Abstract
Despite recent advances in the understanding of clinical aspects of sex differences in Alzheimer’s disease (AD), the underlying mechanisms, for instance, how sex modifies AD risk and why the female brain is more susceptible to AD, are not clear. The purpose of this study is to elucidate sex disparities in brain aging profiles focusing on 2 major areas—energy and amyloid metabolism—that are most significantly affected in preclinical development of AD. Total RNA isolated from hippocampal tissues of both female and male 129/C57BL/6 mice at ages of 6, 9, 12, or 15 months were comparatively analyzed by custom-designed Taqman low-density arrays for quantitative real-time polymerase chain reaction detection of a total of 182 genes involved in a broad spectrum of biological processes modulating energy production and amyloid homeostasis. Gene expression profiles revealed substantial differences in the trajectory of aging changes between female and male brains. In female brains, 44.2% of genes were significantly changed from 6 months to 9 months and two-thirds showed downregulation. In contrast, in male brains, only 5.4% of genes were significantly altered at this age transition. Subsequent changes in female brains were at a much smaller magnitude, including 10.9% from 9 months to 12 months and 6.1% from 12 months to 15 months. In male brains, most changes occurred from 12 months to 15 months and the majority were upregulated. Furthermore, gene network analysis revealed that clusterin appeared to serve as a link between the overall decreased bioenergetic metabolism and increased amyloid dyshomeostasis associated with the earliest transition in female brains. Together, results from this study indicate that: (1) female and male brains follow profoundly dissimilar trajectories as they age; (2) female brains undergo age-related changes much earlier than male brains; (3) early changes in female brains signal the onset of a hypometabolic phenotype at risk for AD. These findings provide a mechanistic rationale for female susceptibility to AD and suggest a potential window of opportunity for AD prevention and risk reduction in women.
Keywords: Alzheimer’s disease, Sex differences, Metabolic aging, Energy metabolism, Amyloid metabolism, Prevention, Risk reduction
1. Introduction
As the leading cause of dementia and rated as the most feared human disease by the American public, Alzheimer’s disease (AD) currently affects approximately 35 million people worldwide, including 5.1 million Americans (Thies and Bleiler, 2013). These numbers are predicted to triple by 2050, with one new case of AD expected to develop every 33 seconds, or nearly a million new cases per year (Thies and Bleiler, 2013). There is no cure currently available, and no success has been found from more than 100 human trials conducted over the last decade in an attempt to find an effective treatment for mid- to late-stage AD (McBride, 2012; Schnabel, 2013). These unprecedented challenges underscore the significance and priority for increased research efforts aimed at defining the risk factors and the underlying mechanisms that would allow institution of AD prevention and early intervention, when a treatment continues to be sought (Mullard, 2012; Rice, 2014).
AD affects women and men differently on many levels (Carter et al., 2012; Regitz-Zagrosek and Seeland, 2012). Of the current AD cases, nearly two-thirds are women (Brookmeyer et al., 2011). After the age of 65, the lifetime risk of AD is 1 in 6 for women (16.7%), whereas it is 1 in 11 for men (9.1%) (Thies and Bleiler, 2013). In addition, sex has been demonstrated to play a major role in the pathogenesis, progression, and clinical manifestation of AD. For instance, depression is associated with a 2-fold increased risk for AD in women but not in men; in contrast, stroke is associated with a 3-fold increased risk for AD in men but not in women (Artero et al., 2008). Women with AD tend to exhibit a broader spectrum of dementia-related behavioral symptoms and experience greater cognitive deterioration than men in the progression of the disease (Barnes et al., 2005; Chapman et al., 2011; Hall et al., 2012; Irvine et al., 2012; Schmidt et al., 2008). Moreover, several studies presented at the recent Alzheimer’s Association International Conference provide new evidence supporting the long-held belief that women’s brains are more vulnerable than men’s brains to AD (Alzheimer’s Association, 2015; Hamilton, 2015).
Increasing evidence indicates that sex also modulates the impact of genetic factors in the etiology of AD. Cognitively normal individuals with a maternal family history of AD were found to express greater phenotypic changes in AD-vulnerable brain regions suggesting a higher risk for developing AD as compared with those with a paternal history or no family history (Berti et al., 2011; Honea et al., 2011; Mosconi et al., 2010). Furthermore, some genetic variants appear to interact with sex to modify the risk for AD. As an example, the ε4 allele of the apolipoprotein E gene (APOE4), the strongest genetic risk factor for late-onset AD, has been associated with a far more pronounced risk for AD in women than in men (Altmann et al., 2014; Bretsky et al., 1999; Farrer et al., 1997; Mortensen and Hogh, 2001; Payami et al., 1996; Ungar et al., 2014). In contrast, the ε2 allele of the APOE gene (APOE2), which is considered as a neuroprotective variant, has been shown to confer a greater protection against AD in men than in women (Altmann et al., 2014; Johnson et al., 1998; Ungar et al., 2014).
Despite recent advances in the understanding of clinical aspects of sex differences in AD, the underlying mechanisms, for instance, how sex modifies AD risk and why the female brain is more susceptible to AD, are not clear. In this study, using custom-designed Taqman low-density arrays (TLDAs) and Ingenuity pathway analysis (IPA) computing tools, we sought to determine sex disparities in brain aging profiles focusing on 2 major areas—energy and amyloid metabolism—that are most significantly affected in preclinical development of AD. The data revealed substantial differences in the overall trajectory of aging changes between female and male brains. Early changes indicative of the onset of a hypo-metabolic phenotype in female brains could serve as an important mechanistic rationale for female susceptibility to AD; and timely intervention at this transition could potentially halt the progression of metabolic deficits and ultimately reduce the risk of developing AD in women.
2. Materials and methods
2.1. Animals
The use of animals was approved by the Institutional Animal Care and Use Committee at the University of Southern California and followed NIH guidelines for the care and use of laboratory animals. Animas were bred and maintained under controlled conditions of temperature (22 °C), humidity, and light (14 hours light, 10 hours dark) with water and food available ad libitum. Both females and males, 4 age groups (5–6, 9–10, 12–13, and 15–16 months), 5 mice per sex × age group, a total of 40,129/C57BL/6 mice were used in this study. Hippocampal tissues collected from one hemisphere of mice brains were used for TLDA gene expression analysis; tissues collected from the other hemisphere were used for Western blot protein expression analysis. The gene network was generated using the IPA bioinformatics program. A detailed description of the methods for both TLDAs and IPA studies can be found in our recent report (Zhao et al., 2012).
2.2. TLDA gene expression profiling
TLDAs with 2 custom designs in 96-format (91 target genes and 5 candidate control genes were included on each design) were manufactured at Applied Biosystems (Foster City, CA). Mouse hippocampal RNA were isolated using the PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA). RNA quantity and quality were analyzed using the Experion RNA StdSens Analysis Kit on an Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA). RNA to cDNA synthesis was prepared using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems) on a MyCycler Thermal Cycler (Bio-Rad). Taqman quantitative real-time polymerase chain reactions (qRT-PCRs) were performed on 50-ng cDNA samples mixed with the TaqMan Universal PCR Master Mix 2X (Applied Biosystems), under the thermal cycling conditions: stage 1: AmpErase UNG activation at 50°C/2 minutes/100% ramp; stage 2: AmpliTaq gold DNA polymerase activation at 94.5 °C/10 minutes/100% ramp; stage 3: melt at 97 °C/30 seconds/50% ramp, followed by anneal/extend at 59.7 °C/1 minute/100% ramp, for 40 cycles. Fluorescence was detected on an ABI 7900HT Fast Real-Time PCR System equipped with the Sequence Detection System Software, version 2.3 (Applied Biosystems). Data were analyzed using the RQ Manager, version 1.2, and DataAssist, version 2.0 (Applied Bio-systems). Gene expression fold changes relative to the comparison group were calculated by the comparative Ct (ΔΔCt) method, with Ct denoting threshold cycle (Livak and Schmittgen, 2008). Selection of the endogenous control gene for normalization was based on the control stability measure (M), which indicates the expression stability of control genes on the basis of non-normalized expression levels. Four samples per sex × age group were included in the analysis. For each sample, ΔCt was calculated as the difference in Ct of the target gene and the endogenous control gene. For each group, mean 2−ΔCt was calculated as the geometric mean of 2−ΔCt of the 4 samples in the group. Fold change was then calculated as mean 2−ΔCt (target group)/mean 2−ΔCt (comparison group). Fold change values greater than 0 indicate a positive expression or upregulation relative to the comparison group. Fold change values less than 0 indicate a negative expression or downregulation relative to the comparison group. The 2−ΔCt values for each target gene were statistically compared between the target and comparison group using Student t test. The statistical significance was indicated by * p < 0.05, ** p < 0.01 and *** p <0.001; p-values were unadjusted by multiple testing corrections, thus it is possible that some of the results may not reach significance when a correction is applied. The volcano plot displays fold changes (X-axis) and p-values (Y-axis) enabling identification of genes with both large and small expression changes that are statistically significant. The heat map displays results of unsupervised hierarchical clustering. Distances between assays/samples are calculated based on the ΔCt values using Pearson’s correlation. The ΔCt value of the middle/median expression level is set such that red indicates an increase with a ΔCt value below the middle level, and green indicates a decrease with a ΔCt value above the middle level. In the assay centric map, for each assay, the middle expression level is set as the median of all of the ΔCt values from all samples for that assay. Data points for a given assay can only be compared relative with other data points for that assay.
2.3. IPA gene network profiling
IPA (Ingenuity Systems, www.ingenuity.com), which leverages the Ingenuity Knowledge Base, the largest database housing biological and chemical relationships extracted from the scientific literature, provides a bioinformatics computing tool to interpret the experimental gene expression data in the context of biological processes, pathways, and molecular networks. The IPA network analysis identified biological connectivity among molecules in the gene expression data set that were significantly upregulated or downregulated (these molecules are called “network eligible molecules” or “focus molecules” that serve as “seeds” for generating networks) and their interactions with other molecules present in the Ingenuity Knowledge Base. Network eligible molecules were combined into networks that maximized their specific connectivity. Additional molecules from the Ingenuity Knowledge Base (these molecules are called “interacting molecules”) were used to specifically connect 2 or more smaller networks to merge them into a larger one. A network was composed of direct (requiring direct physical contact between 2 molecules) and indirect (mediated by intermediate factor[s]) interactions among focus molecules and interacting molecules. Generated networks were ranked by the network score according to their degree of relevance to the network eligible molecules from the data set. The network score was calculated with Fisher’s exact test, taking into account the number of network eligible molecules in the network and the size of the network, as well as the total number of network eligible molecules analyzed and the total number of molecules in the Ingenuity Knowledge Base that were included in the network.
2.4. Western blot protein expression analyses
Total protein samples were extracted from mouse hippocampus as previously described (Zhao et al., 2009); 20–40 μg of protein samples were loaded per lane and separated by electrophoresis on a 10%–12% SDS-PAGE. Proteins were then transferred to 0.2-μm polyvinyl difluoride membranes and probed with primary antibodies against insulin-like growth factor 1 (IGF1; 1:1000, Abcam, Cambridge, MA), clusterin (CLU; 1:1000, Santa Cruz Biotechnology, Dallas, TX), estrogen receptor α (1:1000, Santa Cruz Biotechnology), estrogen receptor β (ERβ; 1:500, Santa Cruz Biotechnology), G protein-coupled receptor 30 (1:750, Santa Cruz Technology), progesterone receptors (PRs; 1:1000, Santa Cruz Technology), and progesterone receptor membrane component 1 (1:1000, Cell Signaling Technology, Danvers, MA), at 4°C overnight, and then with horseradish peroxidase-conjugated secondary antibodies (1:3000–10,000, Thermo Scientific Piece, Rockford, IL). β-tubulin (1: 5000, Thermo Scientific Piece), or β-actin (1:3000, Thermo Scientific Piece) was used as the loading control. Bands were visualized by chemiluminescence with an ECL detection kit (Bio-Rad). Relative intensities of the immunoreactive bands were captured by a C-Digit Chemiluminescence Western Blot Scanner and quantitated by Image Studio, version 4.0 (LI-COR, Lincoln, NE). Statistically significant differences between groups (n = 5) were determined by 1-way analyses of variance followed by Student-Newman-Keuls pairwise multiple comparison post-hoc tests. The statistical significance was indicated by * p < 0.05, ** p < 0.01 and *** p < 0.001.
3. Results
Total RNA isolated from hippocampal tissues of both female and male 129/C57BL/6 mice at ages of 6, 9, 12, or 15 months were comparatively analyzed by custom-designed TLDAs for qRT-PCR detection of a focused set of 182 genes involved in a broad spectrum of biological processes modulating energy production and amyloid homeostasis. Among 5 candidate control genes (18S, Actb, Gapdh, Hprt1, and Ipo8), Gapdh exhibited the most stable expression (M) across all samples and thus was used as the control gene for normalization in the following data analysis. Of the 182 target genes assayed, 17 genes that showed low expression with Ct values >35 were excluded from the data set (Supplementary Tables 1 and 2); 165 genes with Ct values <35 are included in Figs. 1, 2A, B, and Supplementary Table 3.
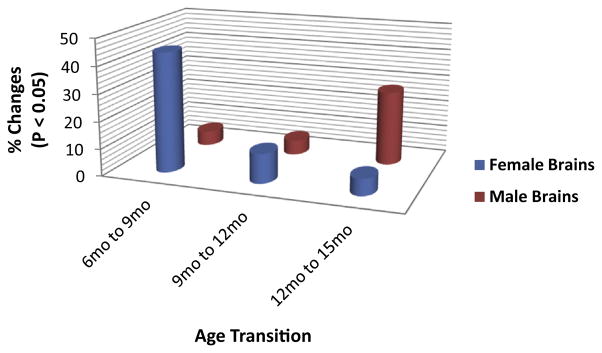
Fig. 1.
Female and male mice hippocampal gene expression changes with age. In female brains, 44.2% (73 genes; 23 increase, 50 decrease), 10.9% (18 genes; 3 increase, 15 decrease), and 6.1% (10 genes; 9 increased, 1 decreased) of genes showed significant changes between the ages of 6 months and 9 months, 9 months and 12 months, and 12 months and 15 months, respectively. In contrast, in male brains, 5.4% (9 genes; 2 increase, 7 decrease), 5.4% (9 genes; 4 increase, 5 decrease), and 27.3% (45 genes; 35 increase, 10 decrease) of genes were significantly changed from the ages of 6 months–9 months, 9 months–12 months, and 12 months–15 months, respectively.
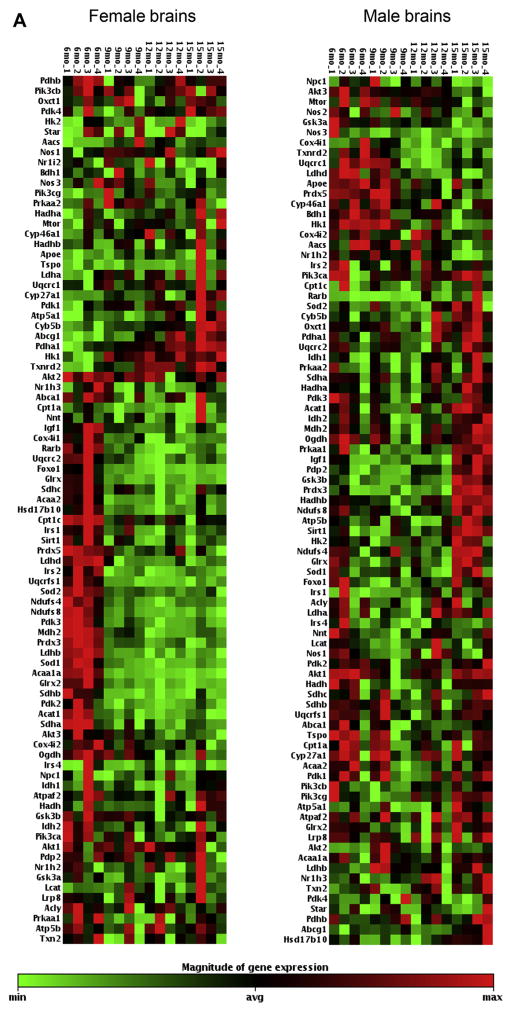
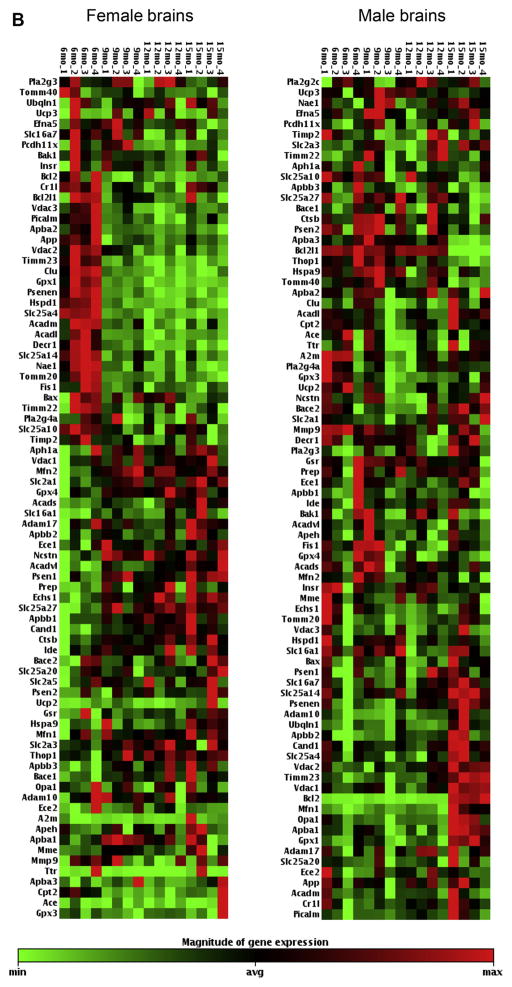
Fig. 2.
(A). Female and male mice hippocampal gene expression aging profile comparison. The heat maps display the relative expression levels of 85 genes with Ct < 35 analyzed by TLDA #1 (list of genes are included in Supplementary Table 1); 4 mice per age group; red indicates high expression, green indicates low expression. (B) Female and male mice hippocampal gene expression aging profile comparison. The heat maps display the relative expression levels of 80 genes with Ct < 35 analyzed by TLDA #2 (list of genes are included in Supplementary Table 2); 4 mice per age group; red indicates high expression, green indicates low expression. TLDA, Taqman low-density array; Ct, threshold cycle. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Gene expression profiles revealed substantial disparities in the overall trajectory of aging changes between female and male brains; and female brains exhibited a much earlier onset of changes compared with male brains (Figs. 1, 2A, B, and Supplementary Table 3). Specifically, in female brains, most changes (73 out of 165 genes; 44.2%; 23 increase and 53 decrease) occurred between the ages of 6 months and 9 months (Figs. 1, 2A, B and 3). Among these, 37 genes involved in energy metabolism exhibited a fold change >1.4 fold, and the majority were downregulation (32 decrease, 5 increase), including: (1) 4 genes involved in insulin/IGF signaling: Foxo1 (FC = −2.36, p = 0.012), IGF1 (FC = −1.45, p = 0.031), Irs2 (FC = −1.83, p = 0.0025), and Prkaa1 (FC = −1.72, p = 0.026); (2) 2 genes involved in glycolysis: Ldhb (FC = −2.01, p = 0.000001), Ldhd (FC = −2.01, p = 0.0024); (3) 4 genes involved in pyruvate dehydrogenase complex and tricarboxylic acid cycle: Pdk2 (FC = −1.70, p = 0.020), Pdk3 (FC = −1.71, p = 0.0052), Sdha (FC = −1.44, p = 0.027), and Sdhb (FC = −1.55, p = 0.0018); (4) 4 genes involved in electron transport chain and oxidative phosphorylation: Ndufs4 (FC = −1.88, p = 0.0011), Ndufs8 (FC = −1.75, p = 0.00073), Uqcrc2 (FC = −1.42, p = 0.0077), and Uqcrfs1 (FC = −1.74, p = 0.00087); (5) 7 genes involved in reduction and oxidation (Redox): Glrx (FC = −2.04, p = 0.0044), Glrx2 (FC = −2.07, p = 0.00053), Gpx1 (FC = −1.96, p = 0.0018), Gpx4 (FC = 1.47, p = 0.037), Prdx3 (FC = −1.47, p = 0.000023), Sod1 (FC = 2.09, p = 0.00077), and Sod2 (FC = −1.43, p = 0.000077); (6) 9 genes involved in transport: Hspd1 (FC = −2.30, p = 0.00013), Slc25a14 (FC = −1.60, p = 0.0017), Slc25a27 (FC = 1.68, p = 0.021), Slc25a4 (FC = −2.21, p = 0.0000003), Slc2a1 (FC = 1.78, p = 0.020), Slc2a3 (FC = 1.44, p = 0.022), Timm23 (FC = −1.99, p = 0.0015), Timm20 (FC = −1.83, p = 0.0037), Vdac2 (FC = −1.48, p = 0.030); (7) 1 gene involved in apoptosis: Bcl2 (FC = −1.58, p = 0.039); (8) 9 genes involved in lipid/ketone metabolism and cholesterol trafficking: Acaa1a (FC = −2.57, p = 0.000011), Acaa2 (FC = −1.70, p = 0.0031), Acadl (FC = −2.19, p = 0.0097), Acadm (FC = −1.49, p = 0.027), Acadvl (FC = 1.60, p = 0.045), Acat1 (FC = −1.58, p = 0.017), Cpt2 (FC = −1.59, p = 0.00090), Decr1 (FC = −1.76, p = 0.0045), and Rarb (FC = −1.69, p = 0.0022). No genes involved in mitochondrial biogenesis and dynamics exhibited a fold change >1.4 fold at the transition from 6 months to 9 months in female brains.
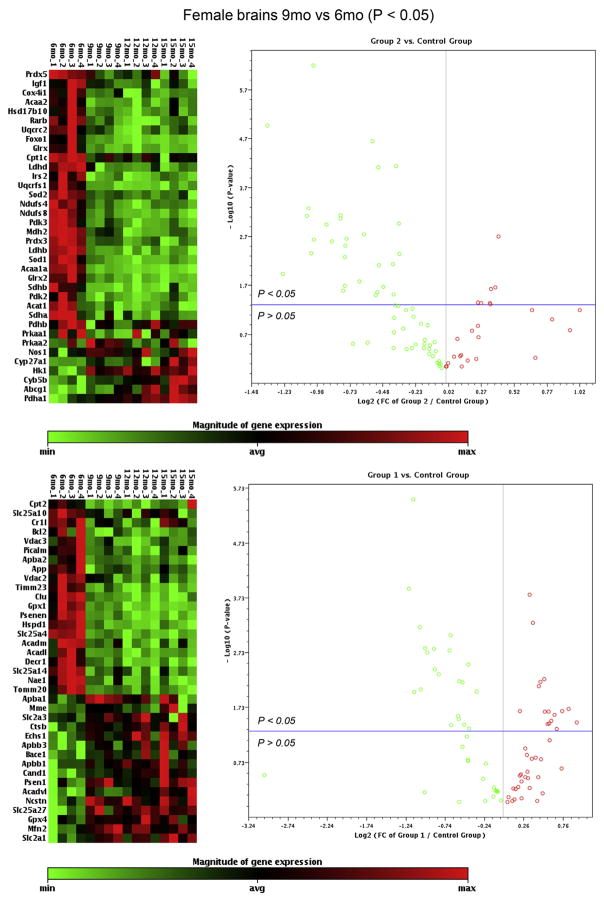
Fig. 3.
Hippocampal gene expression changes between the ages of 6 months and 9 months in female mice. The heat maps display the relative expression levels of genes that were significantly changed from 6 months to 9 months in female mice, including 36 genes from TLAD #1 (upper panel) and 37 genes from TLDA #2 (lower panel); 4 mice per age group; red indicates high expression, green indicates low expression. The volcano plots display fold changes (X-axis) with p-values (Y-axis) from 6 months to 9 months in female mice on all genes analyzed by TLDA #1 (upper panel) and TLDA #2 (lower panel); each dot represents a gene; red indicates upregulated genes, green indicates downregulated genes; dots that fall above the horizontal blue line indicate significantly changed genes (p < 0.05), dots that fall below the blue line indicate nonsignificantly changed genes (p > 0.05). TLDA, Taqman low-density array. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In regards to genes involved in amyloid metabolism, 10 genes exhibited a fold change >1.4 fold at 9 months relative to 6 months in female brains, including 6 that increased in expression: Apbb1 (FC = 1.53, p = 0.032), Apbb3 (FC = 1.91, p = 0.035), Bace1 (FC = 1.57, p = 0.025), Ctsb (FC = 1.50, p = 0.036), Ncstn (FC = 1.44, p = 0.0057), and Psen1 (FC = 1.49, p = 0.022), and 4 that decreased in expression: Apba2 (FC = −1.41, p = 0.038), App (FC = −1.48, p = 0.0061), Nae1 (FC = −2.07 p = 0.0087), and Psenen (FC = −2.09, p = 0.00063). In addition, 2 genes that were recently identified as genetic risk factors for late-onset AD exhibited a decreased expression >1.4 fold, including CLU (FC = −1.84, p = 0.0016) and Picalm (FC = −1.43, p = 0.0065).
Compared with the changes in female brains that occurred at the transition from 6 months to 9 months, there were not only a much smaller number of genes but also most of the them were altered at a much smaller magnitude at transitions from 9 months to 12 months and from 12 months to 15 months. A total of 18 genes (10.9% of the total number of genes analyzed) were significantly changed from 9 months to 12 months, and only 3 of them exhibited a fold change >1.4 fold, including: Foxo1 (FC = −1.40, p = 0.026), Gpx3 (FC = 1.44, p = 0.030), and Pcdh11x (FC = −2.04, p = 0.024). Similarly, a total of 10 genes (6.1% of the total number of genes analyzed) were significantly changed from 12 months to 15 months, and only one of them exhibited a fold change >1.4 fold, Cyp27a1 (FC = 1.85, p = 0.029).
Male brains exhibited 2 major differences compared with female brains. First, male brains underwent substantially less changes between the ages of 6 months and 9 months; 9 genes were significantly changed; 2 increased and 7 decreased, representing 5.4% of the total number of genes analyzed. In addition, the magnitude of changes in male brains was much smaller than those that occurred in female brains; all 9 genes exhibited a fold change <1.3 fold. Similarly, a total of 9 genes exhibited a significant change from 9 months to 12 months, and they all had a magnitude of change <1.3 fold. The second major difference between male and female brains was detected between the ages of 12 months and 15 months. During this age period, female brains exhibited the fewest changes in both the number of genes and the overall magnitude, whereas male brains underwent the most changes involving a total of 45 genes, representing 27.3% of the total number of genes examined. Among them, the majority presented an increase in expression (35 increase, 10 decrease), and 8 genes exhibited a fold change >1.4 fold, including: 5 increases in expression: IGF1 (FC = 1.63, p = 0.00046), Prdx3 (FC = 1.41, p = 0.000024), Mfn1 (FC = 1.54, p = 0.0055), Bcl2 (FC = 6.29, p < 0.0000001), and Bace2 (FC = 1.94, p = 0.038), and 3 decreases in expression: Bcl2l1 (FC = −5.86, p < 0.0000001), Nr1h3 (FC = −1.91, p = 0.016), and Apba3 (FC = −3.52, p = 0.0021) (Fig. 1 and Supplementary Table 3).
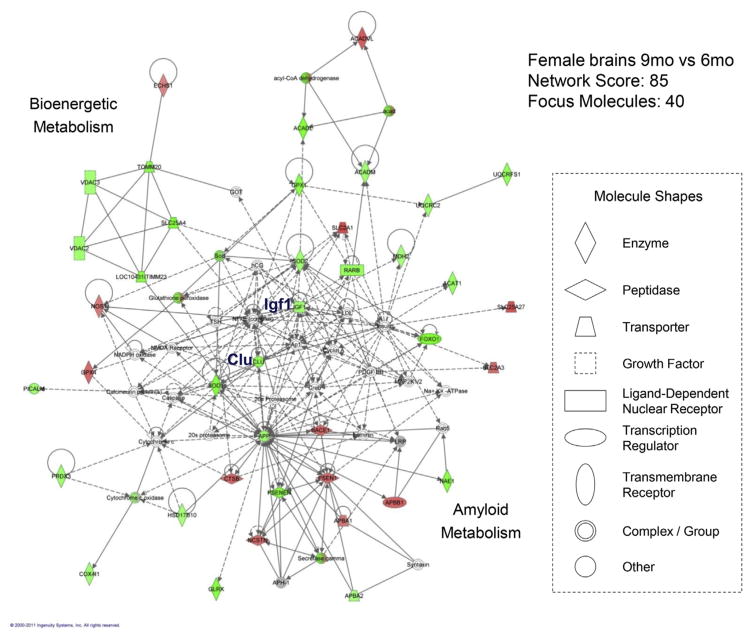
The graph in Fig. 4 displays the IPA network (with a network score of 85), with the most relevance to gene expression changes occurring at the transition from 6 months to 9 months in female brains. Within this large network that includes 40 focus molecules (referring to significantly changed genes included in the data set), 2 subnetworks were revealed. One subnetwork was largely populated with bioenergetic genes and many of them were decreased, in which IGF1 appeared to serve as a central node. The other subnetwork was populated primarily with genes involved in amyloid production and many of them were increased. CLU appeared to serve as a linker between the 2 subnetworks.
Fig. 4.
The IPA-derived molecular network with the most relevance to gene expression changes between the ages of 6 months and 9 months in female mice. Molecules (or nodes) in the network are displayed as various shapes that indicate the functional class. Focus molecules (colored molecules) refer to genes from the data set. Red indicates significantly upregulated genes (p < 0.05), green indicates significantly downregulated genes (p < 0.05), gray indicates nonsignificantly changed genes from the data set (p > 0.05), and white indicates molecules added from the Ingenuity Knowledge Base; color intensity indicates the degree of up or downregulation. Lines (or edges) connecting molecules indicate biological relationships; solid lines indicate direct interaction, dashed lines indicate indirect interactions, and circular lines indicate self-referential relationships that arise from the ability of a molecule to act on itself; the types of arrows indicate specific biological relationships and the directionality of the interaction. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
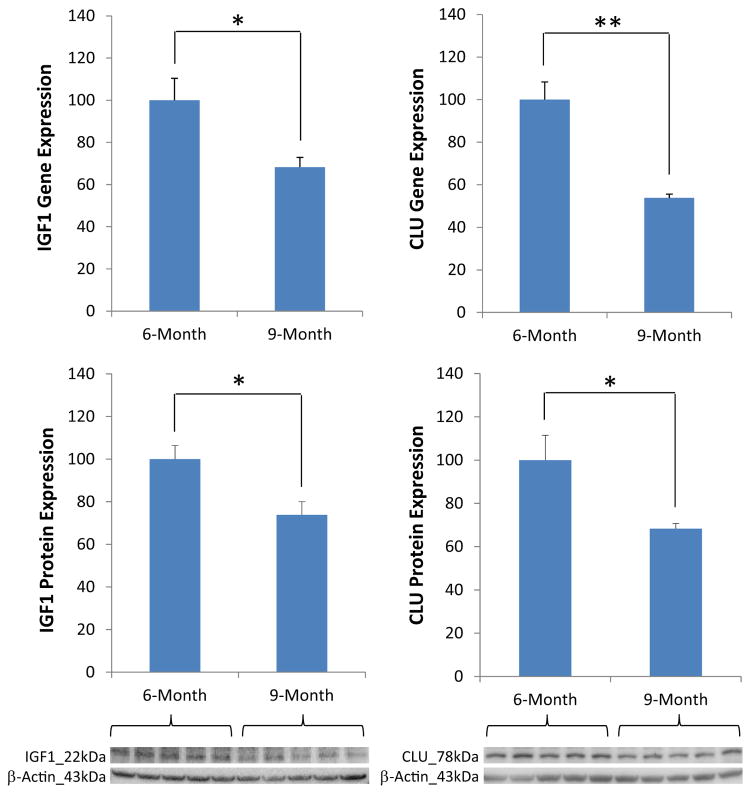
Changes in the expression of IGF1 and CLU at the transition from 6 months to 9 months in female brains were further confirmed by Western blot analyses. Consistent with the gene expression data, the protein levels of both IGF1 (FC = 0.73 9 months vs. 6 months; p = 0.029) and CLU (FC = 0.68 9 months vs. 6 months; p = 0.042) were significantly decreased at 9 months compared with 6 months (Fig. 5). Among the 6 major ERs and PRs analyzed, only ERβ showed a significant change between 6 months and 9 months (FC = 0.70 9 months vs. 6 months; p = 0.047) (Table 1 and Supplementary Fig. 1).
Fig. 5.
Hippocampal insulin-like growth factor 1 (IGF1) and clusterin (CLU) exhibited significant changes at both gene and protein levels at the transition from 6 months to 9 months in female mice. 4–5 mice per age group; * p < 0.05 and ** p < 0.01.
Table 1.
Estrogen and progesterone receptor protein expression changes from 6 months to 9 months in female brainsa
| Receptor | Fold change (9 mo vs. 6 mo) | p-value |
|---|---|---|
| ERα | 0.71 ± 0.14 | 0.073 |
| ERβ | 0.70 ± 0.10 | 0.047* |
| GPR30 | 0.77 ± 0.18 | 0.164 |
| PRA | 0.95 ± 0.11 | 0.397 |
| PRB | 0.85 ± 0.13 | 0.182 |
| PGRMC1 | 1.08 ± 0.17 | 0.345 |
Key: ERα, estrogen receptor α; ERβ, estrogen receptor β; GPR30, G protein-coupled receptor 30; PRA, progesterone receptor A; PRB, progesterone receptor B; PGRMC1, progesterone receptor membrane component 1.
4–5 mice per age group;
p < 0.05. Western blots and graphs are included in Supplementary Fig. 1.
4. Discussion
AD disproportionally affects women more than men. Women have a greater lifetime risk of developing AD and constitute two-thirds of the current AD population (Brookmeyer et al., 2011; Thies and Bleiler, 2013). The prevailing view has been that women’s susceptibility to AD is due to the fact that they live longer and hence have a higher age-associated risk for AD, which is true to a minimal extent. Statistics show that the living age difference between men and women is not as large as one might think. Currently, the average life expectancy worldwide for all people is 71 years, 69 years for men, and 73 years for women, a difference of 4 years (www.statista.com, Statista, 2015), whereas on average people with AD can live 8 to 10 years from the diagnosis (http://www.alzheimers.org.uk, Alzheimer’s Soceity, 2015). Results from a meta-analysis of 7 sex-specific studies concluded that women were 1.5 times more likely to develop AD than age-matched men (Gao et al.,1998), which is consistent with the data derived from the Cache County Study that showed a clear higher incidence of AD in women (Zandi et al., 2002). These findings suggest that age does not solely account for the sex disparity in the prevalence of AD, and there must be other factors that play an even greater role in predisposing females to a higher level of risk for AD. Understanding the biological bases underlying such sex differences in AD could potentially lead to new directions for preventing or reducing the risk of developing the disease, especially in the high-risk population of older women.
In this study, using custom-designed TLDA gene array profiling and IPA bioinformatics computing tools, we analyzed the expression aging profile and interactive network of a focused set of genes in the hippocampus of both female and male mice. The target genes were selected based on their involvement in energy and amyloid metabolism, 2 major areas that are most significantly affected in preclinical development of AD (Brinton, 2009; Mosconi et al., 2006; Swerdlow et al., 2013; Yao et al., 2009). The results revealed substantial disparities in the trajectory of changes with age, most significantly, the onset of changes between female and male brains. In female brains, 44.2% of genes showed significant changes between the ages of 6 months and 9 months, and the changes were overall indicative of decreased bioenergetic function and increased amyloid dyshomeostasis. Protein analysis revealed that among 6 major ERs and PRs, only ERβ exhibited a significant change at this early transition in female brains. In stark contrast, in male brains, only 5.4% of genes were significantly changed in the same period. Subsequent changes in female brains were relatively small; however, in male brains, most changes occurred between the ages of 12 months and 15 months and most of the changes were upregulated, suggesting a possible adaptive response to the aging process. Taken together, these data indicate that female and male brains follow profoundly disparate paths as they age, and female brains appear to undergo a much earlier aging process than male brains, in which ERβ may play a role.
Among the genes involved in energy metabolism, significant changes associated with the earliest transition in female brains spanned across all functional domains examined, including insulin/IGF signaling, glycolysis, pyruvate dehydrogenase and tricarboxylic acid cycle, electron transport chain and oxidative phosphorylation, redox, lipid and ketone metabolism, mitochondrial biogenesis, mitochondrial dynamics, transport, and apoptosis, indicating a system-level bioenergetic decline. Bioinformatics analysis revealed that IGF1 appeared to serve as a central node in the overall decreased bioenergetic network associated with the transition from the age of 6 months–9 months in female brains, which is consistent with the well-characterized properties of IGF1 as a master regulator of metabolism in both central and peripheral systems (McRory and Sherwood, 1997; Wit and Walenkamp, 2013). In the brain, IGF1 plays a significant role in diverse neural activities from early development to adult neurogenesis, to neuronal survival, and to cognition (Benarroch, 2012; Torres-Aleman, 2010). In particular, IGF1 has been well recognized for its role in the regulation of cellular bioenergetics and energy production. IGF1 signaling regulates multiple targets in glucose metabolism, including facilitation of the translocation of glucose transporters from the cytosol to the plasma membrane, thereby facilitating cellular glucose uptake and subsequent ATP generation (Bassil et al., 2014; Yin et al., 2013). Moreover, IGF1 polymorphisms have been associated with increased risk for AD (Vargas et al., 2011; Wang et al., 2012), further highlighting the importance of IGF1 signaling in the maintenance of neurological health. Thus, it can be expected that sustaining IGF1 signaling and the downstream metabolic cascades through physical or pharmacological interventions holds promise to maintain the brain under a bioenergetically robust state and, as a result, offers a therapeutic benefit against the onset of metabolic deficits that could lead to the development of AD.
In addition to IGF1 signaling, another major change of particular significance was CLU, which exhibited a 1.84-fold decrease in expression at the 6–9-month transition in female brains. Bioinformatics analysis revealed that CLU may play a role in the regulation of both bioenergetic and amyloid metabolism in the brain. CLU, also known as apolipoprotein J, is a highly conserved glycoprotein that has been demonstrated as a prosurvival regulator in a variety of pathological events, including cancer progression and neurodegeneration (Bertram and Tanzi, 2010; Nuutinen et al., 2009; Rizzi and Bettuzzi, 2010; Sala et al., 2009). Recently, 2 independent genome-wide association studies and many follow-up studies have demonstrated a strong association between CLU genetic variants and increased risk for late-onset AD (Harold et al., 2009; Lambert et al., 2009). Moreover, recent studies have shown that CLU may play a role in the regulation of glucose metabolism (Daimon et al., 2011; Kim et al., 2011) and serve as a cellular biosensor of oxidative stress (Trougakos, 2013). CLU has also been shown to regulate amyloid metabolism by limiting amyloid peptide misfolding and facilitating their transport across the blood-brain barrier (Charnay et al., 2012; Yu and Tan, 2012). These earlier studies, along with our current findings, warrant further in-depth investigations of the roles of CLU in both normal neurological health and in the etiology of AD as well as its potential as a therapeutic target for AD prevention and early intervention.
The major question that remains to be determined is what could have caused the earliest transition in female brains. Could it have resulted from a reproductive status change, in particular, a reproductive transition similar to what occurs in humans, that is, the transition from premenopause to perimenopause? In general, reproductive cycle irregularity in lab rodents, including both rats and mice, begins after 6 months (the age of “retired breeders”) and is significantly increased after 8 months, although some animals may be still actively cycling until 15 months of age (Gosden et al., 1983; Nelson et al., 1981). Regrettably, the animals used in this study were not monitored for their cycling profiles, so a conclusive correlation could not be drawn between the changes in the brain and the reproductive phenotype. Nevertheless, based on our recent analysis in which we found that approximately 50% of lab rodents at 9–10 months of age are irregular cyclers, it is reasonable to postulate that the 9–10-month female mice group used in our analysis were likely to be a heterogeneous population composed of both regular and irregular cyclers and they could have been at different estrous stages within the cycle. Despite the possibility that these mice differed in their reproductive profiles when tissues were collected, their brains exhibited greatly consistent expression patterns as shown in Fig. 2 and Fig. 3, suggesting that the changes observed between the ages of 6 months and 9 months in female mice brains cannot be simply related to the reproductive status or levels of classical sex hormones, such as 17β-estradiol and progesterone. With respect to the possible role of sex hormone receptors, it remains to be determined how significantly a decrease in ERβ could contribute to the changes at this earliest transition in female brains. Together, these uncertainties warrant further in-depth investigations of potential contributors beyond the traditional concepts of reproductive aging and classical sex hormones that could play a major role in modulating sex-specific brain aging processes.
In summary, the purpose of this study was to determine how female and male brains age differently, specifically in 2 major areas that are most significantly affected in preclinical development of AD. The study presents several strengths and novelties. Technically, the state-of-the-art qRT-PCR–based TLDA gene profiling technology provided us with not only a throughput but also a robust approach to investigate our hypotheses-driven questions at a system level. Scientifically, different from several prior studies that explored a similar subject, including the study conducted by Xu et al., in which the researchers focused on late aging in mice (6, 16, and 24 months) (Xu et al., 2007), our study targeted an early aging window in mice (6, 9, 12, and 15 months). Our findings provide strong evidence indicating that female and male brains follow profoundly dissimilar trajectories as they age. The earliest transition indicative of the onset of a hypometabolic phenotype in female brains could serve as an important mechanistic rationale for female susceptibility to AD; and timely intervention at this transition could potentially halt the progression of metabolic deficits and ultimately reduce the risk of developing AD in women.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute on Aging (P01AG026572–Analytic Core), the Alzheimer’s Association (IIRG-10–172459), and the University of Kansas start-up funds to LZ.
Appendix A. Supplementary data
Supplementary data associated with this article can be found at the online version at http://dx.doi.org/10.1016/j.neurobiolaging.2016.02.011.
Footnotes
Disclosure statement
All authors declare that there is no conflict of interests.
References
- Altmann A, Tian L, Henderson VW, Greicius MD. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol. 2014;75:563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Association. [Accessed August 10, 2015];Women with Mild Cognitive Impairment Decline Twice as Fast as Men with the Condition. 2015 Available at: www.alz.org.
- Alzheimer’s Society. [Accessed April 2, 2015];The progression of Alzheimer’s disease and other dementias. 2015 Available at: http://www.alzheimers.org.uk.
- Artero S, Ancelin ML, Portet F, Dupuy A, Berr C, Dartigues JF, Tzourio C, Rouaud O, Poncet M, Pasquier F, Auriacombe S, Touchon J, Ritchie K. Risk profiles for mild cognitive impairment and progression to dementia are gender specific. J Neurol Neurosurg Psychiatry. 2008;79:979–984. doi: 10.1136/jnnp.2007.136903. [DOI] [PubMed] [Google Scholar]
- Barnes LL, Wilson RS, Bienias JL, Schneider JA, Evans DA, Bennett DA. Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch Gen Psychiatry. 2005;62:685–691. doi: 10.1001/archpsyc.62.6.685. [DOI] [PubMed] [Google Scholar]
- Bassil F, Fernagut PO, Bezard E, Meissner WG. Insulin, IGF-1 and GLP-1 signaling in neurodegenerative disorders: targets for disease modification? Prog Neurobiol. 2014;118:1–18. doi: 10.1016/j.pneurobio.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. Insulin-like growth factors in the brain and their potential clinical implications. Neurology. 2012;79:2148–2153. doi: 10.1212/WNL.0b013e3182752eef. [DOI] [PubMed] [Google Scholar]
- Berti V, Mosconi L, Glodzik L, Li Y, Murray J, De Santi S, Pupi A, Tsui W, De Leon MJ. Structural brain changes in normal individuals with a maternal history of Alzheimer’s. Neurobiol Aging. 2011;32:2325.e17–2325.e26. doi: 10.1016/j.neurobiolaging.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Alzheimer disease: new light on an old CLU. Nat Rev Neurol. 2010;6:11–13. doi: 10.1038/nrneurol.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretsky PM, Buckwalter JG, Seeman TE, Miller CA, Poirier J, Schellenberg GD, Finch CE, Henderson VW. Evidence for an interaction between apolipoprotein E genotype, gender, and Alzheimer disease. Alzheimer Dis Assoc Disord. 1999;13:216–221. doi: 10.1097/00002093-199910000-00007. [DOI] [PubMed] [Google Scholar]
- Brinton RD. Estrogen-induced plasticity from cells to circuits: predictions for cognitive function. Trends Pharmacol Sci. 2009;30:212–222. doi: 10.1016/j.tips.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Evans DA, Hebert L, Langa KM, Heeringa SG, Plassman BL, Kukull WA. National estimates of the prevalence of Alzheimer’s disease in the United States. Alzheimers Dement. 2011;7:61–73. doi: 10.1016/j.jalz.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CL, Resnick EM, Mallampalli M, Kalbarczyk A. Sex and gender differences in Alzheimer’s disease: recommendations for future research. J Womens Health (Larchmt) 2012;21:1018–1023. doi: 10.1089/jwh.2012.3789. [DOI] [PubMed] [Google Scholar]
- Chapman RM, Mapstone M, Gardner MN, Sandoval TC, McCrary JW, Guillily MD, Reilly LA, Degrush E. Women have farther to fall: gender differences between Normal elderly and Alzheimer’s disease in verbal memory engender better detection of Alzheimer’s disease in women. J Int Neuropsychol Soc. 2011;17:654–662. doi: 10.1017/S1355617711000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnay Y, Imhof A, Vallet PG, Kovari E, Bouras C, Giannakopoulos P. Clusterin in neurological disorders: molecular perspectives and clinical relevance. Brain Res Bull. 2012;88:434–443. doi: 10.1016/j.brainresbull.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Daimon M, Oizumi T, Karasawa S, Kaino W, Takase K, Tada K, Jimbu Y, Wada K, Kameda W, Susa S, Muramatsu M, Kubota I, Kawata S, Kato T. Association of the clusterin gene polymorphisms with type 2 diabetes mellitus. Metabolism. 2011;60:815–822. doi: 10.1016/j.metabol.2010.07.033. [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Gao S, Hendrie HC, Hall KS, Hui S. The relationship between age, sex, and the incidence of dementia and Alzheimer disease: a meta-analysis. Arch Gen Psychiatry. 1998;55:809–815. doi: 10.1001/archpsyc.55.9.809. [DOI] [PubMed] [Google Scholar]
- Gosden RG, Laing SC, Flurkey K, Finch CE. Graafian follicle growth and replacement in anovulatory ovaries of ageing C57BL/6J mice. J Reprod Fertil. 1983;69:453–462. doi: 10.1530/jrf.0.0690453. [DOI] [PubMed] [Google Scholar]
- Hall JR, Vo HT, Johnson LA, Wiechmann A, O’Bryant SE. Bonston naming test: gender differences in older adults with and without Alzheimer’s dementia. Psychology. 2012;3:485–488. [Google Scholar]
- Hamilton J. [Accessed August 10, 2015];Women’s Brains Appear More Vulnerable to Alzheimer’s than Men’s. 2015 Available at: www.npr.org.
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea RA, Swerdlow RH, Vidoni ED, Burns JM. Progressive regional atrophy in normal adults with a maternal history of Alzheimer disease. Neurology. 2011;76:822–829. doi: 10.1212/WNL.0b013e31820e7b74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K, Laws KR, Gale TM, Kondel TK. Greater cognitive deterioration in women than men with Alzheimer’s disease: a meta analysis. J Clin Exp Neuropsychol. 2012;34:989–998. doi: 10.1080/13803395.2012.712676. [DOI] [PubMed] [Google Scholar]
- Johnson JK, McCleary R, Oshita MH, Cotman CW. Initiation and propagation stages of beta-amyloid are associated with distinctive apolipoprotein E, age, and gender profiles. Brain Res. 1998;798:18–24. doi: 10.1016/s0006-8993(98)00363-1. [DOI] [PubMed] [Google Scholar]
- Kim G, Kim GH, Oh GS, Yoon J, Kim HW, Kim MS, Kim SW. SREBP-1c regulates glucose-stimulated hepatic clusterin expression. Biochem Biophys Res Commun. 2011;408:720–725. doi: 10.1016/j.bbrc.2011.04.111. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P European Alzheimer’s Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analyzing real-time PCR data using the comparative Ct method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- McBride R. [Accessed September 14, 2012];Pharma Counts Just 3 Alzheimer’s Drug Wins in 13 Years (101 Losses) Available at: http://www.fiercebiotechcom/story/pharma-counts-just-3-alzheimers-drug-wins-13-years-101-losses/2012-09-14.
- McRory JE, Sherwood NM. Ancient divergence of insulin and insulin-like growth factor. DNA Cell Biol. 1997;16:939–949. doi: 10.1089/dna.1997.16.939. [DOI] [PubMed] [Google Scholar]
- Mortensen EL, Hogh P. A gender difference in the association between APOE genotype and age-related cognitive decline. Neurology. 2001;57:89–95. doi: 10.1212/wnl.57.1.89. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Sorbi S, de Leon MJ, Li Y, Nacmias B, Myoung PS, Tsui W, Ginestroni A, Bessi V, Fayyazz M, Caffarra P, Pupi A. Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer’s disease. J Nucl Med. 2006;47:1778–1786. [PubMed] [Google Scholar]
- Mosconi L, Berti V, Swerdlow RH, Pupi A, Duara R, de Leon M. Maternal transmission of Alzheimer’s disease: prodromal metabolic phenotype and the search for genes. Hum Genomics. 2010;4:170–193. doi: 10.1186/1479-7364-4-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Sting of Alzheimer’s failures offset by upcoming prevention trials. Nat Rev Drug Discov. 2012;11:657–660. doi: 10.1038/nrd3842. [DOI] [PubMed] [Google Scholar]
- Nelson JF, Felicio LS, Osterburg HH, Finch CE. Altered profiles of estradiol and progesterone associated with prolonged estrous cycles and persistent vaginal cornification in aging C57BL/6J mice. Biol Reprod. 1981;24:784–794. doi: 10.1095/biolreprod24.4.784. [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer’s disease. Brain Res Rev. 2009;61:89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Payami H, Zareparsi S, Montee KR, Sexton GJ, Kaye JA, Bird TD, Yu CE, Wijsman EM, Heston LL, Litt M, Schellenberg GD. Gender difference in apolipoprotein E-associated risk for familial Alzheimer disease: a possible clue to the higher incidence of Alzheimer disease in women. Am J Hum Genet. 1996;58:803–811. [PMC free article] [PubMed] [Google Scholar]
- Regitz-Zagrosek V, Seeland U. Sex and gender differences in clinical medicine. Handb Exp Pharmacol. 2012:3–22. doi: 10.1007/978-3-642-30726-3_1. [DOI] [PubMed]
- Rice S. [Accessed January 25, 2014];As Drug Trials Fail, Alzheimer’s Researchers Look toward Prevention. Available at: http://www.modernhealthcarecom/article/20140125/MAGAZINE/301259969#. [PubMed]
- Rizzi F, Bettuzzi S. The clusterin paradigm in prostate and breast carcinogenesis. Endocr Relat Cancer. 2010;17:R1–R17. doi: 10.1677/ERC-09-0140. [DOI] [PubMed] [Google Scholar]
- Sala A, Bettuzzi S, Pucci S, Chayka O, Dews M, Thomas-Tikhonenko A. Regulation of CLU gene expression by oncogenes and epigenetic factors implications for tumorigenesis. Adv Cancer Res. 2009;105:115–132. doi: 10.1016/S0065-230X(09)05007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Kienbacher E, Benke T, Dal-Bianco P, Delazer M, Ladurner G, Jellinger K, Marksteiner J, Ransmayr G, Schmidt H, Stogmann E, Friedrich J, Wehringer C. Sex differences in Alzheimer’s disease. Neuropsychiatr. 2008;22:1–15. [PubMed] [Google Scholar]
- Schnabel J. [Accessed July 8, 2013];Why Do All the Large Alzheimer’s Drug Trials Fail? Available at: http://www.danaorg/News/Why_Do_All_the_Large_Alzheimer_s_Drug_Trials_Fail_/
- [Accessed July 5, 2015];Average life expectancy at birth in 2014 by continent (in years) 2015 Statista. Available at: www.statista.com.
- Swerdlow RH, Burns JM, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta. 2013;1842:1219–1231. doi: 10.1016/j.bbadis.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies W, Bleiler L. 2013 Alzheimer’s disease facts and figures. Alzheimers Dement. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Torres-Aleman I. Toward a comprehensive neurobiology of IGF-I. Dev Neurobiol. 2010;70:384–396. doi: 10.1002/dneu.20778. [DOI] [PubMed] [Google Scholar]
- Trougakos IP. The molecular chaperone apolipoprotein J/Clusterin as a sensor of oxidative stress: implications in therapeutic approaches–a mini-review. Gerontology. 2013;59:514–523. doi: 10.1159/000351207. [DOI] [PubMed] [Google Scholar]
- Ungar L, Altmann A, Greicius MD. Apolipoprotein E, gender, and Alzheimer’s disease: an overlooked, but potent and promising interaction. Brain Imag Behav. 2014;8:262–273. doi: 10.1007/s11682-013-9272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas T, Martinez-Garcia A, Antequera D, Vilella E, Clarimon J, Mateo I, Sanchez-Juan P, Rodriguez-Rodriguez E, Frank A, Rosich-Estrago M, Lleo A, Molina-Porcel L, Blesa R, Gomez-Isla T, Combarros O, Bermejo-Pareja F, Valdivieso F, Bullido MJ, Carro E. IGF-I gene variability is associated with an increased risk for AD. Neurobiol Aging. 2011;32:556.e3–556.e11. doi: 10.1016/j.neurobiolaging.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Wang W, Yu JT, Tan L, Liu QY, Wang HF, Ma XY. Insulin-like growth factor 1 (IGF1) polymorphism is associated with Alzheimer’s disease in Han Chinese. Neurosci Lett. 2012;531:20–23. doi: 10.1016/j.neulet.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Wit JM, Walenkamp MJ. Role of insulin-like growth factors in growth, development and feeding. World Rev Nutr Diet. 2013;106:60–65. doi: 10.1159/000342546. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhan M, Duan W, Prabhu V, Brenneman R, Wood W, Firman J, Li H, Zhang P, Ibe C, Zonderman AB, Longo DL, Poosala S, Becker KG, Mattson MP. Gene expression atlas of the mouse central nervous system: impact and interactions of age, energy intake and gender. Genome Biol. 2007;8:R234. doi: 10.1186/gb-2007-8-11-r234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Jiang T, Cadenas E. Metabolic triad in brain aging: mitochondria, insulin/IGF-1 signalling and JNK signalling. Biochem Soc Trans. 2013;41:101–105. doi: 10.1042/BST20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JT, Tan L. The role of clusterin in Alzheimer’s disease: pathways, pathogenesis, and therapy. Mol Neurobiol. 2012;45:314–326. doi: 10.1007/s12035-012-8237-1. [DOI] [PubMed] [Google Scholar]
- Zandi PP, Carlson MC, Plassman BL, Welsh-Bohmer KA, Mayer LS, Steffens DC, Breitner JC. Hormone replacement therapy and incidence of Alzheimer disease in older women: the Cache County Study. JAMA. 2002;288:2123–2129. doi: 10.1001/jama.288.17.2123. [DOI] [PubMed] [Google Scholar]
- Zhao L, Mao Z, Brinton RD. A select combination of clinically relevant phytoestrogens enhances estrogen receptor beta-binding selectivity and neuroprotective activities in vitro and in vivo. Endocrinology. 2009;150:770–783. doi: 10.1210/en.2008-0715. [DOI] [PubMed] [Google Scholar]
- Zhao L, Morgan TE, Mao Z, Lin S, Cadenas E, Finch CE, Pike CJ, Mack WJ, Brinton RD. Continuous versus cyclic progesterone exposure differentially regulates hippocampal gene expression and functional profiles. PLoS One. 2012;7:e31267. doi: 10.1371/journal.pone.0031267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.