Summary:
Access to high-quality reference data is essential for the development, validation, and implementation of in vitro and in silico approaches that reduce and replace the use of animals in toxicity testing. Currently, these data must often be pooled from a variety of disparate sources to efficiently link a set of assay responses and model predictions to an outcome or hazard classification. To provide a central access point for these purposes, the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods developed the Integrated Chemical Environment (ICE) web resource. The ICE data integrator allows users to retrieve and combine data sets and to develop hypotheses through data exploration. Open-source computational workflows and models will be available for download and application to local data. ICE currently includes curated in vivo test data, reference chemical information, in vitro assay data (including Tox21TM/ToxCast™ high-throughput screening data), and in silico model predictions. Users can query these data collections focusing on end points of interest such as acute systemic toxicity, endocrine disruption, skin sensitization, and many others. ICE is publicly accessible at https://ice.ntp.niehs.nih.gov. https://doi.org/10.1289/EHP1759
Introduction
Traditional tests to evaluate chemicals for their potential impact on human and environmental health are performed using animal models, with varying degrees of success in accurately identifying hazards. Advances in science and technology offer the potential to develop more effective approaches based on combining higher-throughput testing methods, human cell–based systems, small model organisms, and computational models. Development and acceptance of new approaches that reduce and replace animal use for better predictivity of human health outcomes require that regulators, test method developers, and computational modelers have access to high-quality toxicity data. Ideally, these data and relevant analysis tools would be available in a user-friendly format that facilitates data integration and exploration, encourages hypothesis generation, and allows for computational processing and modeling. In reality, progress in developing and evaluating new approaches is often hindered by the lack of open-source central access points for curated data on which models can be built or tested. In addition, the data available are often of unknown quality; constructing models or validating new methodologies on such data produces models and test systems with high uncertainty and suboptimal performance. Ultimately, locating and accessing the types of data needed—data in a structured format with metadata or annotation sufficient to gauge their quality and applicability—becomes a huge time sink for researchers and a barrier to innovation.
Existing sources of toxicity test data have advantages and limitations. The European Chemical Agency’s registered substances list (ECHA 2017) contains detailed information and test results on thousands of substances obtained from chemical dossiers submitted by manufacturers. Although this resource has depth of information, the data contained in it do not fully support computational analysis owing to the database’s restrictions on data use, to its limited batch query capability, and to formatting variations of the data fields. The U.S. Environmental Protection Agency (EPA) has several web-based resources providing information on chemical properties and toxicity data available in a computational-friendly format without restrictions on use. These include ToxCast™ data available through the tcpl package (Filer et al. 2016), Aggregated Computational Toxicology Resource (ACToR) (Judson et al. 2008, 2012), and the Chemistry Dashboard (U.S. EPA 2017). These U.S. EPA resources are extremely valuable because they provide detailed information on chemical properties and activities from various sources. However, building and evaluating new approaches require different types of data to be combined in a structured format that is informed by relevant biology or toxicity end points. This task is best facilitated by having the data organized by test guidelines or regulatory end points, a feature that is not currently provided by any of these U.S. EPA resources. Similar challenges with respect to organization exist with other resources such as PubChem (Kim et al. 2016; Wang et al. 2014) and the Toxicological Data Network from the U.S. National Library of Medicine (NLM 2016), as well as the Chemical Effects in Biological Systems (CEBS) database from the National Institute of Environmental Health Sciences (NIEHS) (Lea et al. 2017), all of which provide rich content but may not meet data structure needs.
The National Toxicology Program’s Integrated Chemical Environment (ICE) is a web-based resource (https://ice.ntp.niehs.nih.gov) that provides access to curated data and tools that can be used in the development, validation, and implementation of in vitro and in silico approaches that reduce and replace the use of animals in toxicity testing. The ICE data integrator function allows users to query high-quality in vivo and in vitro test results by chemical and by end point. ICE also includes reference chemical lists and supporting data sets, as well as computational predictions for properties such as physicochemical parameters and clearance rates, which are often needed in model development.
ICE Overview
The current release of ICE features in vivo and in vitro data sets covering regulatory test end points such as acute oral and dermal toxicity, skin irritation and sensitization, eye corrosion, and endocrine disruption as well as in silico predictions of test end points and chemical properties (Table 1). These data sets include data assembled and curated by the National Toxicology Program Interagency Center for Evaluation of Alternative Toxicological Methods (NICEATM) during past evaluations of new test methods and reference chemical lists generated from these efforts. ICE also includes a curated version of the high-throughput screening (HTS) data from Tox21TM and ToxCast™ (Huang 2016; Kavlock et al. 2012; Richard et al. 2016; Tice et al. 2013), with data filtered on the basis of chemical quality control information from the National Center for Advancing Translational Sciences and flags generated by the U.S. EPA’s tcpl pipeline (Filer et al. 2016; U.S. EPA 2016;). Additional data sets will be added with future releases.
Table 1.
Data types included in 2017 ICE releases and example end points.
| Data types | Availability | Type | End point examples |
|---|---|---|---|
| Acute dermal toxicity | October 2017 (tentative) | In vivo | Rodent LD50 |
| Acute inhalation toxicity | October 2017 (tentative) | In vivo | Rodent LC50 |
| Acute oral toxicity | March 2017 | In vivo | Rodent LD50 |
| Acute oral toxicity | March 2017 | In vitroa | Basal cytotoxicity IC50 |
| Androgenic activity | March 2017 | In vitro | Androgen receptor binding and transactivation (agonist and antagonist activity) |
| Androgenic activity | July 2017 (tentative) | In vivo | Lowest effect level in the rodent Hershberger assay |
| Androgenic activity | March 2017 | In silico | Androgen receptor pathway model scores |
| Curated HTS | March 2017 | In vitro | Assay ACC, AC50 |
| Dermal irritation | March 2017 | In vivo | Skin irritation/corrosion classification categories |
| Dermal sensitization | March 2017 | In vivo | Mouse LLNA EC3 and human patch test lowest effect level |
| Dermal sensitization | March 2017 | In vitro | KeratinoSens™, DPRA, hCLAT assay results |
| Dermal sensitization | July 2017 (tentative) | In silico | Binary sensitizer/nonsensitizer call |
| Estrogenic activity | March 2017 | In vivo | Lowest effect level in the rodent uterotrophic assay |
| Estrogenic activity | March 2017 | In silico | Estrogen receptor pathway model scores |
| Ocular irritation | March 2017 | In vivo | Eye irritation/corrosion classification categories |
| Physicochemical property predictions | March 2017 | In silico | LogP, logVP, logBCF, logS, melting point, boiling point |
Notes: AC50, concentration that increases activity by 50%; ACC, activity concentration at cutoff, a measure of the activity threshold for an assay response based on curve-fitting models; EC3, in the LLNA, a test chemical concentration that produces a stimulation index of 3; hCLAT, human cell line activation test; IC50, concentration that inhibits activity (in this context, decreases cell viability) by 50%; LC50, inhalation concentration expected to produce lethality in 50% of animals tested; LD50, dose expected to produce lethality in 50% of animals tested; LLNA, local lymph node assay; physicochemical properties characterized as log values are log 10; logBCF, log of the bioconcentration factor; logP, octanol-water partition coefficient; logVP, the vapor pressure; logS, log of the solubility in water.
In vitro data were used to develop a nonanimal method for setting starting doses for in vivo acute oral toxicity studies.
The query function of ICE, the integrator, facilitates combining the different data sets within ICE, allowing users to merge data points from the different data sets to simultaneously examine end points based on all available substance data or by using an input list of chemicals. Upon submission of the query, the user is provided with an overview of the query results along with several exportable data views, supporting both computational manipulation and manual interaction.
To assist the user with developing and interpreting queries, ICE provides a “Help” page that walks the user through building a search and understanding search output. Other user support resources include a “Home” page where updates and other news will be announced, an “About” page with background information, and a “Data Sets” page with information about the sources of the data in ICE. This page includes links that lead to more detailed information about each data set, including references and metadata for context about the data end points. Some information about the studies available in the source data sets is not available in ICE (for example, detailed clinical observations or some study protocol details), but it is available via the provided links and will also be accessible via CEBS (Lea et al. 2017) to facilitate integration with other data resources from the NIEHS.
User Stories
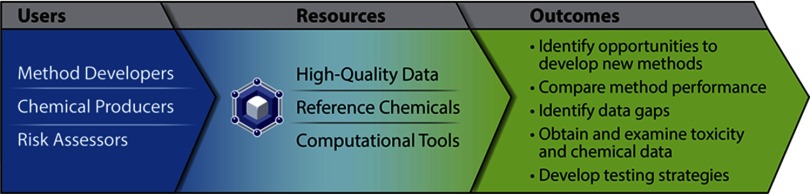
We developed ICE to address needs that have been expressed frequently by NICEATM stakeholders. As such, we tailored the design of ICE around user stories that developed from those needs. Here, we describe stories from three different user groups (method developer, chemical producer, risk assessor) and how ICE can help meet these users’ needs (Figure 1).
Figure 1.
Integrated Chemical Environment (ICE) users, resources, and outcomes. ICE was developed to support three main user roles: method developers, chemical producers, and risk assessors. The center panel lists the resources ICE provides to help these user groups complete some of the major tasks listed in the right panel.
Method Developer
Method developers are developing new in vitro or in silico methods to prioritize substances for further toxicity testing, or to test a substance for potential health impacts. The availability of data anchored to regulatory end points of interest from ICE facilitates exploration of the current state of methods, and such curated end point data can be compared with user-generated test data. These capabilities help method developers improve the performance of existing methods or target new method development to areas where the currently available test methods leave room for improvement. Furthermore, by having in vivo data organized with the in vitro and in silico data, it is easier to identify data gaps that may be targeted using in silico approaches.
In vitro test method developers often need reference chemicals, which are chemicals that cause specific, well-characterized biological effects and can therefore be used to assess the performance of a test method designed to measure that effect. These reference chemicals, along with the supporting data, are helpful in both in vitro method development and validation efforts. The reference chemical lists included in ICE provide a starting place to identify test materials and associated data.
For those developing in silico approaches, good quality data are important for training and testing predictive models. Method developers will find that the data in ICE are well annotated, cleaned, and formatted to support in silico modeling, allowing them to focus on method optimization versus data preparation. ICE currently provides many of the parameters needed as input for these models, and open-source workflows for in vitro to in vivo extrapolation and chemical property prediction will be added to ICE later in 2017.
Chemical Producer
For developers of new products for which toxicity testing will be required, typical testing needs include prioritizing substances to move forward through the development process and generating data needed to meet regulatory requirements. The ICE integrator allows the chemical producer to input a list of chemicals and obtain available testing data, which may help with evaluating the potential for adverse impacts during lead agent prioritization.
For chemical producers seeking to replace animals required for testing, the ICE integrator allows end point–oriented searches to compare data sets across all available substances. This capability allows the chemical producer to compare the results coming from different test methods relating to a specific end point. A query that returns in vitro and in silico test results paired with in vivo animal data may allow the chemical producer to better identify which nonanimal methods will best meet their information needs. This information may provide the chemical producer with the confidence in those nonanimal methods that is needed to change in-house testing approaches for ones that use fewer animals, save money, and better predict the end point of interest.
Risk Assessor
Similar to the chemical producer, the risk assessor may want to obtain available animal or human test data on a list of chemicals or review available data from nonanimal methods. Resources currently in ICE can help risk assessors with prioritizing chemicals for further toxicity testing and with identifying the most informative tests for that purpose in addition to providing data on the likely health impacts of a substance. The ICE data explorer view provides user-friendly data interaction and an easy-to-use snapshot of the types of data and end points. This information can be useful in prioritization or in identification of data gaps. The risk assessor may also be interested in comparing the performance of nonanimal test data with existing guideline animal test results across a wide range of chemicals, which is easily done using the ICE integrator.
ICE supports the exporting of data, which facilitates more detailed comparisons of end point variability. Exported data are preformatted and ready for analysis, enabling the risk assessor to move easily from query output into the subsequent analysis workflows. Transparency and reproducibility of data and process are important to the risk assessor; therefore, ICE provides reference details on the Data Sets page and clear referencing from the query output.
Next Steps
Computational tools will be included in ICE by the fall of 2017. These programs and workflows developed by NICEATM staff and external partners will enable researchers to conduct analyses locally using either their own data or data downloaded from ICE. Currently in the development queue are machine learning models for six physicochemical properties (Zang et al. 2017), predictive signatures based on HTS data such as embryonic vascular disruption leading to adverse prenatal outcomes (Knudsen and Kleinstreuer 2011), and in vivo–to–in vitro extrapolation (IVIVE) workflows (Chang et al. 2015; Wetmore et al. 2012). In addition to downloadable predictions and workflows, interactive tools such as those that facilitate biological pathway–informed IVIVE are needed to help link in vitro activity concentrations to relevant in vivo exposures and outcomes.
Future versions of ICE will also include tutorials aimed at helping users better understand the tools available in ICE and their applications in regulatory safety testing. These tutorials will address questions that frequently arise about appropriate use of computational tools and models on topics such as characterizing the domain of applicability and model uncertainty. Development of these resources is currently underway with a tentative 2018 launch date.
Acknowledgments
This project was funded in part with federal funds from the National Institute of Environmental Health Sciences, National Institutes of Health under contract no. HHSN273201500010C to Integrated Laboratory Systems (ILS) and its subcontractor, Sciome, in support of the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM). The views expressed in this manuscript do not necessarily represent the official positions of any federal agency.
References
- Chang X, Kleinstreuer N, Ceger P, Hsieh J-H, Allen D, Casey W. 2015. Application of reverse dosimetry to compare in vitro and in vivo estrogen receptor activity. Appl Vitro Toxicol 1:33–44, 10.1089/aivt.2014.0005. [DOI] [Google Scholar]
- ECHA (European Chemicals Agency). Registered Substances database. Available: https://echa.europa.eu/information-on-chemicals/registered-substances [accessed 23 January 2017].
- Filer DL, Kothiya P, Setzer RW, Judson RS, Martin MT. 2016. tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics, 10.1093/bioinformatics/btw680. [DOI] [PubMed] [Google Scholar]
- Huang R. 2016. A quantitative high-throughput screening data analysis pipeline for activity profiling. Methods Mol Biol 1473:111–122, PMID: 27518629, 10.1007/978-1-4939-6346-1_12. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix D, Houck K, Elloumi F, Martin M, et al. 2008. ACToR–Aggregated Computational Toxicology Resource. Toxicol Appl Pharmacol 233:7–13, PMID: 18671997, 10.1016/j.taap.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Judson RS, Martin MT, Egeghy P, Gangwal S, Reif DM, Kothiya P, et al. 2012. Aggregating data for computational toxicology applications: The U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) system. Int J Mol Sci 13:1805–1831, PMID: 22408426, 10.3390/ijms13021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, et al. 2012. Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem Res Toxicol 25:1287–1302, PMID: 22519603, 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. 2016. PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213, PMID: 26400175, 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen TB, Kleinstreuer NC. 2011. Disruption of embryonic vascular development in predictive toxicology. Birth Defects Res C Embryo Today 93:312–323, PMID: 22271680, 10.1002/bdrc.20223. [DOI] [PubMed] [Google Scholar]
- Lea IA, Gong H, Paleja A, Rashid A, Fostel J. 2017. CEBS: A comprehensive annotated database of toxicological data. Nucleic Acids Res 45:D964–D971, PMID: 27899660, 10.1093/nar/gkw1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NLM (National Library of Medicine). 2016. Fact Sheet. TOXNET®: Toxicology Data Network. Available: https://www.nlm.nih.gov/pubs/factsheets/toxnetfs.html [accessed 23 January 2017].
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, et al. 2016. ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem Res Toxicol 29:1225–1251, PMID: 27367298, 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121:756–765, PMID: 23603828, 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2016. ToxCast Chemicals: Data Management and Quality Considerations Overview. Available: https://www.epa.gov/chemical-research/toxcast-chemicals-data-management-and-quality-considerations-overview [accessed 18 January 2017].
- U.S. EPA (U.S. Environmental Protection Agency). Chemistry Dashboard. Available: comptox.epa.gov [accessed 23 January 2017].
- Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T, et al. 2014. PubChem BioAssay: 2014 update. Nucleic Acids Res 42:D1075–D1082, PMID: 24198245, 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, et al. 2012. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol Sci 125:157–174, PMID: 21948869, 10.1093/toxsci/kfr254. [DOI] [PubMed] [Google Scholar]
- Zang Q, Mansouri K, Williams AJ, Judson RS, Allen DG, Casey WM, et al. 2017. In silico prediction of physicochemical properties of environmental chemicals using molecular fingerprints and machine learning. J Chem Inf Model 57:36–49, PMID: 28006899, 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- ECHA (European Chemicals Agency). Registered Substances database. Available: https://echa.europa.eu/information-on-chemicals/registered-substances [accessed 23 January 2017].