Abstract
In metazoans, histone mRNAs are not polyadenylated but end in a conserved stem-loop. Stem-loop binding protein (SLBP) binds to the stem-loop and is required for all steps in histone mRNA metabolism. The genes for the five histone proteins are linked. A histone locus body (HLB) forms at each histone gene locus. It contains factors essential for transcription and processing of histone mRNAs, and couples transcription and processing. The active form of U7 snRNP contains the HLB component FLASH (FLICE-associated huge protein), the histone cleavage complex (HCC), and a subset of polyadenylation factors including the endonuclease CPSF73. Histone mRNAs are rapidly degraded when DNA replication is inhibited by a 3′ to 5′ pathway that requires extensive uridylation of mRNA decay intermediates.
Replication-Dependent Histone mRNAs: A Novel Set of Cell Cycle-Regulated mRNAs
Histone mRNAs are tightly regulated and are present in high levels only in S-phase to provide the histone proteins necessary for packaging the newly replicated DNA. Histones are among the most evolutionarily conserved proteins in eukaryotes. They form the fundamental unit of chromatin, the nucleosome, which packages the newly replicated chromosomal DNA. There are two major classes of histone proteins, the canonical replication-dependent histones and the histone variants. The replication-dependent histones are synthesized during S-phase, and comprise the bulk of the histones in the chromatin in multicellular organisms. The metazoan replication-dependent histone mRNAs are not polyadenylated but end instead in a conserved stem-loop, while in plants, and most single-cell eukaryotes, the replication-dependent histone mRNAs are polyadenylated [1]. After extensive deep sequencing analysis, the metazoan replication-dependent histone mRNAs remain the only known eukaryotic cellular mRNAs that are not polyadenylated.
There are also histone variants, for example H3.3, H2a.Z, macroH2a, and H10, which have specific functions and are typically constitutively expressed from polyadenylated mRNAs. An exception is histone H2a.X, which in vertebrates is synthesized in large amounts during S-phase from an mRNA ending in a stem-loop, but the same gene expresses a longer polyadenylated mRNA outside S-phase [2,3].
SLBP Binds to the 3′ Ends of Histone mRNAs
Stem-loop binding protein (SLBP; see Glossary) provides the general functions of the poly(A) tail, including participating in translation and mRNA degradation (Figure 1, Key Figure). SLBP is a novel RNA-binding protein, and replication-dependent histone mRNAs are its only known target [4]. The structure of the SLBP–stem-loop RNA complex has been determined [5] (Box 1) and this complex at the 3′ end of the mRNA is involved in all steps of histone mRNA metabolism (i.e., processing, nuclear export, translation, and degradation of histone mRNA). There are no homologs of SLBP in species whose histone mRNAs are polyadenylated [6].
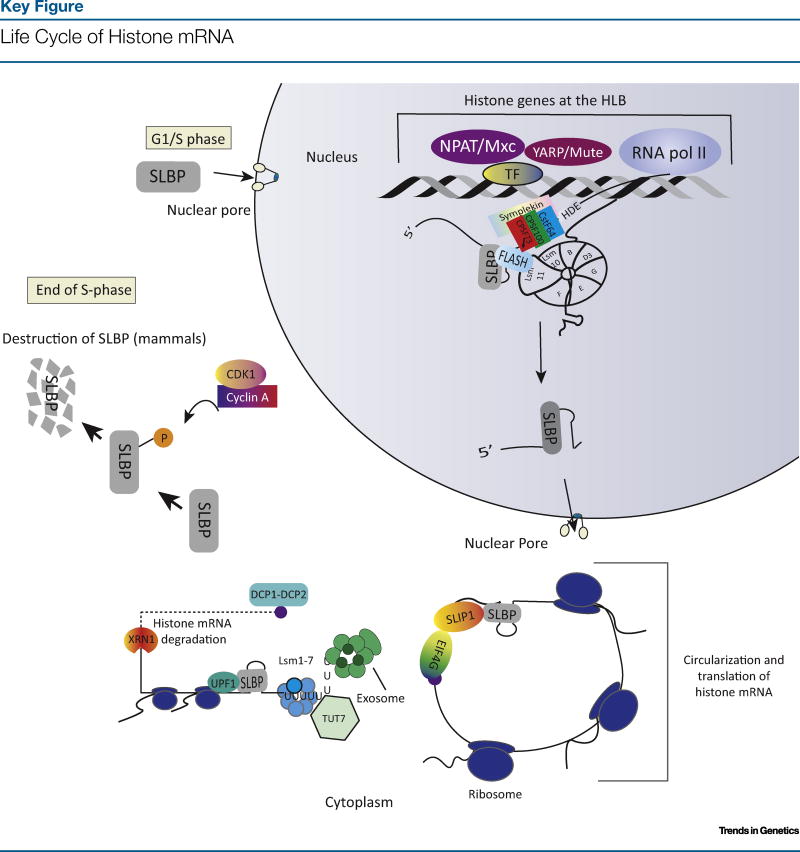
Figure 1.
A schematic of the life cycle of histone mRNAs and SLBP in mammalian cells. Note that SLBP participates in every part of the histone mRNA life cycle, and is also cell cycle-regulated. Abbreviations: SLBP, stem-loop binding protein; P, phosphorylation; Pol II, polymerase II
Box 1. Roles of the Stem-Loop Binding Protein.
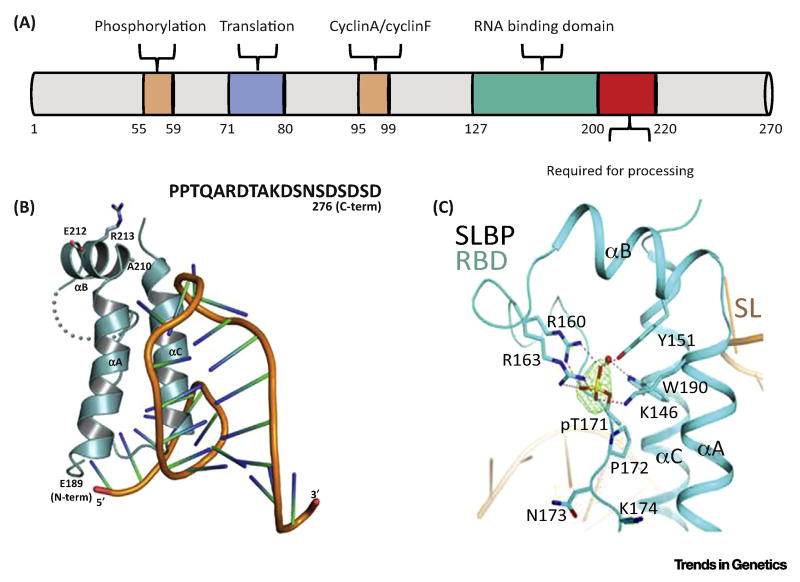
SLBP was isolated 20 years ago using the yeast three-hybrid system [64,65]. SLBP has a conserved RNA binding domain (RBD) of ~75 aa. Figure I A shows a schematic of human SLBP, indicating the RBD, the region required for processing, the region required for translation, and the cyclin-binding sites and phosphorylation sites that are required for degradation.
Crystal structures of the Drosophila RBD bound to the stem-loop (SL) RNA show that SLBP binds to the 5′ side of the stem-loop and to the 5 nt before the stem [27] (Figure I B). The RBD is composed of three α-helices (A, B, and C). Helices A and C contact the RNA, and helix B projects away from the RNA in a position where it could interact with processing factors. Conserved amino acids, A210, E212 and R213, in helix B are required for processing. The C-terminal region required for processing and part of the loop between helices B and C is disordered in the structure. The structure is stabilized by phosphate on T171 which contacts the three helices in human SLBP (Figure I C). The phosphorylated form of SLBP is the form found in vivo [66].
In vivo 3′hExo and SLBP are bound to the stem-loop (Figure 4). Strikingly, binding of either SLBP or 3′hExo to the stem-loop results in a similar deformation of the loop. The 3′hExo can trim 2–3 nt off the 3′ side of the stem-loop in vitro, and further degradation is blocked by SLBP [67]. SLBP is also required for translation of histone mRNA, by interacting with SLIP1 that in turn binds to translation initiation factors [68]. In vertebrates SLBP is rapidly degraded at the end of S-phase, as a result of cyclin A/Cdk2 phosphorylating two threonines in the SFTTP motif. The ubiquitin ligase responsible for SLBP degradation at the end of S-phase is not clear, and both cyclin F [69] and DCAF11 [70] have been implicated in its degradation.
Figure I.
Structure of SLBP.
The SLBP RNA-binding domain can readily be identified by informatic approaches. Likely SLBP homologs have been identified in several single-cell eukaryotes including Volvox, Dictyostelium, and Chlamydomonas. These species also have stem-loops close to the 3′ end of their histone mRNAs, suggesting that histone mRNAs ending in stem-loops may have been present in some single-cell eukaryotes, and thus this property has been lost in evolution [6].
Organization and Coordinate Expression of the Histone Genes
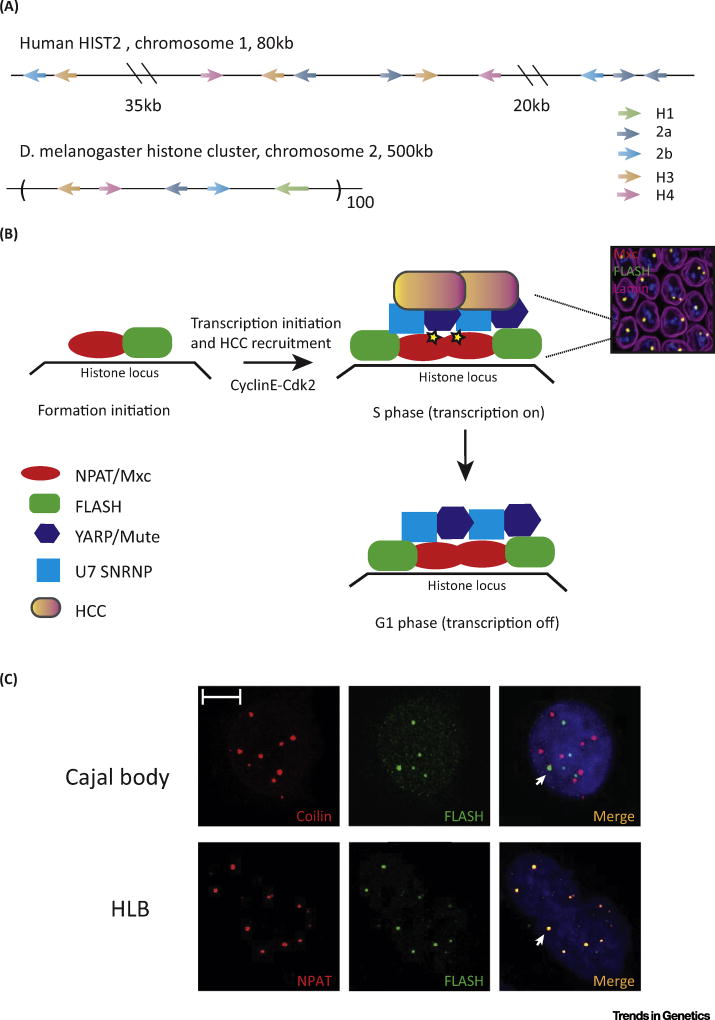
The high demand for histone protein in S-phase is met by the coordinated expression of multiple histone genes. In metazoans, the genes for the five replication-dependent histone proteins are tightly linked. These clusters may occur as tandem repeats containing a copy of each canonical histone gene (e.g., Drosophila and embryonic sea urchin histone genes) or in jumbled clusters where the genes have no repeating organization (e.g., the HIST1 cluster, ~60 genes) and the HIST2 cluster (~ 10 genes) in mammals (Figure 2A). D. melanogaster, which has a genome 1/30 the size of that of mammals, contains ~ 100 copies of a tandemly arrayed 5 kb repeat with each repeat containing one copy of the genes encoding the canonical histones (H2a, H2b, H3, and H4) and the linker histone H1. By contrast, none of the genes for histone variants H3.3, H2a.Z, and H10 are physically linked but are present as isolated single-copy genes.
Figure 2. Histone genes and the HLB.
(A) Organization of the histone genes in the human HIST2 cluster and in Drosophila melanogaster. (B) Model of HLB assembly and changes during the cell cycle in Drosophila. The image shows the merged image of HLBs in every nucleus of the Drosophila syncytial embryo stained with Mxc and FLASH. (C) Staining of HeLa cells with FLASH and Coilin (top) and NPAT and FLASH (bottom). Note that the HLBs are distinct from the Cajal body in these cells [76]. Adapted from Duronio and Marzluff [87]. Abbreviations: HCC, histone cleavage complex; HLB, histone locus body.
The replication-dependent histone genes are present in a specialized nuclear domain, the HLB (Box 2). The HLB creates an optimal environment for efficient transcription and histone pre-mRNA processing (3′-end formation) by concentrating factors necessary for histone mRNA biogenesis [7]. Formation of the HLB requires nuclear protein at the ataxia-telangiectasia locus (NPAT; Mxc in Drosophila) which is essential for the expression of all five classes of histone genes [8,9]. FLICE-associated huge protein (FLASH) and U7 small nuclear ribonucleoprotein (U7 snRNP), both of which are required for processing, are also found in the HLB.
Box 2. The Histone Locus Body.
The HLB is present at the replication-dependent histone genes and has been characterized in Drosophila and mammals. In mammalian cells it was observed that U7 snRNA is localized near the histone genes, and that it colocalizes with coilin, suggesting that it is present in a subset of Cajal bodies [71]. A novel protein, NPAT, was discovered as a cyclin E substrate that localizes in a nuclear body near the histone genes in mammalian cells, and like U7 snRNA is not present in other Cajal bodies [10,11]. In 2006, Joe Gall and colleagues defined the HLB and showed that in Drosophila the HLB and the Cajal body are two distinct bodies. Using the small Cajal body (sca) RNA U85, and U2 and U7 snRNAs, as probes they found in nurse and follicle cells that U85 and U2 localize into a discrete nuclear focus, the Cajal body, that is distinct from the focus containing U7 snRNA, that they termed the HLB [72].
Transgenic DNA containing only the Drosophila histone H3–H4 promoter is sufficient to nucleate a HLB [73]. NPAT/Mxc is the defining member of the HLB and is phosphorylated by cyclin E/Cdk2 to activate histone gene expression. Knockdown of NPAT is cell-lethal [74] and results in loss of HLBs and reduction in the levels of all histone RNAs, suggesting that NPAT is a global regulator of histone expression [9]. Multi sex combs (mxc) was identified as the Drosophila ortholog of NPAT using a genome-wide RNAi screen for HLB formation [75].
Other components of the HLB are U7 snRNP, FLASH, and Mute (YARP in mammals), all of which are present in the HLB throughout the cell cycle. FLASH is directly involved in histone mRNA 3′-end formation. Mute (muscle wasted) was identified in a screen for genes required for myogenesis in Drosophila [76]. Mutations in Mute displayed progressive muscle degeneration late in embryogenesis and misregulation of histone gene expression, with increased histone mRNA levels [13].
NPAT, Mute, and FLASH are large proteins (>1500 aa in mammals), with few identifiable domains. NPAT self-associates through its N terminus, and this interaction is essential for HLB formation and viability [8]. Both FLASH and Mute interact with the C-terminal region of NPAT/Mxc [38]. NPAT does not bind to DNA and likely acts as a coactivator. It is complexed with factors required for histone gene transcription, including HinF-P for H4 genes [77] and Oct-1 for H2b genes [78].
NPAT was discovered as a cyclin E substrate which is localized at the histone loci in mammals and is essential for histone gene expression [10,11]. Activation of histone gene expression requires phosphorylation of NPAT by cyclin E/CDK2. The Drosophila ortholog of NPAT was identified as the previously described homeotic gene, multi sex combs (mxc) [12]. Like NPAT, Mxc is also phosphorylated by cyclin E in S-phase [12] and is essential for histone gene expression [8]. A fourth component of the HLB, muscle wasted (Mute), appears to act as a negative regulator of histone gene expression in Drosophila [13,14]. The HLB, containing these four core factors, is present throughout the cell cycle and is dissembled at mitosis.
Biosynthesis of Histone mRNAs
Replication-dependent histone mRNAs do not contain introns and require only one RNA processing step, endonucleolytic cleavage after the stem-loop, to form the 3′ end of histone mRNA. The 5′ caps on histone mRNAs contain 6-methyl (me) adenosine in a novel cap structure, 7MeGpppmeAO-meNO-me [15]. The significance of the 6-meA on the histone mRNA cap is not clear but, with the current excitement about multiple roles for 6-meA in RNA metabolism, this unusual cap may play a role in histone mRNA metabolism. Cleavage to form the 3′ end is defined by two sites in the RNA; the stem-loop bound by SLBP 5′ of the cleavage site, and the histone downstream element (HDE) which basepairs with U7 snRNP 3′ of the cleavage site (Box 3). Cleavage occurs very rapidly after transcription, and in both mammals and Drosophila transcription terminates shortly after the processing signal [16,17].
Box 3. Histone mRNA 3′-End Formation.
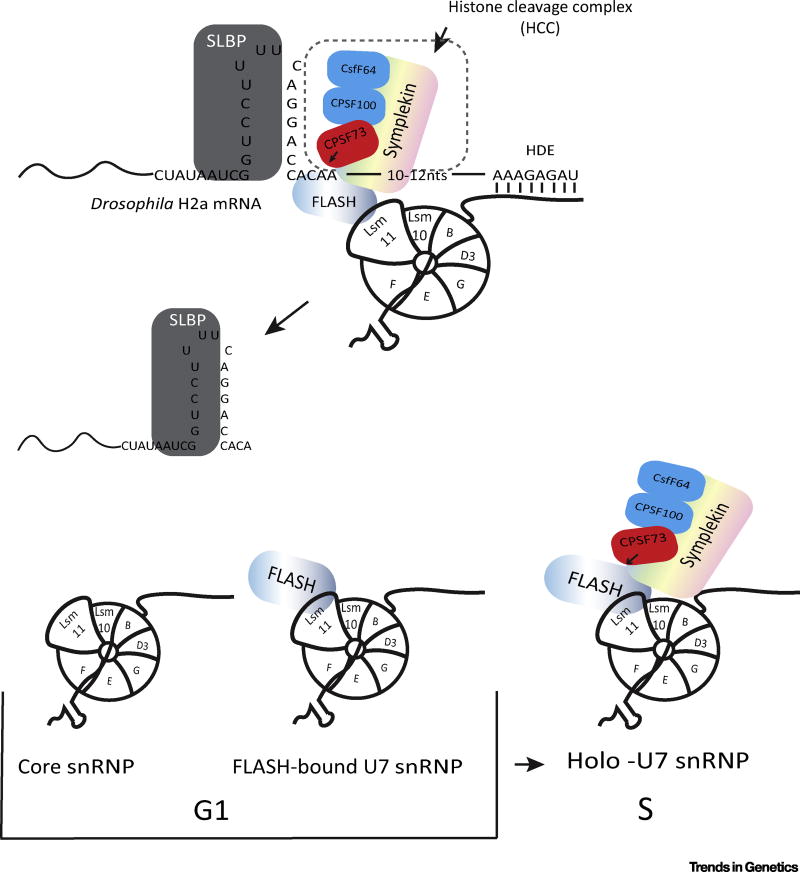
Formation of mature histone mRNA requires endonucleolytic cleavage either 4 nt (invertebrates) or 5 nt (vertebrates) after the stem-loop. The cis elements required are the stem-loop (a 26 nt sequence starting 5 nt before the stem and ending 5 nt after the stem-loop) that binds to SLBP, and a purine-rich region with the core consensus AAAGAG located about 10 nt after the cleavage site, and which basepairs with the 5′ end of U7 snRNA, a component of U7 snRNP. Cleavage is catalyzed by CPSF73 [79], the same protein that cleaves other mRNAs in cleavage/polyadenylation [80]. U7 snRNA is similar to the spliceosomal snRNAs, with a 2,2,7 tri-Me-G cap and a binding site for the Sm ring. The U7 snRNP contains two proteins, Lsm10 and Lsm11, which replace SmD1 and SmD2 in the Sm ring.
The N-terminal region of FLASH binds to the N-terminus of Lsm11, a protein in the heptameric ring of U7 snRNP, and together they create an interface for recruitment of the HCC [81], which contains a subset of polyadenylation factors. In addition to this crucial role, FLASH is also required for SLBP to stabilize the binding to the histone pre-mRNA [25
U7 snRNA is a small (<70 nt) RNA containing a 2,2,7 trimethyl G cap. Like the spliceosomal snRNPs, U7 snRNP can exist as a core snRNP and a larger holo-snRNP which contains factors essential for processing. The core U7 snRNP consists of U7 snRNA bound to five Sm proteins, B, D3, E, F, and G, that are found in spliceosomal snRNAs, and to Lsm10 and Lsm11 which replace SmD1 and SmD2 [18,19]. Lsm11 is much larger than the other Sm proteins, 360 amino acids (aa) in mammals and 256 aa in Drosophila, and the N terminus of Lsm11 plays a crucial role in histone pre-mRNA processing [20].
The FLASH/Lsm11 complex assembles the histone cleavage complex (HCC), a complex of polyadenylation factors that catalyze cleavage, on the U7 snRNP. Only the 100 N-terminal amino acids of the >1600 aa of FLASH are required for processing in vitro [21]. Strikingly, when the U7 snRNP is purified from nuclear extracts using an antisense oligonucleotide that binds to the 5′ end of U7 snRNA, much of the U7 snRNP is bound to the HCC, suggesting that this holo-U7 snRNP is the ‘active’ form of U7 snRNP. The core of the HCC consists of symplekin, CstF64, CPSF100, and CPSF73, the endonuclease that cleaves the pre-mRNA. CstF64 binds to symplekin using the same site in CstF64 that binds to CstF77 [22,23], explaining why only CstF64 and not the other CstF subunits are present in the HCC. Disruption of the binding of symplekin and CstF64 reduces histone pre-mRNA processing in vivo [23,24].
SLBP plays a direct role in processing by stabilizing the binding of U7 snRNP to the histone pre-mRNA [25]. SLBP has a 73 aa RNA-binding domain (RBD) that binds to the stem-loop, and 20 additional amino acids C-terminal of the RBD that are required for processing [26]. The RBD consists of three α-helices (A, B, and C) and a large loop which interacts with all three helices [5,27]. Helices A and C interact directly with the RNA, and helix B extends away from the RNA (Box 1). Specific residues in helix B are necessary to stabilize the binding of U7 snRNP to the pre-mRNA, as are the C-terminal 20 aa [25]. Formation of a stable complex of U7 snRNP on a noncleavable histone pre-mRNA in vitro also requires FLASH, but not the HCC, suggesting that SLBP may interact directly with FLASH and/or the FLASH/Lsm11 complex [25]. Thus, FLASH plays two independent roles in processing: the formation of a stable complex of U7 snRNP on the histone pre-mRNA, and recruitment of the HCC to the U7 snRNP.
Genetic Studies of Histone Gene Expression
Histone synthesis is essential for proliferation, making it difficult to study genetically in most organisms (e.g., in C. elegans, SLBP mutants die at the two-cell stage [28], and mouse embryos that are unable to transcribe the histone H4 genes die very early in embryogenesis [29]). Drosophila provides an amenable system for studying the genetics of histone gene expression. The large maternal deposition of histone mRNAs and proteins into the egg is sufficient for development into an embryo with several thousand cells. In addition, Drosophila histone genes differ from other metazoan histone genes in that there are cryptic polyadenylation sites 3′ of the normal processing site in each histone gene. As a result, mutations that block histone gene expression develop until cycle 15 using maternal histone protein and mRNA, and mutations that block histone pre-mRNA processing result in the production of polyadenylated histone mRNAs when zygotic transcription is activated. Mutations in Drosophila SLBP and U7 snRNP result in the production of polyadenylated histone mRNAs, and development proceeds normally through the larval stages [30,31].
The ease in detecting the Drosophila HLB owing to the presence of a single 500 kb cluster makes it possible to study the genetics of HLB formation and function. Mutations in HLB factors mute and mxc in Drosophila affect development and perturb histone gene expression. Hypomorphic alleles of mxc resulting from deletions of different amounts of sequence from the C terminus synthesize less histone mRNA [8]. By contrast, mutations in mute result in increased accumulation of histone mRNA, suggesting that Mute functions to turn down histone gene expression [13]. It is attractive to hypothesize that the changes in the levels of replication-dependent histone mRNA contribute to the observed phenotypes, possibly by altering the relative amounts of histone variants incorporated into chromatin. Note that the replacement histones H2a.V (the Drosophila homolog of H2a.Z) and H3.3 are expressed constitutively from polyadenylated mRNAs. Thus, when there is reduced synthesis of the replication-dependent histones, increased levels of H3.3 and H2A.V may be assembled into chromatin, resulting in inappropriate function. The absence of Mute may result in increased expression of H3 and H2a, reducing the amounts of H3.3 and H2a.V. The importance of the proper balance of H2a.V and H2a.1 histones has been demonstrated by recent studies using mutants in SLBP to reduce the deposition of H2a.1 histone protein in the egg, and different copy numbers of the H2a.V gene to alter the H2a.V concentrations. Either too much or too little H2a.V relative to H2a.1 resulted in embryonic lethality [32].
Cell-Cycle Regulation of Histone mRNAs
In all eukaryotes histone mRNAs are cell cycle-regulated with high concentrations of histone mRNA being present only in S-phase. This tight control is achieved by regulating histone gene transcription, processing, and the half-life of histone mRNAs. We discuss multiple control points in the life of histone mRNAs below.
Transcriptional Regulation of Histone mRNAs During the Cell Cycle
Despite the fact that histone mRNAs are coordinately expressed, genes for different histone proteins display a surprising lack of common transcription regulatory elements. The histone gene promoters contain TATAA boxes and transcription starts 10–40 nt 5′ of the initiation codon. There are binding sites in the histone H2b gene promoters for Oct-1 and in the mammalian H4 genes for Hinf-P, but these factors are also required for the expression of many other genes in the genome. There are no binding sites for the well-characterized E2F factors which are important for most genes whose transcription rate is increased in S-phase.
A comprehensive computational analysis of chromatin immunoprecipitation and deep sequencing (ChIP-Seq) data for over 50 transcription factors from human embryonic stem cells identified some factors associated with the histone genes [33]. These experiments were carried out on exponentially growing cells, and cells in S-phase have active histone genes, whereas histone genes in G1 and G2 cells are less active or inactive. Despite the absence of E2F binding sites in most histone genes, E2F1 was found at histone genes in ChIP-Seq studies from all cell types, as was the repressive E2F4, consistent with the possibility that these factors are involved in cell-cycle regulation of histone genes. A typical E2F-regulated gene binds E2F1 in late G1/S-phase to activate the gene, and E2F4 in G1 and G2 to inactivate the gene [34].
How E2F is recruited to histone genes remains unknown. Another protein implicated in the activation of histone gene transcription is YY1. All mammalian histone genes contain YY1 binding sites within the open reading frame (ORF) [35]. Although YY1 has been reported to interact with E2F1 [36], and is present at the histone genes in neural progenitor cells, YY1 is not found at histone genes in ES cells, suggesting that it is a cell type-specific regulator [33]. E2F must be recruited by a YY1-independent mechanism in ES cells.
A recent study carried out ChIP-Seq studies at different cell-cycle stages. They identified a repressive complex containing SFMBT1, a histone demethylase (LSD1), and coREST that was associated with histone genes in G1-phase cells, but not in S-phase cells [37]. This complex recognizes chromatin, and they observed an increase in dimethylated histone H3 lysine 4 (H3K4me2) at histone genes in G1 and G2 cells, consistent with the demethylase acting to repress the rate of histone gene transcription. How these complexes are recruited to the histone genes is not known, but they could be specifically recruited to the HLB.
Role of the HLB is Coordinating Transcription and Processing During the Cell Cycle
One function of the HLB is to coordinate transcription of the entire cluster of histone genes. The HLB also functions to couple histone transcription and processing. It likely exists in two states – ‘off’ when NPAT/Mxc is not phosphorylated and ‘on’ after phosphorylation by cyclin E/Cdk2. Because there are not obvious common cis-acting elements in the five histone gene promoters, coordinate regulation may be achieved by factors in the HLB, acting as coactivators. NPAT and the other constitutive HLB proteins FLASH and Mute are large proteins that are predicted to be largely unstructured, and could interact with coactivators or corepressors to modulate the expression of histone genes. How the specificity of NPAT for histone genes is determined remains an important unanswered question. Mxc self-associates through multiple sites in its N-terminal region, and these interactions are required for HLB formation [8]. It also directly binds to FLASH and Mute/Yin Yang 1-associated protein-related protein (YARP), and this interaction is essential for the formation of an HLB [38] containing histone-processing factors [39]. Phosphorylation of NPAT on multiple sites by cyclin E/Cdk2 is essential for increased histone mRNA accumulation and increased histone gene transcription as mammalian cells enter S-phase [9]. It is likely that phosphorylation of NPAT alters the interactions within the HLB that in turn may result in the recruitment of specific factors (e.g., the HCC) to activate U7 snRNP for processing, as well as transcription coactivators, and these factors are lost when NPAT is dephosphorylated. Other factors such as TBP, Hinf-P, and Oct1 could remain bound outside of S-phase.
The HLB also promotes coupling of processing and termination. Mutants in FLASH that are unable to localize to the HLB, but are active in processing, do not completely restore histone mRNA processing to a FLASH mutant. In Drosophila FLASH mutants, ‘read-though’ transcripts, that fail to terminate transcription at the normal site, are detected at the histone locus, and a small amount of polyadenylated histone mRNA is also formed [39]. The failure to terminate some histone transcripts normally supports the idea that the HLB promotes rapid histone pre-mRNA processing followed by transcription termination. This coupling is less efficient when FLASH is present in the nucleoplasm. In addition, a FLASH protein which had only 10–20% activity in vitro was found to be more than 90% active in vivo when it was localized to the HLB, but not when it was in the nucleoplasm [39]. These results suggest that concentrating processing factors in the HLB promotes efficient processing and coupling to transcription termination.
Data from mammalian cells suggest that the HLB may also couple transcription, processing, and termination. Knockdown of the transcription elongation factors, NELF [40] and pTEF-B [41], as well as of Ars2 [42], a protein that directly interacts with FLASH and is important for cell cycle progression [43], results in small amounts of misprocessing of histone mRNA. Knockdown of a small ncRNA, Ro-RNA Y3*, has a similar effect, resulting in accumulation in read-through RNAs and alteration of HLB structure [44]. These results suggest that proper transcription elongation, together with factors in the HLB, also promotes coupling of processing and termination in mammalian cells.
Post-Transcriptional Regulation of Histone mRNAs
Both 3′-end processing and the half-life of histone mRNAs are regulated during the cell cycle, with processing being activated as cells enter S-phase, and the mRNA being degraded at the end of S-phase [45]. In metazoans the stem-loop at the 3′ end of histone mRNAs provides a cis-element that allows the cell to readily distinguish the histone mRNAs from all other cellular mRNAs. A heterologous mRNA driven by a constitutive promoter and ending with the histone 3′ end is cell cycle-regulated [45,46] at the level of processing and mRNA degradation.
Early work provided evidence that processing was regulated during the cell cycle by a heat-labile factor in mammalian cells, and that U7 snRNP was constitutively present [47]. The heat-labile factor required for processing was subsequently identified as symplekin [48]. In Drosophila symplekin is not present in the HLB in cells that are not cycling, and is preferentially enriched in the HLB in S-phase cells [39]. This result suggests that recruitment of the HCC to the U7 snRNP may be a cell cycle-regulated event that contributes to regulating the processing of histone pre-mRNA. Although both FLASH and U7 snRNP are constitutively present in the HLB, this colocalization is not a result of the interaction between FLASH and Lsm11 that is essential for processing [39]. Thus, one possibility is that the interaction between FLASH and Lsm11 is cell cycle-regulated, and does not occur in G1 cells. This interaction may be activated as cells enter S-phase, resulting in recruitment of the HCC to the HLB and formation of the HCC on U7 snRNP (Figure 3).
Figure 3. Processing of Histone Pre-mRNAs.
(Top) Schematic of the histone pre-mRNA processing reaction. (Bottom) Possible forms of U7 snRNP that are present during the cell cycle. The active form (holo-U7) snRNP is present in S-phase. Based on staining of Drosophila HLBs [82], the HCC is not present bound to U7 snRNP in non-dividing cells. We cannot distinguish whether the U7 snRNP and FLASH are separate within the HLB, or whether the U7 snRNP/FLASH complex is present and not able to recruit the HCC. Abbreviations: HCC, histone cleavage complex; HLB, histone locus body.
In addition to the possible regulation of the activity of U7 snRNP, the levels of SLBP protein are cell cycle-regulated in mammalian cells. SLBP accumulates shortly before cells enter S-phase and is rapidly degraded at the end of S-phase [49]. Because SLBP is bound to histone mRNA throughout its entire life, participating in both mRNA synthesis and degradation, the amount of histone mRNAs present at any one time is limited by the amount of SLBP. Thus regulation of SLBP levels during the cell cycle is a crucial component of cell-cycle regulation of mammalian histone mRNA levels.
Regulation of Histone mRNA Half-Life
Histone mRNAs, like SLBP, are also degraded rapidly at the end of S-phase. This requires the stem-loop at the 3′ end of histone mRNA [49] and the mRNAs must be translated [50] (Box 4). Histone mRNAs are also degraded rapidly when DNA replication is inhibited in S-phase cells, although SLBP levels do not change under these conditions. This suggests that the signal to degrade histone mRNAs is not a cell-cycle signal, but the cell responding to reduced demand for histone proteins.
Box 4 Histone mRNA Degradation.
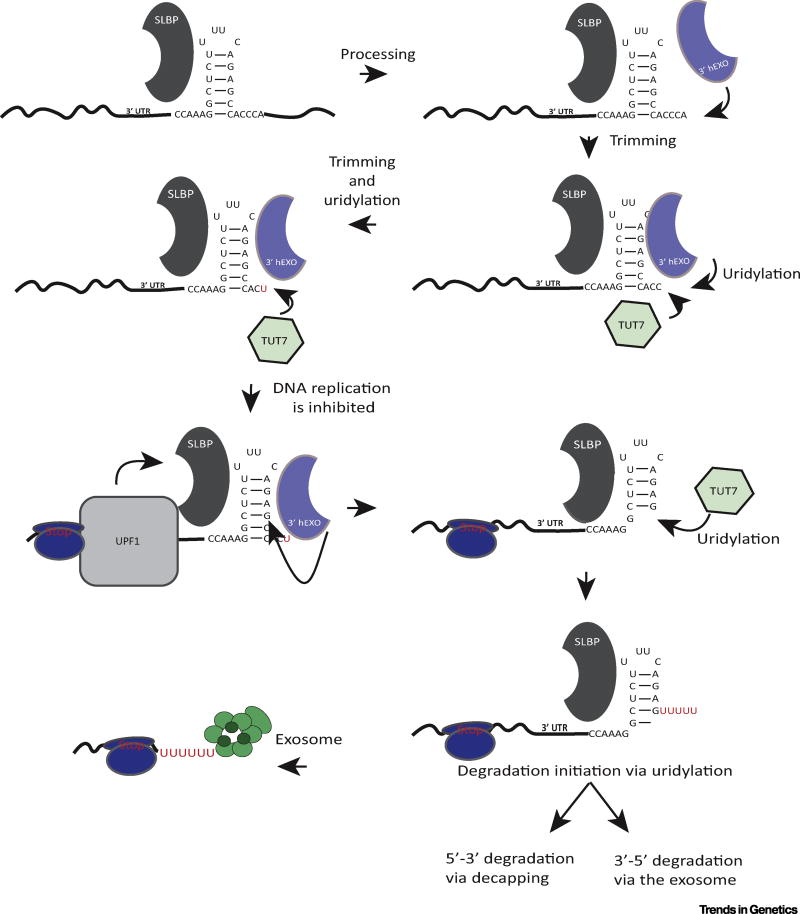
Histone mRNAs are rapidly degraded at the end of S-phase or when DNA replication is inhibited. Degradation requires that the histone mRNA is being translated [82]. The stem-loop at the 3′ end of the histone mRNA is necessary and sufficient for degradation [83]. The stem-loop must be located close to the stop codon, starting 25–100 nt from the stop codon, and this distance between the SL and the stop codon has been conserved in all metazoan histone genes [82]. These requirements suggest that translation termination may be regulated to initiate histone mRNA degradation [84]. Upf1, but not other nonsense-mediated decay factors, is essential for histone mRNA degradation, consistent with the requirement for translation and a potential role of translation termination [85]. In 2008 it was reported that histone mRNA degradation requires uridylation of the 3′ end of histone mRNA [56]. Subsequently it was shown that 3′hExo (see Figure 4 in main text) is also required for the initial step of histone mRNA degradation, degrading the mRNA partly into the stem, and that these intermediates are uridylated by TUT7 [53]. The Lsm1–7 ring, which binds to the oligo(U) tail, is required for degradation of histone mRNA [56]. The molecular mechanisms that alter the structure of the 3′end of the histone mRNP to allow 3′hExo to initiate degradation of the stem-loop are not understood, but could involve modification of SLBP, and may also involve the interaction of the C-terminal tail of Lsm4 directly with both SLBP and 3′hExo [86].
A 3′ to 5′ pathway for rapid degradation of histone mRNAs (Figure 4) was recently defined using a high-throughput sequencing strategy designed to determine the 3′ ends of all histone mRNA molecules, including the presence of non-templated nucleotides [50]. Using this system, an unexpected modification of the 3′ end of cytoplasmic histone mRNAs was found. Non-templated uridines were added to the 3′ end to maintain the normal length of the 3′ end in S-phase [51]. Following processing of the mRNA in the nucleus, leaving an ACCCA tail, the mRNA is shortened by 2–3 nt through the action of histone 3′ exonuclease (3′hExo/ERI1), which forms a ternary complex on the 3′ end of histone mRNA together with SLBP [5,52] (Figure 4). If further degradation occurs by 3′hExo, the length of the tail is restored to 3 nt by uridylation, resulting in mRNAs that end in ACC, ACU, or AUU [51,53].
Figure 4. Uridylation and Degradation of Histone mRNA.
After processing the histone mRNA is trimmed by 3′hExo. If trimming proceeds to within 1–2 nt of the stem the mRNA is repaired by TUT7 to the proper length. When DNA replication is inhibited, Upf1 is recruited to the histone mRNA, and 3′hExo can degrade into the stem. When the nuclease stalls the mRNA is then uridylated and degradation can resume. Abbreviations: SLBP, stem-loop binding protein; UTR, untranslated region.
When degradation is initiated by inhibition of DNA replication, 3′hExo initiates degradation into the stem-loop, removing 5–7 nt [54] (Figure 4). This intermediate is heavily uridylated, and likely still contains SLBP bound to the histone mRNA. Once 3′hExo degrades 3–4 nt into the stem it can no longer bind to the RNA. Removal of SLBP, possibly by the helicase activity of UPF1, then allows 3′ to 5′ degradation of the histone mRNA by the exosome, until it reaches the stalled ribosome. Intermediates accumulate 15 nt 3′ of the stop codon, consistent with the position of the stalled ribosome. These intermediates are also uridylated before they are degraded further [50]. Decapping, which also can be stimulated by Lsm1–7 bound to oligo(U) tails, seems to play a relatively minor role, although it certainly also can occur [55,56]. Histone mRNA degradation intermediates which are decapped and have been degraded from both the 5′ end 3′ ends have been observed [56].
Uridylation of the stem-loop is carried out by terminal uridylyl transferase TUT7 both in S-phase to form histone mRNAs ending in ACU and AUU and to initiate degradation, based on RNAi knockdown experiments [53]. The mechanism of histone mRNA degradation is likely to be conserved in all metazoans. Intermediates similar to those in mammals have been observed in Drosophila embryos [51].
Regulating both histone mRNA synthesis and histone mRNA degradation is crucial for the appropriate production of the amounts of histone proteins that are necessary to package the newly replicated DNA. The extent of transcriptional regulation may vary, provided that the regulation of processing is able to restrict histone mRNA production. For example, in mammals, histone gene transcription varies only 3–5-fold and is not dramatically regulated, based on nuclear run-on assays [57,58], but it is the tight regulation of SLBP levels that effectively restricts histone mRNA accumulation to S-phase. In other species histone gene transcription might be much more dramatically regulated.
Why Do Replication-Dependent Histone mRNAs in Metazoans End in a Stem-Loop and Not a Poly(A) Tail?
Even after a decade of high-throughput sequencing analysis, histone mRNAs remain the only known cellular eukaryotic mRNAs that are not polyadenylated. This property is not essential for viability because plants and most single-cell eukaryotes have polyadenylated histone mRNAs, whose levels are also tightly cell cycle-regulated. The evolutionary analysis of Samuelsson and coworkers [6] strongly suggests that histone mRNAs may have differed from other mRNAs in some single-cell eukaryotic lineages, consistent with the stem-loop being ancient and having been lost from many lineages, together with SLBP, NPAT, FLASH, and U7 snRNP. In the immediate precursors of metazoans, histone mRNAs likely had stem-loops. They have evolved a sophisticated system for regulating histone mRNA levels, both via the degradation of histone mRNA to prevent inappropriate synthesis of histone proteins, and the unique mechanism for processing histone mRNAs, both of which require SLBP. Part of this mechanism includes sequestering histone genes into the HLB. In C. elegans an alternative mechanism for 3′-end formation [59] has evolved which replaced the initial U7 snRNA-dependent mechanism. This also resulted in loss of the tight linkage of all the histone genes, and of the HLB (NPAT, FLASH, and U7 snRNP). However, the mechanism for regulating histone mRNA degradation has been retained, and SLBP still plays a central role in histone mRNA metabolism, likely binding to the histone pre-mRNA to stabilize the nascent histone mRNA, as well as participating in translation and degradation of histone mRNA.
Expression of Histone mRNAs in Non-Dividing Tissues
Cells in adult differentiated tissues live for months and even years without dividing. Although the half-life of histone proteins in nucleosomes is several weeks or months, there is still a need for synthesis of some histones at a very low rate. In mammals, the variant histone H3.3 replaces much of the H3 histones as tissues age, but there are no known replacement variants for the other nucleosomal histones. Recently it was found that a subset of nucleosomal histone genes, that are normally expressed as replication-dependent histone mRNAs ending in the stem-loop, are expressed as polyadenylated mRNAs in adult non-dividing tissues [60]. The same set of mRNAs is expressed in multiple human tissues, and many of the same genes are expressed in mouse and cow tissues. These mRNAs encode the same minor histone H2b and H2a protein variants previously reported to be increased in adult mouse tissues. The switch to expressing these genes as polyadenylated mRNAs (many of which are also spliced) is likely a component of the terminal differentiation program [60,61].
Concluding Remarks and Future Perspectives
Properly regulated synthesis of histone proteins is a crucial component of successful chromosome replication. The regulation of histone genes provides an excellent example of regulating multiple steps in RNA metabolism (transcription, processing, and half-life) to achieve and maintain the appropriate level of histone mRNAs. In addition, reversible modification of a key regulator, NPAT, and controlled degradation of another key factor, SLBP, and a system to degrade excess histone proteins [62], demonstrates the utilization of a complete suite of post-transcriptional and translational mechanisms to control histone protein levels. The multiple copies of histone genes allowed visualization of the factors assembled at the locus under the light microscope. With recent advances in high-resolution microscopy, it will be possible to probe the changes in structure of the HLB as the histone genes are turned on and off. It will also be possible to determine directly the relationship between the histone gene chromatin and the HLB. The insights gained from these studies are likely to be relevant to single-copy genes which cannot be visualized using current technology.
Reconstituting complex RNA-processing events (e.g., splicing and 3′-end formation) has been very challenging. For 3′-end formation the observation that CPSF73 is recruited as part of an inactive complex, which then must be activated, makes sense in protecting other RNAs from these enzymes. How this activation occurs is not known, but there is clearly a commonality between cleavage/polyadenylation and histone mRNA 3′-end formation that likely extends to the formation of the 3′ ends of snRNAs, another set of polymerase II transcripts.
The regulated degradation of histone mRNAs (and the fact that a large number of histone mRNAs are affected simultaneously) has allowed the determination of a major pathway of degradation, using high-throughput sequencing to precisely identify the spectrum of degradation intermediates and the role of oligouridylation in histone mRNA metabolism. Recent results (reviewed in Gagliardi [63]) show that deadenylated mRNAs also have uridines added to the remaining short A tail, and suggest that this serves to stabilize those molecules, similarly to uridylation of histone mRNA in S-phase. It remains to be seen whether similar degradation pathways exist for cytoplasmic mRNAs, and whether 3′hExo may play a role in these pathways (see Outstanding Questions).
Outstanding Questions.
How is NPAT/Mxc specifically recruited to the histone gene locus? In Drosophila, the cis-elements for HLB formation are in the histone tandem repeat. Because Mxc does not specifically bind to DNA, either factors bound to the DNA or some aspect of the chromatin at the histone locus must recruit Mxc. The cis-elements required for HLB formation in mammals have not yet been investigated. NPAT is likely the factor that initiates formation of the HLB. Because there are now methods to determine the overall organization of specific regions of chromatin in the nucleus, it may be possible to identify candidates for organizing sequences in the histone loci in mammalian cells.
Is the HLB intimately associated with histone gene chromatin or is it localized near the histone genes to store the necessary factors where they will be needed. Because a DNA sequence in the histone locus is capable of forming an HLB, it is likely that the HLB directly contacts the histone chromatin, but high-resolution imaging experiments will be necessary to determine whether this is the case.
What structural changes occur within the HLB as a result of phosphorylation of NPAT/Mxc by cyclin E? The HLB components (FLASH, NPAT, and Mute) likely contain binding sites for a large number of factors, and understanding these interactions and changes that result from phosphorylation will be necessary to understand the detailed mechanisms resulting in activating transcription and processing.
What is the structure of the HCC? In particular, what regions of symplekin result in formation of the HCC rather than of the cleavage/polyadenylation complex, and can we generate symplekin separation-of-function mutants that function in one reaction and not the other. Both the cleavage/polyadenylation complex and the HCC can be recruited to the pre-mRNA substrate, but cleavage requires the subsequent action of other factors. How is CPSF73 activated to cleave pre-mRNAs?
What are the biochemical changes at the 3′ ends of histone mRNPs that result in the transition from maintaining the proper length of histone mRNAs, by the action of 3′hExo and TUT7, to degradation into the stem of histone mRNA, and the addition of long U-tails to those intermediates?
The overall mechanism for histone gene expression is conserved in flies and mammals. The ability to carry out sophisticated genetics experiments in Drosophila, the accessibility of the embryo to imaging, together with the serendipitous finding in Drosophila that mutation of processing factors still allows expression of sufficient histones for continued cell growth, has allowed not only RNAi screens for factors affecting histone mRNA synthesis but also, most importantly, the ability to test whether interactions discovered in vitro are also relevant in vivo. Thus far all factors identified in one species have been present in the other, suggesting this approach will continue to be very fruitful.
Trends.
The genes for all five replication-dependent histone mRNAs are linked in metazoans. They encode the only eukaryotic cellular mRNAs that are not polyadenylated.
SLBP, which binds to the stem-loop at the 3′ end of histone mRNA, is required for all steps of histone mRNA metabolism.
Factors for coordinating expression of the genes for the five histone proteins and the processing of histone mRNAs are concentrated in the HLB. The HLB is present constitutively and histone gene expression is activated by phosphorylation of NPAT, a crucial factor for HLB formation, by cyclin E/Cdk2.
The active form of U7 snRNP contains a novel set of factors, including FLASH and a complex of polyadenylation factors, the HCC.
Uridylation of histone mRNA maintains the proper length of the 3′ end. When DNA replication is completed or inhibited, histone mRNA is rapidly degraded by a 3′ to 5′ pathway initiated by 3′hExo, a component of the histone mRNP. Degradation is dependent on translation, and requires Upf1 as well as uridylation of the degradation intermediates by TUT7.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants GM58921 to W.F.M. and R.J. Duronio, and GM29832 to W.F.M. and Z. Dominski. K.K. was partially supported by NIH training grant 5T32 GM007092.
Glossary
- FLICE-associated huge protein (FLASH)
an HLB protein necessary for histone pre-mRNA processing.
- Histone cleavage complex (HCC)
a complex of polyadenylation factors required for histone pre-mRNA processing. Components include symplekin, CstF64, CPSF100, CPSF160, and the nuclease CPSF73.
- Histone downstream element (HDE)
a purine-rich sequence in the pre-mRNA that basepairs with the U7 snRNA.
- Histone 3′ exonuclease (3′hExo/ERI1)
a 3′ exonuclease that forms a ternary complex on the stem-loop with SLBP.
- Multi sex combs (Mxc)
the Drosophila ortholog of NPAT.
- Muscle wasted (Mute)
a protein required for muscle development that is a component of the Drosophila HLB where it serves as a putative repressor of histone gene expression.
- Nuclear protein at the ataxia-telangiectasia locus (NPAT)
in mammals, NPAT is required for HLB formation and histone gene expression.
- Stem-loop binding protein (SLBP)
binds to the 3′ end of histone mRNA and is required for histone mRNA metabolism (processing, nuclear export, translation, degradation).
- Terminal uridylyl transferases
members of the family of noncanonical poly(A) polymerases that can add oligo(U) tails to RNAs.
- U7 small nuclear ribonuclear protein (U7 snRNP)
composed of a U7 snRNA and a novel heptameric Sm ring made up of Sm proteins B', D3, E, F, G, and Sm-like proteins (LSm) 10 and 11.
- Yin Yang 1-associated protein-related protein (YARP)
a transcriptional repressor and a homolog of Drosophila Mute.
References
- 1.Marzluff WF, et al. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonner WM, et al. Histone H2A.X gene transcription is regulated differently than transcription of other replication-linked histone genes. Mol. Cell. Biol. 1993;13:984–992. doi: 10.1128/mcb.13.2.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mannironi C, et al. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks L, III, et al. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA. 2015;21:1943–1965. doi: 10.1261/rna.053389.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan D, et al. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′ hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez MD, Samuelsson T. Early evolution of histone mRNA 3′ end processing. RNA. 2008;14:1–10. doi: 10.1261/rna.782308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JL, et al. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terzo EA, et al. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol. Biol. Cell. 2015;26:1159–1174. doi: 10.1091/mbc.E14-10-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye X, et al. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma TL, et al. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao JY, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 12.White AE, et al. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulchand S, et al. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci. 2010;123:2697–2707. doi: 10.1242/jcs.063172. [DOI] [PubMed] [Google Scholar]
- 14.McKay DJ, et al. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell. 2015;32:373–386. doi: 10.1016/j.devcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B, et al. Histone mRNAs contain blocked and methylated 5′ terminal sequences but lack methylated nucleosides at internal positions. Cell. 1977;10:113–120. doi: 10.1016/0092-8674(77)90145-3. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, et al. Regulation of RNA polymerase II termination by phosphorylation of Gdown1. J. Biol. Chem. 2014;289:12657–12665. doi: 10.1074/jbc.M113.537662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anamika K, et al. RNA polymerase II pausing downstream of core histone genes is different from genes producing poly-adenylated transcripts. PLoS One. 2012;7:e38769. doi: 10.1371/journal.pone.0038769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillai RS, et al. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillai RS, et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burch BD, et al. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 2011;17:1132–1147. doi: 10.1261/rna.2566811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang XC, et al. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 2011;31:1492–1502. doi: 10.1128/MCB.00979-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruepp MD, et al. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell. 2011;22:91–104. doi: 10.1091/mbc.E10-06-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeo V, et al. CstF64: cell cycle regulation and functional role in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. Biol. 2014;34:4272–4284. doi: 10.1128/MCB.00791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skrajna A, et al. U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein. RNA. 2017;23:938–951. doi: 10.1261/rna.060806.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominski Z, et al. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol. 2001;21:2008–2017. doi: 10.1128/MCB.21.6.2008-2017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, et al. Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2937–E2946. doi: 10.1073/pnas.1406381111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettitt J, et al. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 2002;115:857–866. doi: 10.1242/jcs.115.4.857. [DOI] [PubMed] [Google Scholar]
- 29.Ghule PN, et al. Maternal expression and early induction of histone gene transcription factor Hinfp sustains development in pre-implantation embryos. Dev. Biol. 2016;419:311–320. doi: 10.1016/j.ydbio.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan E, et al. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godfrey AC, et al. TheDrosophila U7 snRNP proteins Lsm10 and Lsm11 play an essential role in development independent of histone pre-mRNA processing. RNA. 2009;15:1661–1672. doi: 10.1261/rna.1518009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, et al. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr. Biol. 2014;24:1485–1491. doi: 10.1016/j.cub.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gokhman D, et al. Multilayered chromatin analysis reveals E2f, Smad and Zfx as transcriptional regulators of histones. Nat. Struct. Mol. Biol. 2013;20:119–126. doi: 10.1038/nsmb.2448. [DOI] [PubMed] [Google Scholar]
- 34.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 35.Bowman TL, et al. An H3 coding region regulatory element is common to all four nucleosomal classes of mouse histone-encoding genes. Gene. 1996;176:1–8. doi: 10.1016/0378-1119(96)00198-9. [DOI] [PubMed] [Google Scholar]
- 36.Schlisio S, et al. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 2013;27:749–766. doi: 10.1101/gad.210963.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XC, et al. A conserved interaction that is essential for the biogenesis of histone locus bodies. J. Biol. Chem. 2014;289:33767–33782. doi: 10.1074/jbc.M114.616466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatomer DC, et al. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J. Cell Biol. 2016;213:557–570. doi: 10.1083/jcb.201504043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narita T, et al. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Pirngruber J, et al. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruber JJ, et al. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiriyama M, et al. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 2009;29:4729–4741. doi: 10.1128/MCB.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohn M, et al. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 2015;29:1998–2003. doi: 10.1101/gad.266486.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris ME, et al. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two post-transcriptional steps. Mol. Cell. Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stauber C, et al. A signal regulating mouse histone H4 mRNA levels in a mammalian cell cycle mutant and sequences controlling RNA 3′ processing are both contained within the same 80-bp fragment. EMBO J. 1986;5:3297–3303. doi: 10.1002/j.1460-2075.1986.tb04643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gick O, et al. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slevin MK, et al. Deep sequencing shows multiple oligouridylations are required for 3′ to 5′ degradation of histone mRNAs on polyribosomes. Mol. Cell. 2014;53:1020–1030. doi: 10.1016/j.molcel.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welch JD, et al. EnD-Seq and AppEnD: sequencing 3′ ends to identify nontemplated tails and degradation intermediates. RNA. 2015;21:1375–1389. doi: 10.1261/rna.048785.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang XC, et al. Characterization of 3′ hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- 53.Lackey PE, et al. TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA. 2016;22:1663–1672. doi: 10.1261/rna.058107.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoefig KP, et al. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 55.Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heintz N, et al. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeLisle AJ, et al. Regulation of histone mRNA production and stability in serum stimulated mouse fibroblasts. Mol. Cell. Biol. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Avgousti DC, et al. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012;31:3821–3832. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyons SM, et al. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016;44:9190–9205. doi: 10.1093/nar/gkw620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kari V, et al. A subset of histone H2B genes produces polyadenylated mRNAs under a variety of cellular conditions. PLoS One. 2013;8:e63745. doi: 10.1371/journal.pone.0063745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gunjan A, et al. The emergence of regulated histone proteolysis. Curr. Opin. Genet. Dev. 2006;16:112–118. doi: 10.1016/j.gde.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 63.Scheer H, et al. Uridylation earmarks mRNAs for degradation … and more. Trends Genet. 2016;32:607–619. doi: 10.1016/j.tig.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z-F, et al. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 65.Martin F, et al. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borchers CH, et al. Combined top-down and bottom-up proteomics identifies a phosphorylation site in stem-loop binding proteins that contributes to high-affinity RNA binding. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3094–3099. doi: 10.1073/pnas.0511289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang XC, et al. Studies on the 50 exonuclease and endonuclease activity of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 2009;29:21–32. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Moeller H, et al. Structural and biochemical studies of SLIP1–SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res. 2013;14:7960–7971. doi: 10.1093/nar/gkt558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dankert JF, et al. Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2. Mol. Cell. 2016;64:507–519. doi: 10.1016/j.molcel.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Djakbarova U, et al. DDB1 and CUL4 associated factor 11 (DCAF11) mediates degradation of stem-loop binding protein at the end of S phase. ABBV Cell Cycle. 2016;15:1986–1996. doi: 10.1080/15384101.2016.1191708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu JL, et al. Nuclear bodies in the Drosophila germinal vesicle. Chromosome Res. 2006;14:465–475. doi: 10.1007/s10577-006-1062-5. [DOI] [PubMed] [Google Scholar]
- 73.Salzler HR, et al. A sequence in the Drosophila H3-H4 promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev. Cell. 2013;24:623–634. doi: 10.1016/j.devcel.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei Y, et al. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White AE, et al. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang XC, et al. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miele A, et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng L, et al. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 79.Dominski Z, et al. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang XC, et al. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 2013;33:28–37. doi: 10.1128/MCB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Graves RA, et al. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 83.Pandey NB, Marzluff WF. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol. Cell. Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 86.Lyons SM, et al. The C-terminal tail of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA. 2014;20:88–102. doi: 10.1261/rna.042531.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duronio RJ, Marzluff WF. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017;14:726–738. doi: 10.1080/15476286.2016.1265198. [DOI] [PMC free article] [PubMed] [Google Scholar]