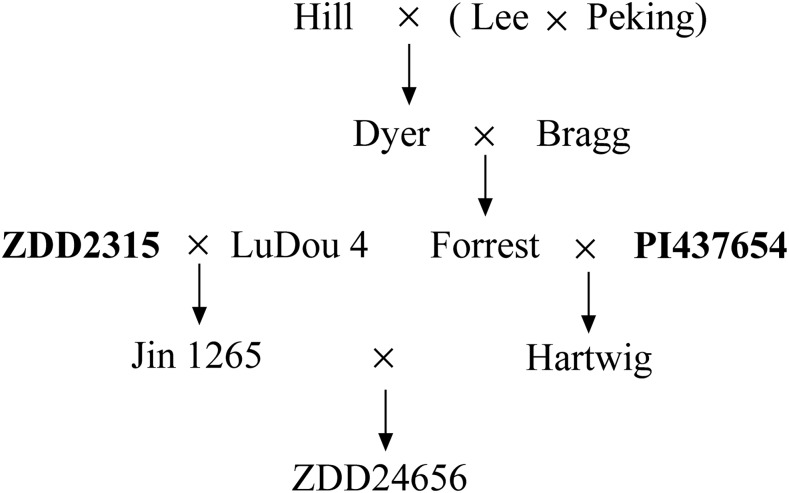
Seeds of indicator lines for the race and HG type tests, Lee68 (susceptible check), ZDD2315, and ZDD24656 were taken from the germplasm resources at the Henan Academy of Agricultural Science. The resistance genes of ZDD24656 come from ZDD2315 and PI437654 and this line has been authorized for release in China because of its higher yield and stable high resistance to races 1 and 2 (Liu et al., 2008). The pedigree chart of ZDD24656 is shown in Fig. 1.
Fig. 1.
Pedigree chart of ZDD24656. ZDD24656 derived from PI437654 and ZDD2315. Highly resistant to race 1 and race 2.
Bioassay method:
ZDD2315, ZDD24656, four indicator lines for race (Peking, Pickett, PI88788, PI90763), and seven indicator lines for the HG type test (Peking, PI88788, PI90763, PI437654, PI 209332, PI 89772, and PI548316) were evaluated for race X12 infestation following an environmental chamber bioassay at the Henan Academy of Agricultural Science, Zhengzhou, China. Briefly, plastic cups (6 cm × 12 cm) were filled with soil infected by race X12. Variety Lee68 was planted in the cups and sufficient numbers of eggs for the following bioassay were reared. Nine plants from each test line, indicator lines for the race test and the HG type test, and Lee68 were arranged in a randomized complete block design. Three days after transplantation, seedlings were inoculated with about 2,500 eggs from race X12. The experiments were maintained at 70% to 80% relative humidity, 25°C to 22°C (L/D), and a photoperiod of 16 hr : 8 hr (L:D) and were watered daily. Thirty days postinoculation, nematode cysts were washed from each plant roots, uniformly distributed to a piece of black cloth, and nematode cysts images were captured with a common digital camera. Carestream molecular imaging software was used to obtain counts of the number of cysts (Wang et al., 2014).
DNA extraction:
Several cysts were transferred into 1.5-ml eppendorf tubes containing 8 μl of distilled water and 10 μl of nematode lysis buffer (500 mM KCl, 100 mM Tris-HCl pH 8.0, 15 mM MgCl2, 1.0 mM DTT, 4.5% Tween 20) and crushed with a microhomogeniser Vibro Mixer for 3 min. Two microliters of proteinase K (600 μg·ml−1) was added and the tubes were incubated for 1 hr at 65°C and 10 min at 95°C consecutively. Then, the tube was centrifuged for 1 min at 16,000 g. The DNA suspension was used for further study.
PCR reaction:
Two microliter DNA suspension was added to 18-μl PCR reaction mixture containing: 2.0 μl 10× Qiagen PCR buffer, 1.6 μl 25 mM MgCl2, 0.8 μl 10 mM deoxynucleotides, 0.2 μl of each primers (1.0 μg/μl) (synthesized by ShengGong Company, Shanghai, China), 0.2 μl of Taq DNA Polymerase (5 U/μl) (Taq PCR Core Kit; Qiagen, Hilden, Germany) and 12.6 μl of distilled water.
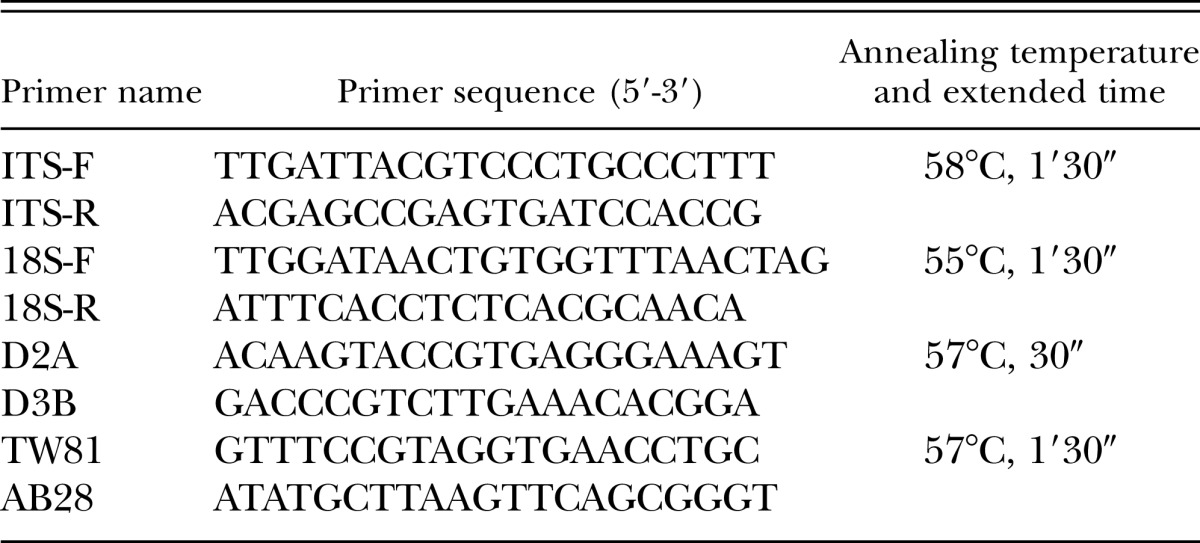
Four sets of nematode universal PCR primers of ITS, 18S, and D2/D3 regions were used in the PCR reaction to amplify fragments from race X12 (Subbotin et al., 2000a, 2000b; Mimee et al., 2014). The PCR reaction key factors are listed in Table 1.
Table 1.
Universal primer sequences and amplification conditions.
Two other sets of primers were used to identify the race X12 as SCN H. glycines. The first set contained the universal D3A (5′-GACCCGTCTTGAAACACGGA-3′) and D3B (5′-TCGGAAGGAACCAGCTACTA-3′) primers (Al-Banna et al., 1997; Thomas et al., 1997), which amplified a 345 bp fragment of the D3 expansion region from 28S rDNA gene, which indicates the presence of template nematode DNA in the sample and the quality of performance of the PCR. The second set contained species-specific primers GlyF1 (5′-TTACGGACCGTAACTCAA-3′) and rDNA2 (5′-TTTCACTCGCCGTTACTAAGG-3′), a universal primer, which amplified a 181 bp fragment from the ITS2 region and part of the 28S gene. The primers GlyF1 and rDNA2 have been shown to be species-specific to H. glycines populations (Subbotin et al., 2001). The PCR mixtures with DNA templates from race 2, 4, and 5, respectively, were run as positive controls. The PCR mixtures without DNA template were also run as negative control. The experiments for H. glycines cysts identification were repeated three times.
Results
Race X12 morphological characteristics of cysts and J2:
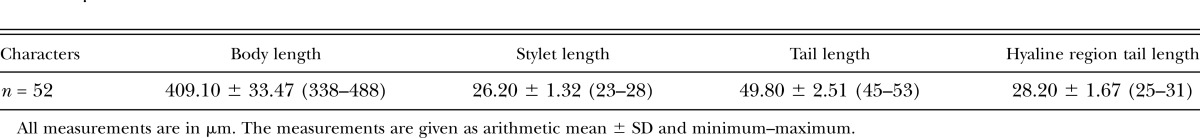
The cysts of race X12 were brown and lemon-shaped with a protruding neck and cone, ambifenestrated, underbridged, and had a strongly developed bullae. Second-stage juveniles (J2) were vermiform with a hyaline region in the tail terminus. The J2 (n = 52) were typical for SCN with a body length of 409.10 ± 33.47 μm, stylet length of 26.20 ± 1.32 μm, tail length of 49.80 ± 2.51 μm, and hyaline region tail length of 28.20 ± 1.67 μm (Table 2).
Table 2.
Morphometrical characters of juveniles (J2) of Heterodera glycines from race X12.
Race X12 cysts molecular characteristics:
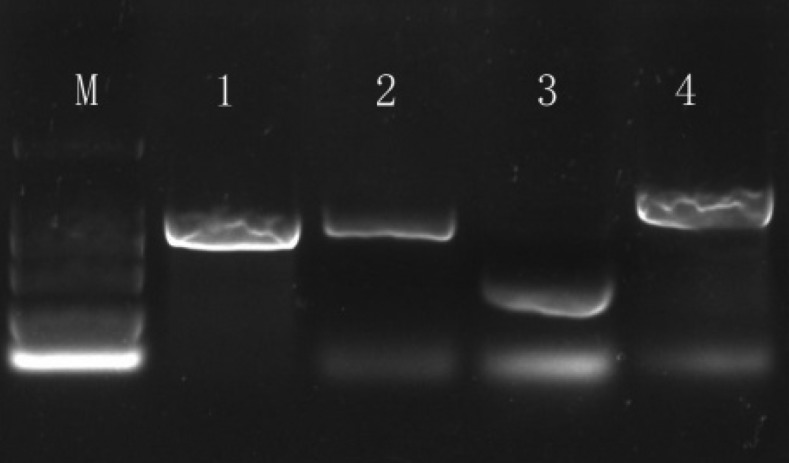
The PCR amplifications were performed using the race X12 DNA as template. The expected sizes of fragments were amplified, and the amplification products were about 750 bp with primer ITS-F/ITS-R, about 800 bp with primer TW81/AB28 from ITS, about 760 bp with primer 18S-F/18S-R from 18S and about 300 bp with primer D2A/D3B from 28S (Fig. 2).
Fig. 2.
PCR amplification with universal primers of Heterodera glycines. Expected fragments were yield with four sets primers amplification from race X12 DNA. M, DL2000; 1, Primer ITS-F+ITS-R; 2, Primer 18S-F+18S-R; 3, Primer D2A+D3B; 4, Primer TW81+AB28.
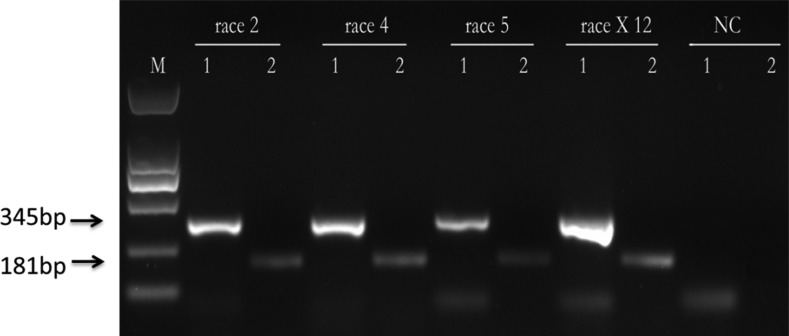
We further identified race X12 as H. glycines using a species-specific primer combination. All the tested populations including race 2, 4, 5, and race X12 yielded two distinct fragments (345 bp, a universal fragment to nematode species and 181 bp, a species-specific fragment to H. glycines populations), but these fragments were not found in the negative control (Fig. 3).
Fig. 3.
PCR with the GlyF1 species-specific primer. The amplified products are obtained from single cyst samples of race 2, 4, 5, and X12, respectively. Lane M: DL2000; Lane 1: PCR amplification with universal primers D3A/D3B and approximately 345 bp fragments were yield from 28S gene of Heterodera glycines except NC; 2: PCR amplification with a species-specific primer GlyF1 and a universal primer DNA2 in combination and 181bp fragments were yield from ITS2 region and a part of the 28S gene of Heterodera glycines except NC. NC = negative control.
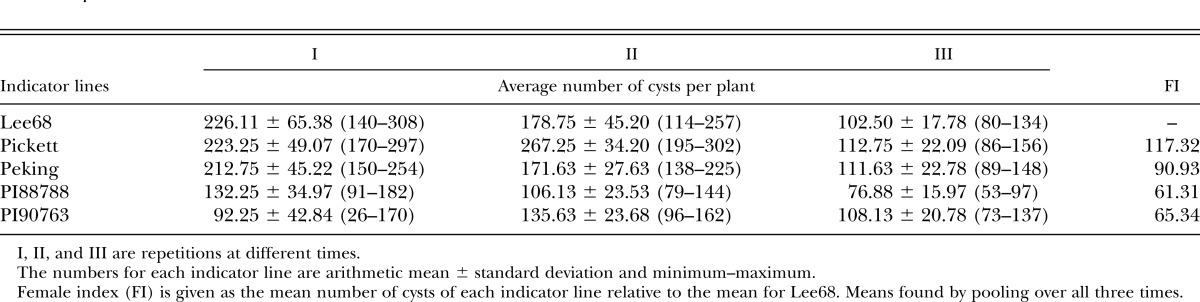
Virulence evaluation of race X12 using race and HG type tests:
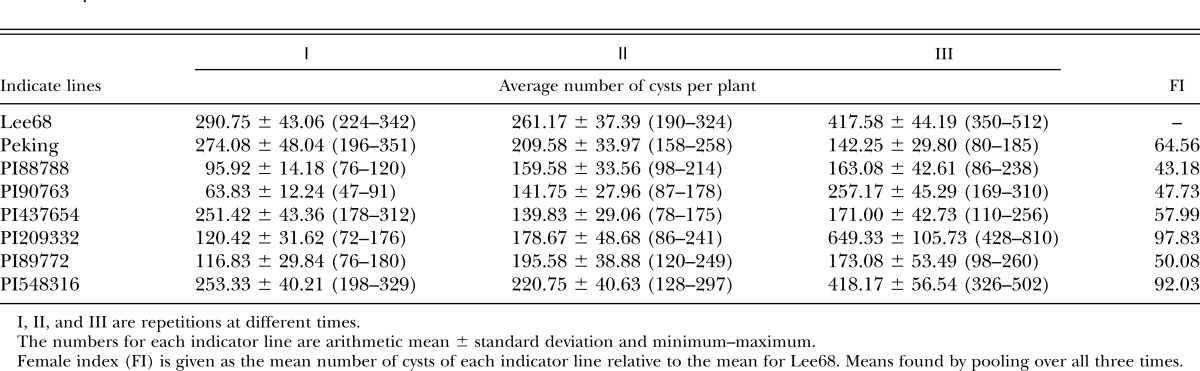
To show the virulence of race X12, we evaluated it with indicator lines of both race and HG type tests. Results showed that race 4 could parasitize on Pickett, Peking, PI88788, and PI90763. The populations produced from race X12 could also parasitize on these same four varieties. The average number of females on the Lee68 was 171.40 with the lowest FI 61.31 on PI88788 and the highest FI 117.32 on Pickett (Table 3). When tested with HG type, the results showed that the populations could reproduce on all seven indicate lines with the lowest FI 43.18 on PI88788 and the highest FI 97.83 on PI548316. The average number of females on Lee68 was 323.17 (Table 4).
Table 3.
Evaluation race X12 with race test.
Table 4.
Evaluation race X12 with HG type test.
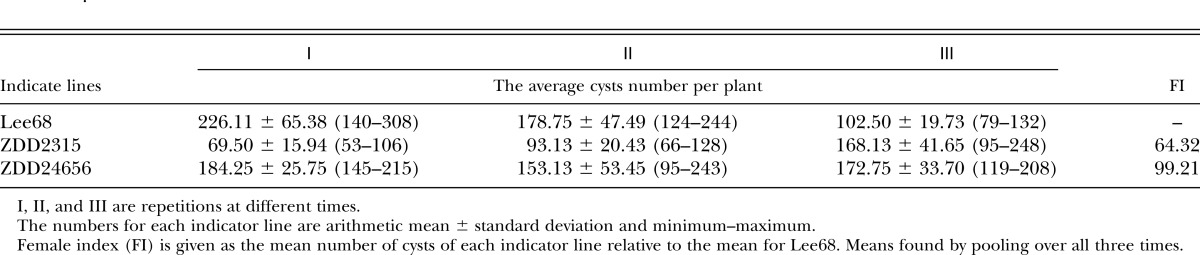
Virulence evaluation of race X12 using ZDD2315 and ZDD24656:
ZDD2315 is highly resistant to race 1, race 2, and race 4 with FI 0, 0, and 1.5, respectively (Table 5). ZDD2315 and ZDD24656 are highly sensitive to race X12 with FI 64.32 and 99.21, respectively (Table 6). Race X12 could reproduce on ZDD2315 successfully, whereas race 4 could hardly reproduce on it. Also, ZDD2315 is highly resistant to race 1 and race 2 but highly sensitive to race X12. That is to say, race X12 is more virulent than other races.
Table 5.
Test the resistance of ZDD2315 to race 1, race 2 or race 4.
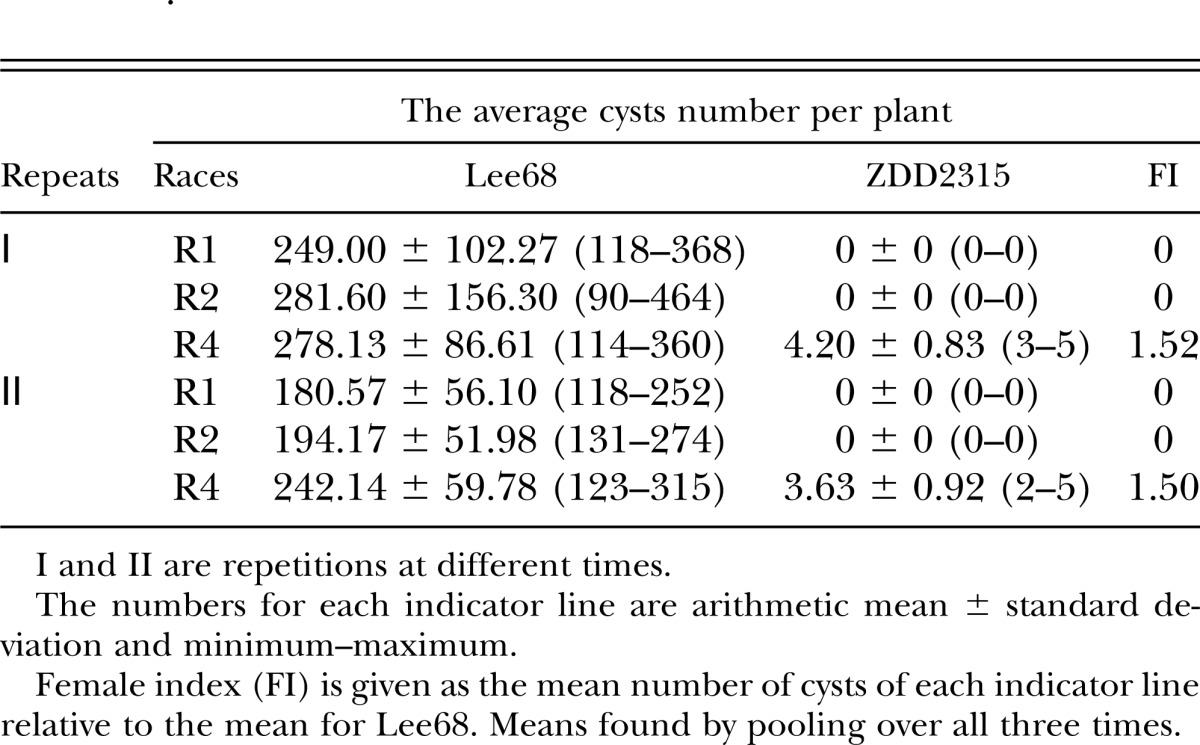
Table 6.
Testing race X12 with elite resistant sources.
Discussion
A new race of SCN H. glycines:
Race X12, was detected in a soybean field heavily infected with SCN in Shanxi province, China, during our survey on the diversity and distribution of H. glycines races in the Huang-Huai Rivers Valley (Lian et al., 2006). In practice, cyst forming nematodes are assumed to be H. glycines if found infesting a field with a soybean history (Riggs and Niblack, 1993). Morphological studies of cysts and J2 also confirmed that the nematode population of race X12 is SCN of H. glycines because the morphology of the cysts and J2 of race X12 were consistent with those of H. glycines (Subbotin et al., 2001). Studies showed that the SCN fragments do not vary in length significantly from other nematodes (Al-Banna et al., 1997; Thomas et al., 1997). Race X12 has the molecular character of a nematode, and we found all the expected size fragments by PCR. Amplifying a 181 bp fragment from race X12 using species-specific primer combination of nematode H. glycines further confirmed that race X12 was H. glycines.
The present situation of SCN races:
In the United States, the most common races are 1, 2, and 3 in frequency in Missouri, accounting for 44.4%, 22.2%, and 22.2% of the populations sampled, respectively, in 2005 with virulence phenotypes of H. glycines shifting from 1998 (Mitchum et al., 2007). Races 3, 1, and 2 (in frequency) accounted for 86% of H. glycines populations during a survey in Missouri in the years 1998 to 1999 (Niblack et al., 2003). HG types 0, 2.5.7, and 7 accounted for 81% of all the HG types determined for the samples tested during a survey in South Dakota (Acharya et al., 2016). In Canada, the SCN was only reported in the province of Ontario before 2014. However, since then the SCN was detected in the province of Quebec and has been spreading in a north and northeast direction along the St. Lawrence River (Mimee et al., 2014). In China, the SCN was restricted to the northeast and the Huang-Huai River Valley, the two principal soybean production areas. Races 2, 5, and 4 (in frequency) accounted for 83.9% of H. glycines populations in a race distribution survey in Huang-Huai Rivers Valley from the year 2012 to 2015 (Lian et al., 2016). Our survey also showed that the predominant race H. glycines have changed from race 1 in 2001 to 2003 (Lu et al., 2006a) to race 2 in 2012 to 2015 in Huang-Huai Rivers Valley. Now, the SCN has spread into the Xinjiang Uygur Autonomous Region, Guangxi, Guizhou, Jiangxi, Gansu, and Ningxia province which were not infected areas before (Peng et al., 2015; Wang et al., 2015). To a large extent, overutilization of resistant cultivars for H. glycines as a management tool results in decreased resistance to a given race over time. It is believed that continuous planting of the same resistant cultivar can lead to genetic changes in the SCN so that they eventually develop and reproduce on those cultivars (Grabau and Chen, 2014; Jain et al., 2016).
SCN with increasing infestation tendency in China:
In Huang-Huai Rivers Valley, SCN takes place much more seriously in Shanxi than in other provinces. Therefore, most of the elite resistant germplasms come from this province, like ZDD2315 (Lu et al., 2006b), PI437654, Peking, etc. (searching in USDA http://www.ars-grin.gov/npgs/index.html). ZDD2315 has been widely recognized as the most resistant soybean line in China for its high resistance to all the races. It is also reasonable to find much more diversity of SCN population in Shanxi province because of the soybean-SCN interaction. Race X12 is more virulent than race 4 because our study showed that ZDD2315 had high sensitivity to race X12 but high resistance to race 4. The results indicated that there might be some new virulent genes in race X12. The presence of race X12 showed that the SCN has an increasing infestation tendency, and race X12 is a potential threat to soybean production especially in Huang-Huai Rivers Valley in China.
The impact of new race on soybean:
Planting resistant cultivars have formed the core management strategy for the SCN for many years. However, the emergence of race X12 is a problem in that and the current resistant varieties are susceptible to it. This may result in the increase of the prevalence of SCN. The resistant varieties include three main sources for genetic resistance to the SCN: PI88788, Peking, and PI437654 (Hartwig and CystX). The resistance genes, such as Rhg1 and Rhg4, represent the major sources of resistance for the SCN in soybean cultivars, and many quantitative trait loci harboring minor genes have been identified for SCN resistance (Cook et al., 2012, 2014; Liu et al., 2012; Jiao et al., 2015; Kadam et al., 2016) are no longer resistance genes for race X12. The SCN is the most economically important soybean pathogen in the United States because it causes more yield loss than any other disease (Koenning and Wrather, 2010). Race X12 is likely to cause immeasurable losses to soybean production in the future. Therefore, effective measures should be taken to limit its spread into other areas and reduce the female numbers in infected soil.
Management of race X12:
The SCN needs a period of years to build up to damaging levels in fields and it almost cannot be eradicated from infected fields because the SCN persists in the soil for many years. Consequently, periodic monitoring for population densities and devising management strategies to reduce SCN numbers and slow SCN spreading speed should take first priority. Management strategies including (i) Screening new resistant germplasm and using it in soybean breeding programs to develop H. glycines resistant soybean cultivars. (ii) Identification of molecular markers by DNA sequencing that is associated with race X12 resistance for developing resistant varieties. (iii) Rotation with nonhost crop, such as corn, alfalfa, and grain sorghum is necessary to reduce the SCN densities. (iv) Cultural practices, such as early planted soybean, because the population density declines from April to June and no-till farming practices which could suppress cyst densities. (v) Chemical control should be considered for controlling the new race even though there may be negative economic returns for growers because of the high cost required in nematicides combined with the low price of soybeans.
Literature Cited
- Acharya K, Tande C, Byamukama E. Determination of Heterodera glycines virulence phenotypes occurring in South Dakota. Plant Disease. 2016;100:2281–2286. doi: 10.1094/PDIS-04-16-0572-RE. [DOI] [PubMed] [Google Scholar]
- Al-Banna L, Williams V, Gardner SL. Phylogenetic analysis of nematodes of the genus Pratylenchus using nuclear 26S rDNA. Molecular Phylogenetics and Evolution. 1997;7:94–102. doi: 10.1006/mpev.1996.0381. [DOI] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science. 2012;338:1206–1209. doi: 10.1126/science.1228746. [DOI] [PubMed] [Google Scholar]
- Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF. Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiology. 2014;165:630–647. doi: 10.1104/pp.114.235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabau ZJ, Chen SY. Efficacy of organic soil amendments for management of Heterodera glycines in greenhouse experiments. Journal of Nematology. 2014;46:267–274. [PMC free article] [PubMed] [Google Scholar]
- Jain S, Chittem K, Brueggeman R, Osorno JM, Richards J, Nelson BD. Comparative transcriptome analysis of resistant and susceptible common bean genotypes in response to soybean cyst nematode infection. PLoS One. 2016;11(7):e0159338. doi: 10.1371/journal.pone.0159338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Vuong TD, Liu Y, Meinhardt C, Liu Y, Joshi T, Cregan PB, Xu D, Shannon JG, Nguyen HT. Identification and evaluation of quantitative trait loci underlying resistance to multiple HG types of soybean cyst nematode in soybean PI 437655. Theoretical and Applied Genetics. 2015;128:15–23. doi: 10.1007/s00122-014-2409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Vuong TD, Qiu D, Meinhardt CG, Song L, Deshmukh R, Patil G, Wan J, Valliyodan B, Scaboo AM, Shannon JG, Nguyen HT. Genomic-assisted phylogenetic analysis and marker development for next generation soybean cyst nematode resistance breeding. Plant Science. 2016;242:342–350. doi: 10.1016/j.plantsci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- Kim KS, Vuong TD, Qiu D, Robbins RT, Grover Shannon J, Li Z, Nguyen HT. Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean. Theoretical and Applied Genetics. 2016;129:2295–2311. doi: 10.1007/s00122-016-2816-x. [DOI] [PubMed] [Google Scholar]
- Koenning SR, Wrather JA. Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Progress. 2010 doi: 10.1094/PHP-2010-1122-01-RS. [Google Scholar]
- Lian Y, Wang JS, Li HC, Wei H, Li JY, Wu YK, Lei CF, Zhang H, Wang SF, Guo JQ, Li YX, Li ZH, Jin QL, Xu SX, Zhang ZM, Yang CY, Yu HY, Geng Z, Shu WT, Lu WG. Race distribution of soybean cyst nematode in the main soybean producing area of Huang-Huai Rivers Valley. Acta Agronomica Sinica. 2016;42:1479–1486. [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492:256–260. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- Liu ZX, Lu WG, Chang RZ, Qiu LJ. Creation of new soybean SCN 4-resistant lines. Soybean Science. 2008;27(6):911–914. [Google Scholar]
- Lu WG, Gai JY, Li WD. Sampling survey and identification of races of soybean cyst nematode (Heterodera glycines Ichinohe) in Huang-Huai Valleys. Scientia Agricultura Sinica. 2006a;39:306–312. [Google Scholar]
- Lu WG, Gai JY, Zheng YZ, Li WD. Construction of a soybean genetic linkage map and mapping QTLs resistant to soybean cyst nematode (Heterodera glycines Ichinohe) Acta Agronomica Sinica. 2006b;3:1272–1279. [Google Scholar]
- Mimee B, Peng H, Popovic V, Yu Q, Duceppe MO, Tétreault MP, Belair G. First report of soybean cyst nematode (Heterodera glycines ichinohe) on soybean in the province of Quebec, Canada. Plant Disease. 2014;98:429. doi: 10.1094/PDIS-07-13-0782-PDN. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wrather JA, Heinz RD, Shannon JG, Danekas G. Variability in distribution and virulence phenotypes of Heterodera glycinesin in Missouri during 2005. Plant Disease. 2007;91:1473–1476. doi: 10.1094/PDIS-91-11-1473. [DOI] [PubMed] [Google Scholar]
- Niblack TL, Colgrove AL, Colgrove K, Bond JP. Shift in virulence of soybean cyst nematode is associated with use of resistance from PI 88788. Plant Health Progress. 2008 doi:10.1094/PHP-2008-0118-01-RS. [Google Scholar]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL. A revised classification scheme for genetically diverse populations of Heterodera glycines. Journal of Nematology. 2002;34:279–288. [PMC free article] [PubMed] [Google Scholar]
- Niblack TL, Wrather JA, Heinz RD, Donald PA. Distribution and virulence phenotypes of Heterodera glycines in Missouri. Plant Disease. 2003;87:929–932. doi: 10.1094/PDIS.2003.87.8.929. [DOI] [PubMed] [Google Scholar]
- Peng DL, Peng H, Wu DQ, Huang WK, Ye WX, Cui JK. First report of soybean cyst nematode (Heterodera glycines) on soybean from Gansu and Ningxia China. Plant Disease. 2015;100:229. [Google Scholar]
- Riggs RD, Niblack TL. 1993. Nematode pest of oilseed crops and grain legumes. Pp. 209–258 in K. Evans, D. L. Trudgill, and J. M. Webster, eds. Plant parasitic nematodes in temperate agriculture. Wallingford, UK: CAB International.
- Riggs RD, Schmitt DP. Complete characterization of the race scheme for Heterodera glycines. Journal of Nematology. 1988;20:392–395. [PMC free article] [PubMed] [Google Scholar]
- Subbotin SA, Peng DL, Moens M. A rapid method for the identification of the soybean cyst nematode Heterodera glycines using duplex PCR. Nematology. 2001;3:365–371. [Google Scholar]
- Subbotin SA, Waeyenberge L, Moens M. Molecular characterization of Chinese Heterodera glycines and H. avenae populations based on RFLPs and sequences of rDNA-ITS regions. Russian Journal of Nematology. 2000a;8:109–113. [Google Scholar]
- Subbotin SA, Waeyenberge L, Moens M. Identification of cyst forming nematodes of the genus Heterodera (Nematoda: Heteroderidae) based on the ribosomal DNA-RFLP. Nematology. 2000b;2:153–164. [Google Scholar]
- Thomas WK, Vida JT, Frisse LM, Mundo M, Baldwin JG. DNA sequences from formalin-fixed nematodes: Integrating molecular and morphological approaches to taxonomy. Journal of Nematology. 1997;29:250–254. [PMC free article] [PubMed] [Google Scholar]
- Wang D, Duan YX, Wang YY, Zhu XF, Chen LJ, Liu XY, Chen JS. First report of soybean cyst nematode, Heterodera glycines, on soybean from Guangxi, Guizhou, and Jiangxi Provinces, China. Plant Disease. 2015;99:893. [Google Scholar]
- Wang JS, Lu WG, Li JY, Lian Y, Wei H, Li HC, Lei CF. 2014. China Patent No. 2014SR060158.
- Woo MO, Beard H, MacDonald MH, Brewer EP, Youssef RM, Kim H, Matthews BF. Manipulation of two α-endo-β-1,4-glucanase genes, AtCel6 and GmCel7, reduces susceptibility to Heterodera glycines in soybean roots. Molecular Plant Pathology. 2014;15:927–939. doi: 10.1111/mpp.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]