SUMMARY
Mechanisms that control cell cycle dynamics during tissue regeneration require elucidation. Here we find in zebrafish that regeneration of the epicardium, the mesothelial covering of the heart, is mediated by two phenotypically distinct epicardial cell subpopulations. These include a front of large, multinucleate leader cells, trailed by follower cells that divide to produce small, mononucleate daughters. By live imaging of cell cycle dynamics, we show that leader cells form by spatiotemporally regulated endoreplication, caused primarily by cytokinesis failure. Leader cells display greater velocities and mechanical tension within the epicardial tissue sheet, and experimentally induced tension anisotropy stimulates ectopic endoreplication. Unbalancing epicardial cell cycle dynamics with chemical modulators indicated autonomous regenerative capacity in both leader and follower cells, with leaders displaying an enhanced capacity for surface coverage. Our findings provide evidence that mechanical tension can regulate cell cycle dynamics in regenerating tissue, stratifying the source cell features to improve repair.
eTOC Blurb
Cao et al. devise genetic tools and live imaging platforms in zebrafish to visualize cell cycle dynamics during regeneration of the epicardial tissue that covers the heart. They uncover evidence that differences in mechanical tension within the tissue sheet create distinct zones of endoreplicating and dividing cells that enable regeneration.
INTRODUCTION
Cells undergoing DNA replication classically progress through cytokinesis to generate two diploid daughter cells. Notable deviations include endocycling, in which DNA synthesis occurs without karyokinesis, and endomitosis, in which nuclei divide with incomplete cell division to generate multinucleate cells (Fox and Duronio, 2013; Losick et al., 2013). The outcome of these two forms of endoreplication is polyploidy, which can increase cell size, can also occur through cell fusion, and exists in many animal tissues (Orr-Weaver, 2015; Losick et al., 2013; Celton-Morizur and Desdouets, 2010; Abmayr et al., 2003; Ravid et al., 2002).
Endoreplication has been recently invoked in mechanisms of tissue repair. In Drosophila, abdominal epithelial cells, hindgut pyloric epithelial cells, and ovarian follicular epithelial cells undergo endoreplication during closure of surface wounds, regeneration of intestinal damage, and follicular epithelia repair, respectively (Losick et al., 2013; Tamori and Deng, 2013). Rodent liver regeneration after major surgical resection proceeds through a diverse mechanism involving hepatocyte hyperplasia, endocycling, and hypertrophy (Chen et al., 2012; Miyaoka et al., 2012; Pandit et al., 2012; Lazzerini Denchi et al., 2006). Adult mammalian cardiomyocytes similarly display a major component of hypertrophy after ischemic heart injury that is often accompanied by endoreplication, along with a minor component of hyperplasia (Tzahor and Poss, 2017). Thus, tissue repair or regeneration contexts do not always require a stem cell reserve or parenchymal cell with high proliferative capacity, but instead may be mediated by cells that increase DNA content and size.
Polyploidy can confer advantages during tissue repair. Multinucleate cardiac and skeletal muscle cells harvest the additional genomes for production of additional sarcomere components, augmenting contractile function. Drosophila cells undergoing endoreplication repress the apoptotic response to DNA damage caused by ionizing radiation or elevated Cdt1 expression (Hassel et al., 2014; Zhang et al., 2014; Mehrotra et al., 2008). Eliminating polyploidy caused by endoreplication and cell fusion in Drosophila abdominal epithelial cells disrupts wound healing (Losick et al., 2013). Mammalian polyploid hepatocytes are reported to have increased resistance to metabolic stress and injury (Duncan et al., 2012; Duncan et al., 2010). Transient participation in tissue repair, or limitations in detection methodology, likely contribute to an underestimation of the perceived occurrence and pro-regenerative roles of endoreplication. Additionally, the signals that dictate endoreplication and cytokinesis during tissue repair and regeneration are poorly understood.
The epicardium, a cardiac form of mesothelial tissue that lines organs and organ cavities, promotes myocardial regeneration in zebrafish and cardiac repair in mice (Wang et al., 2015; Wei et al., 2015; Huang et al., 2012; Riley, 2012; Kikuchi et al., 2011b; Zhou et al., 2011; Lepilina et al., 2006). The epicardium itself is highly regenerative, a capacity that helps maintain the mesothelial lining and likely protects against organ adhesions that form spontaneously or after internal injury. When the zebrafish epicardium is ablated from the cardiac ventricular surface, it repopulates in a wave from the chamber base to the apex from spared epicardial cells (Figure 1A) (Wang et al., 2015).
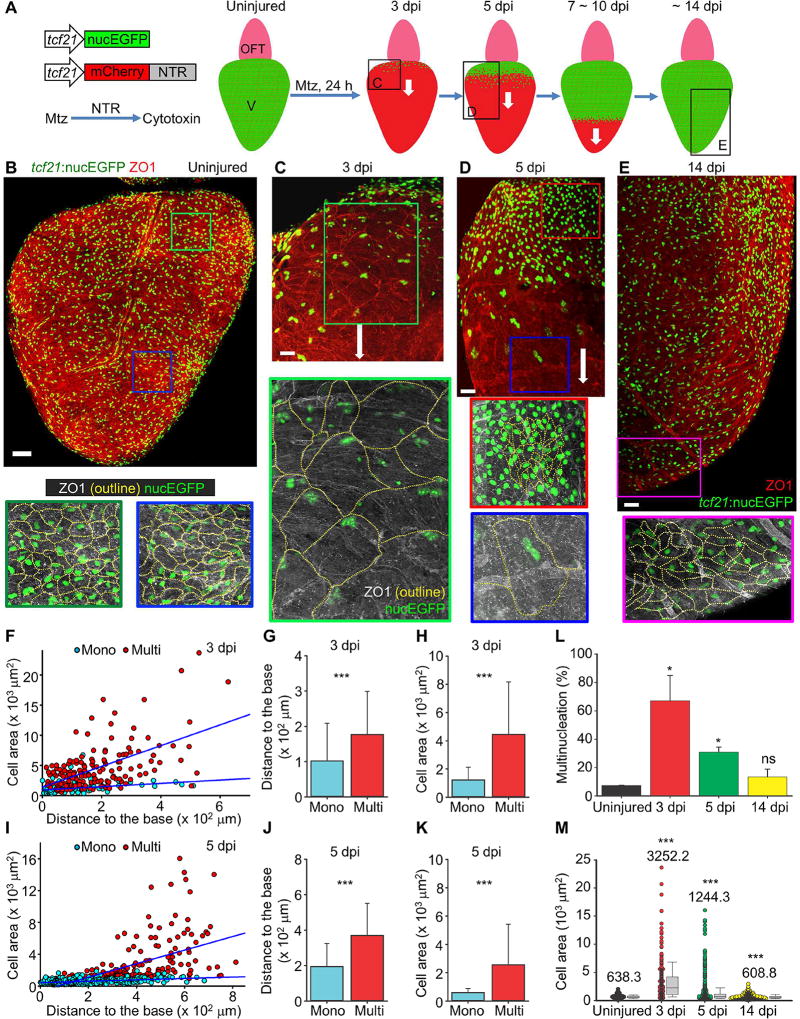
Figure 1. Transient Hypertrophy and Polyploidy in Regenerating Epicardial Cells.
(A) Schematic for epicardial ablation and regeneration in vivo. White arrows indicate the direction of regeneration. OFT, outflow tract; V, ventricle. The frames with letters indicate regions shown in (C–E).
(B) Flattened images of a whole-mounted uninjured heart stained with an anti-ZO1 antibody (red). Framed regions are enlarged below in the same scale as the enlarged panels of (C–E). ZO1 staining is outlined by dashed lines. Scale bar, 100 µm.
(C–E) Flattened images of whole-mounted adult hearts at 3 (C), 5 (D) and 14 dpi (E) stained with an anti-ZO1 antibody (red). Framed regions are enlarged below in the same scale, with epicardial ZO1 staining outlined by dashed lines. White arrows indicate the direction of regeneration. Scale bars, 50 µm.
(F–H) Quantifications of epicardial cell area and distance from the ventricular base at 3 dpi. Mononucleate cells (Mono) are represented by cyan dots (F) or bars (G, H), and multinucleate cells (Multi) by red dots (F) or bars (G, H). n = 94 for Mono and 161 for Multi. The blue lines in (F) show regression results for Mono and Multi, respectively. (F) P < 0.001, ANCOVA. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(I–K) Similar quantifications as (F–H), using samples at 5 dpi. n = 401 for Mono and 198 for Multi. (I) P < 0.001, ANCOVA. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(L) Quantification of multinucleation for uninjured, 3, 5 and 14 dpi hearts. n = 4 (uninjured), 5 (3 dpi), 4 (5 dpi) and 3 (14 dpi) hearts, respectively. * P < 0.05; ns, not significant; Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(M) Quantification of cell area distribution for uninjured, 3, 5 and 14 dpi hearts. n = 449 (uninjured), 255 (3 dpi), 599 (5 dpi) and 1,678 (14 dpi), respectively. Numbers on the plot indicate mean values. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate S.D.
See also Figures S1 and S2, Movie S1.
Here, we created genetic tools to visualize the mechanisms of epicardial regeneration. We identify locally regulated endoreplication events in the regenerating epicardium, forming a tissue front of large, polyploid cells that lead the regeneration process. High mechanical tension is evident at the tissue front, and experimental alterations in tension are sufficient to instruct endoreplication in epicardial cells. Our results reveal paradigms for how mechanical tension spatiotemporally controls cell cycle decisions during regeneration, and how these targeted endoreplication events can increase the efficacy of tissue regeneration.
RESULTS
Regenerating Epicardial Tissue Contains a Front of Hypertrophic, Multinucleate Cells
Recently, we generated a transgenic nitroreductase (NTR) system for inducible death in zebrafish cells activating regulatory sequences of the transcription factor tcf21, a marker of epicardial and epicardial-derived cells (tcf21:NTR) (Wang et al., 2015). Treatment with metronidazole (Mtz) kills most epicardial cells covering adult tcf21:NTR ventricles, whether applied in live animals or explanted hearts cultured ex vivo (Figure 1A) (Cao and Poss, 2016; Wang et al., 2015). To visualize epicardial cell dynamics during regeneration, we partially ablated epicardial tissue in tcf21:NTR; tcf21:nucEGFP zebrafish and examined cell morphology at 3, 5 and 14 days post injury (dpi) by immunostaining for the tight junction marker ZO1. Unexpectedly, we observed many large, multinucleate epicardial cells on the ventricular surface during regeneration (Figures 1B–1E and S1A–1D). At 3 dpi, the average epicardial cell surface area was ~410% larger than that of cells in uninjured hearts, with ~67% of cells on average possessing multiple nuclei (vs. ~7% in vehicle-treated animals) (Figures 1B, 1C, 1F–1H, 1L and 1M). Strikingly, by 5 dpi, we observed two distinct cellular domains, with large, multinucleate cells at the front of the epicardial sheet, trailed by small mononucleate cells closer to the ventricular base (Figures 1D and 1I–1K). Cell nuclei at the front of the sheet tended to localize in trailing regions of these cells at 3 and 5 dpi, indicating polarization along the direction of regeneration (Figures S1E and S1F). At 14 dpi, when the ventricular surface was fully repopulated, only small mononucleate cells were prominent, and the average epicardial cell surface area was comparable to that of uninjured hearts (Figures 1E, 1L and 1M). Flow cytometry and quantification of DNA content in tcf21+ cells at 7 and 14 dpi also indicated ploidy increases during regeneration (Figure S1G). We observed these same patterns of cell size and nuclear number during regeneration of epicardial tissue in hearts cultured ex vivo (Figure S2).
To identify the fates of polyploid cells, we longitudinally imaged the apices of cultured tcf21:NTR ventricles near the completion of regeneration, visualizing epicardial cell chromatin in a new tcf21:H2A-EGFP zebrafish strain at 10-min intervals over 5 days (Figures S1H and S1I, and Movie S1). During this period, we observed fragmentation of ~72% of nuclei arranged in a doublet (23 of 32). One of these nuclei fragmented after going through unconventional karyokinesis (Figure S1K). These observations indicate that epicardial regeneration occurs by assembly of a population of hypertrophic, multinucleate cells that we refer to as “leader cells”, a term used in other contexts of collective cell migration where a group of cells migrates coordinately while maintaining cell-cell adhesions (Mayor and Etienne-Manneville, 2016). Leader cells generally undergo apoptosis when regeneration is completed, leaving behind a more permanent population of smaller, mononucleate “follower” cells.
Emergence of Leader and Follower Cells in ex vivo Epicardial Explants
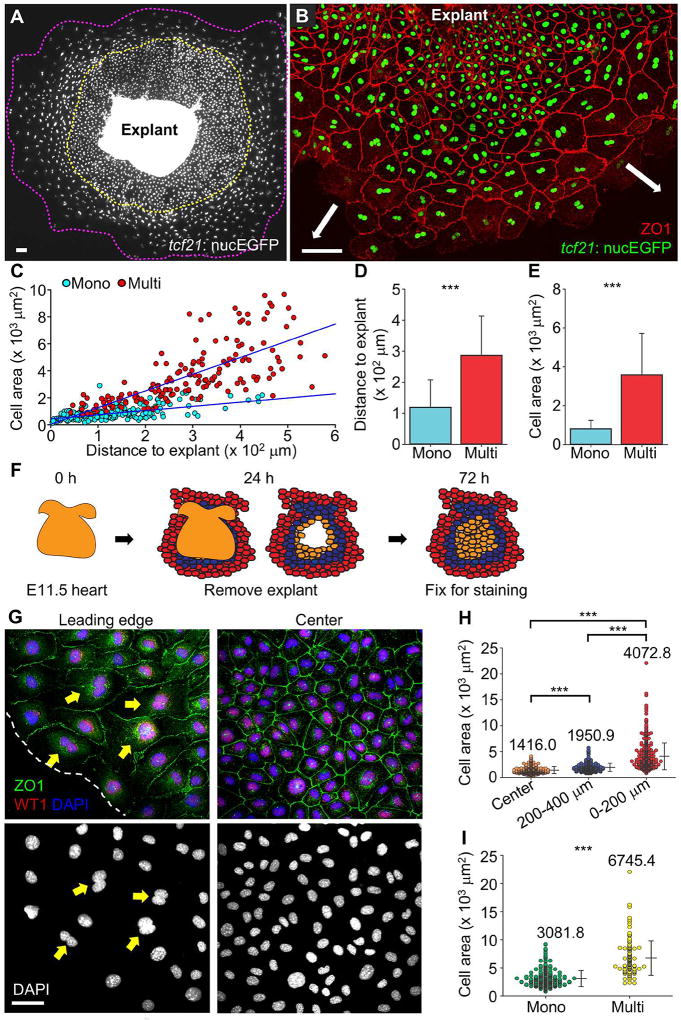
To study these cell populations in a more easily manipulated experimental system, we plated cardiac explants and monitored epicardial cells emerging from the explants. Strikingly, growing epicardial sheets recapitulated the stratified features observed in vivo, with large, multinucleate leader cells and small, mononucleate follower cells (Figures 2A and 2B). Conspicuous differences in cell surface area were evident after 3 days of culture, dependent on distance from the explant (Figures 2B and 2C). Cells > 400 µm and 200–400 µm from the explant were 506% and 248% larger on average, respectively, than cells within 200 µm of the explant. In these cultures, mononucleate cells, with an average area of 801.1 µm2, were positioned on average 118.9 µm from the explant. Multinucleate cells had a 346% larger area on average and were positioned 141% farther from the explant (Figures 2D and 2E). Three-dimensional views of projection images indicated that follower cells have slightly (~10%) greater depth in the Z-plane than leader cells (6.3 µm vs. 5.7 µm; Figures S3A–S3D); thus, we estimate that leader cells are ~304% larger in cell volume on average (20,379 µm3 vs 5,047 µm3). Leader and follower features were comparable in explants with or without the tcf21:nucEGFP transgene (data not shown) and on different ECMs (Figures S3E–S3I). Thus, the formation of leader and follower cell phenotypes is a characteristic that is inherent to epicardial cells and not formally dependent on a specific substrate or in vivo provision.
Figure 2. Emergence of Leader and Follower Cells ex vivo.
(A) tcf21:nucEGFP epicardial cells migrating from an explant after 5 d culture in a coated dish. The front of the leading edge is outlined with a magenta dashed line, and the mononucleate cell domain is outlined with a yellow dashed line. Scale bar, 100 µm.
(B) tcf21:nucEGFP explant culture stained for ZO1. White arrows indicate direction of migration. Scale bar, 100 µm.
(C) Quantification of cell area and distance to the explant for samples in (B). Mononucleate cells are represented by cyan dots and multinucleate cells by red dots. n = 224 (Mono) and 166 (Multi) respectively. The blue lines show regression results for Mono and Multi, respectively. P < 0.001, ANCOVA.
(D, E) Distribution of cell distances to explant (D) and cell areas (E) for mononucleate (Mono) and multinucleate cells (Multi). n = 224 (Mono) and 166 (Multi) respectively. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(F–I) Polyploidy and hypertrophy of murine fetal epicardial cells. (F) Schematic of murine epicardial explant culture. The colors indicate location of regions quantified in (H). (G) A 72-h culture stained for WT1 (red) and ZO1 (green) indicating epicardial cells at the leading edge (left) or the center (right). Nuclear staining is blue in merged images and white in bottom panels. Yellow arrows denote binucleate cells. (H) Quantification of cell areas at three different regions. n = 352 (center), 299 (200–400 µm from the front of leading edge) and 451 (0–200 µm) respectively. Numbers on the plot indicate mean values. (I) Quantification of cell areas at the leading edge (0–200 µm from the front) for mononucleate (Mono, n = 329) and multinucleate (Multi, n = 122) cells. Numbers on the plot indicate mean values. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D. Scale bar, 50 µm.
See also Figure S3.
To test whether these binary cellular phenotypes were characteristic of epicardium in other species, we examined Wt1+ epicardial cells migrating from cultured murine fetal heart explants (Trembley et al., 2016) (Figures 2F and 2G). We observed a similar relationship between cellular size and distance from the tissue source (Figure 2H). Approximately 20% of epicardial cells at the leading edge were multinucleate, with an average size that was 119% larger than mononucleate cells in this region (Figure 2I). Thus, the distance from the leading edge influences the size and nuclear number of epicardial cells in vivo and ex vivo.
Leader Cells Form by Localized Endomitosis and Endocycling
Multinucleate cells can be generated by either cell fusion (e.g. muscle fiber formation from individual myoblasts) or endoreplication (e.g. occurring in cardiomyocytes and hepatocytes) (Duncan, 2013; Abmayr and Pavlath, 2012; Engel et al., 2006). During healing of the punctured adult Drosophila abdominal epithelium, both mechanisms participate (Losick et al., 2013). To dissect how leader cell features emerge in zebrafish epicardial cells, we generated a transgenic line with a tcf21:H2A-mCherry reporter that labels epicardial cell chromatin (Pauls et al., 2001) (Figures S4A–S4C), and a tcf21:LifeAct-EGFP reporter line that labels filamentous actin structures like the cell cortex and stress fibers (Riedl et al., 2008) (Figures S4D and S4E). To test whether epicardial cells readily fuse, we positioned together explants from tcf21:LifeAct-EGFP; tcf21:H2A-mCherry and tcf21:H2A-EGFP; tcf21:mCherry-NTR animals (Figure 3A). Cells from each explant migrated to converge in the middle of the culture dish. Twenty-one explant pairs in 3 independent experiments were monitored for up to 12 days, and no cells expressing both nuclear markers were observed (Figure 3A). This result indicates that epicardial hypertrophy and polyploidy during regeneration are unlikely to be the outcome of epicardial cell fusion.
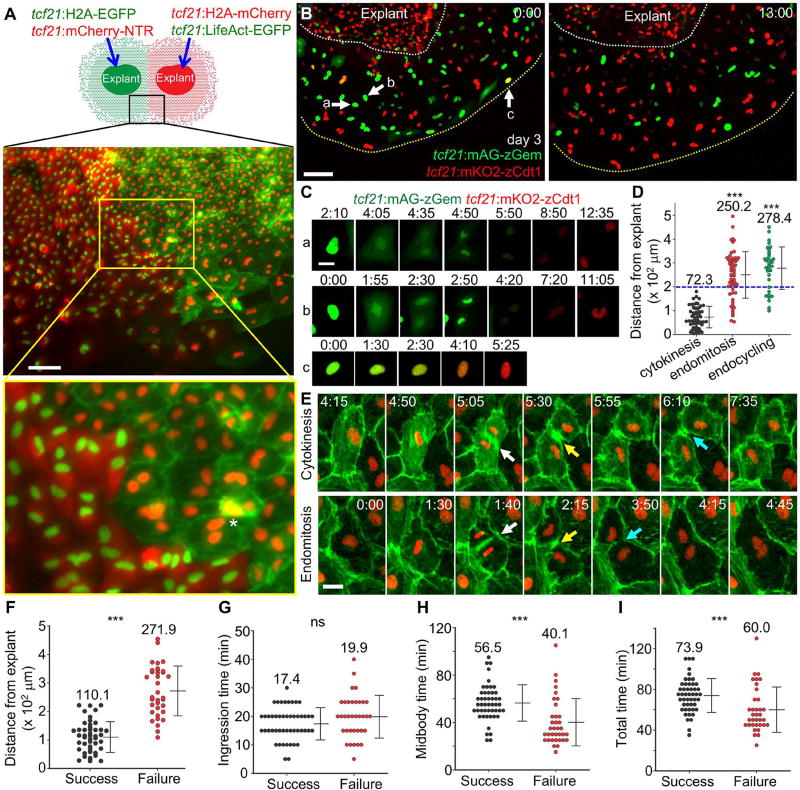
Figure 3. Endomitosis and Endocycling Events Underlie Epicardial Cell Polyploidy.
(A) (Top) Schematic of experimental design to detect epicardial cell fusion. (Middle) Image of the framed region represented in the cartoon acquired after 5 d culture. (Bottom) The region framed in yellow, enlarged to show detail. Asterisk indicates actin meshwork (green) covering a red nucleus, not a fused cell. Scale bar, 100 µm.
(B) Video frames of FUCCI cell cycle analysis of explanted epicardial cells. A 3-d culture of an explant carrying FUCCI reporters was subjected to live imaging for 13 h. White dashed lines outline the explant, and yellow dashed lines outline the leading edge. Arrows and lowercase letters denote nuclei shown in (C). Scale bar, 100 µm.
(C) Video frames of the nuclei highlighted in (B) showing cytokinesis (cell a), endomitosis (cell b) and endocycling (cell c). Scale bar, 20 µm. Timing, hh:mm.
(D) Spatial distribution of cell cycle behaviors. Cell positions were calculated at the point of nuclear membrane breakdown (for cytokinesis and endomitosis) or fluorescent change from green to red (for endocycling). n = 56 (cytokinesis), 59 (endomitosis) and 35 (endocycling), respectively. Numbers on the plot indicate mean values. Blue dashed line marks 200 µm position. *** P < 0.001, Mann-Whitney Rank Sum Test (compared with cytokinesis). Bars indicate mean ± S.D.
(E) Video frames of explanted epicardial cells from a tcf21:LifeAct-EGFP; tcf21:H2A-mCherry line showing normal cytokinesis (top) and cytokinesis failure in an endomitotic cell (bottom). White arrows indicate start of cleavage furrow ingression, yellow arrows indicate formation of midbodies and cyan arrows indicate furrow abscission or regression. Scale bar, 20 µm. Timing, hh:mm.
(F) Spatial distribution of successful and failed cytokinesis. Cell positions were calculated at the point of nuclear membrane breakdown. n = 48 (Success) and 35 (Failure), respectively. Numbers on the plot indicate mean values. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(G–I) Quantifications of the durations for cleavage furrow ingression (G), midbody formation (H), and the total time of both (I), for successful and failed cytokinesis, respectively. Ingression time is defined as the time from the dense LifeAct-EGFP signal emergence between two daughter nuclei (white arrows in (E)) to the formation of the narrowest furrow (yellow arrows in (E)). Midbody time was defined as the time from the narrowest furrow formation to furrow abscission or furrow regression (cyan arrows in (E)). n = 48 (Success) and 35 (Failure), respectively. Numbers on the plot indicate mean values. *** P < 0.001; ns, not significant; Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
To visualize cell cycle phases, we created tcf21:mAG-zGeminin and tcf21:mKO2-zCdt1 transgenic lines that express components of the fluorescent ubiquitination-based cell cycle indicator (FUCCI) system (Sugiyama et al., 2009). Live imaging of these reporters in epicardial explant cultures indicated reciprocal oscillating of green (S/G2/M phases) and red (G1/G0) fluorescence (Movies S2 and S3; Figures S4F and S4G), enabling discrimination of cell cycle decisions. Using FUCCI, cytokinesis events are identifiable as the generation of two uncoupled red nuclei emergent from a single nucleus that has lost green fluorescence. By contrast, endomitosis events visualize as a tight doublet of red nuclei (contacting each other and moving coordinately) emergent from a green nucleus, and endocycling as a direct green-to-red nuclear color change (Figures 3B and 3C; Movie S3). We quantified the spatial distribution of these events in epicardial cells migrating from an tcf21:FUCCI cardiac explant over a period of up to 24 h after 3 days of culture. We found that cycling cells located 200 µm or further away from the explant exclusively underwent endoreplication, which we identified as 63% endomitosis (44 of 70 cells) and 37% endocycling (26 of 70 cells). By contrast, cycling cells within 200 µm of the explant were most likely to complete cell division (70%, 56 of 80 cells). This spatial distribution was represented in the population as a whole, with the probability of undergoing endoreplication, and not cytokinesis, increasing with distance from the explant (i.e., decreasing with distance from the leading edge) (Figure 3D).
To visualize the genesis of endoreplication events, we performed live imaging of tcf21:LifeAct-EGFP; tcf21:H2A-mCherry epicardial cells in explant cultures (Movie S4). Successful cytokinesis occurs in four steps: positioning the division plane, ingression of the cleavage furrow, midbody formation, and abscission of the furrow (Normand and King, 2010). An actomyosin ring compresses the cell membrane to form the cleavage furrow and progresses to encircle the midbody (Normand and King, 2010). For purposes of quantification, we interpreted the narrowest furrow indicated by LifeAct-EGFP signal as the midbody. During epicardial endomitosis, we observed that the first two steps appear normal, with successful positioning of the division plane and furrow ingression (Figures 3E and 3F; Movie S5). Cleavage furrow ingression occurred over a similar average duration in cells undergoing endomitosis (19.9 min) vs. cytokinesis (17.4 min) (Figure 3G). By contrast, midbodies were less stable in endomitotic (40.1 min) than dividing (56.5 min) cells on average (Figure 3H), culminating with midbody dissociation and furrow regression (Figure 3E). The average total time of furrow ingression and midbody duration was also shorter for endomitotic cells (60.0 min vs. 73.9 min; Figure 3I). These results suggest that cytokinesis failure is mainly attributable to an unstable midbody. Once the leader cell domain is established, we noted that leaders in the most peripheral cell layer cycle less frequently than internally located cells in the zone of endoreplication (Movie S4). Together, our results indicate that cells in the leading edge of the regenerating epicardium undergo endomitosis and endocycling to generate large polyploid cells, whereas trailing cells undergo cytokinesis.
Leader Cells Experience Increased Mechanical Tension
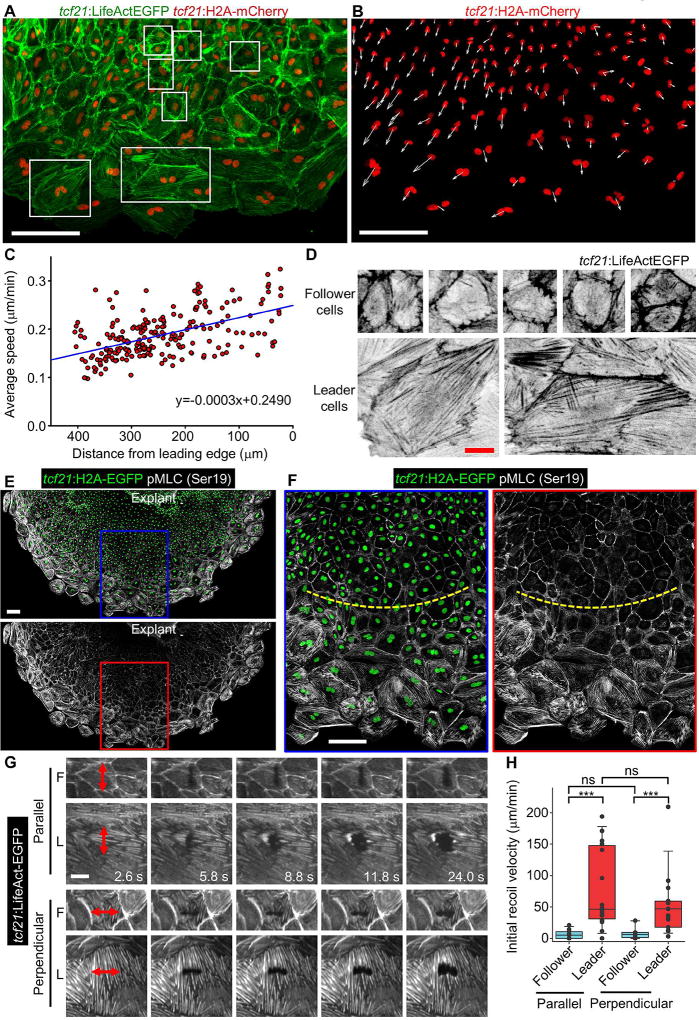
We took advantage of subcellular markers in tcf21:LifeAct-EGFP; tcf21:H2A-mCherry zebrafish to compare additional features of leader and follower cells. By tracking the distance of nuclear displacement over time in explant cultures, we measured the velocities of individual cells within epicardial tissue sheets. In these experiments, distance from the tissue front was inversely correlated with migration velocity, with leader cells migrating on average 29% faster than follower cells (0.22 ± 0.05 µm/min in cells 0–200 µm from the leading edge, vs. 0.17 ± 0.04 µm/min in cells >200 µm from the leading edge, P < 0.001, Mann-Whitney Rank Sum test; Figures 4A–4C; Movie S4). For each 100 µm distance from the front of regeneration, the average velocity of cell migration decreased 0.03 µm/min. We also used live imaging to assess cytoskeletal actin dynamics within leader and follower cells of explants. Migrating leader cells displayed more pronounced stress fibers than follower cells (Figures 4A and 4D; Movie S4), with many of these fibers arranged in parallel configuration. These features resembled those of cultured cells under sheer or stretch force, and they suggested the presence of differential mechanical tension within cells comprising the epicardial sheet (Hoffman et al., 2012; Kaunas et al., 2005; Noria et al., 2004).
Figure 4. Leader Cells Display Higher Migration Velocity and Mechanical Tension than Followers.
(A) Epicardial explant culture showing F-actin and nuclei by tcf21:LifeAct-EGFP;tcf21:H2A-mCherry reporters. The framed regions were enlarged for display in (D). LifeActEGFP shown in green and H2A-mCherry shown in red. Scale bars, 100 µm.
(B) Same experiment in (A) showing velocity vectors (white arrows) for each nucleus (H2A-mCherry, red) in a 5-h window. Scale bars, 100 µm.
(C) Dot plot for each individual nucleus indicating the average speed in 15.5 h over the average distance from the leading edge. The blue line indicates the regression result. n = 224.
(D) Magnified view of the framed regions in (A). LifeActEGFP shown in inverted grayscale. Scale bars, 20 µm.
(E, F) A 5-day tcf21:H2A-EGFP (green) epicardial explant culture was stained with pMLC (Ser19, grayscale). The framed regions in (E) are enlarged to show details in (F). The yellow dashed lines approximately separate follower (top) and leader cell regions (below). Scale bars, 100 µm.
(G) Video frames of explanted epicardial cells from a tcf21:LifeAct-EGFP line showing laser incisions and recoil of the cells. LifeAct-EGFP is shown in grayscale. Red double arrows indicate position and length of incisions. F, follower cells; L, leader cells. Scale bars, 20 µm.
(H) Quantification of the initial recoil velocity of leader and follower cells after cutting in a direction parallel or perpendicular to the migration direction. n = 12 (parallel-follower), 16 (parallel-leader), 14 (perpendicular-follower) and 15 (perpendicular-leader) respectively. *** P < 0.001; ns, not significant; Mann-Whitney Rank Sum Test. Bars indicate S.D.
To assay mechanical tension in migrating epicardial sheets, we first examined the presence of phosphorylated myosin light chain II (pMLC, Ser19) in leader and follower cells. pMLC (Ser19) is a marker of activation of actomyosin contraction, and it has been used as an indicator in tumor metastasis, epithelial cell scattering and neuronal morphology (Madsen et al., 2015; Roland et al., 2014; Pellegrin and Mellor, 2007; de Rooij et al., 2005; Katoh et al., 2001). We observed intense pMLC (Ser19) staining signals in leader cells, labeling stress fibers and actin bundles and suggesting high tension. By contrast, pronounced signals were limited to the cell-cell junctions of follower cells (Figures 4E and 4F).
To directly assess tension in the growing cell sheet, we made laser incisions within cells and measured the initial recoil velocities of the wounds, a property that is proportional to tissue tension (Mayer et al., 2010). We performed single incisions along a 12.8 µm line within each cell, in a direction either parallel or perpendicular to the direction of growth. As predicted from pMLC staining, leader cells displayed a much higher initial recoil velocity than follower cells following incisions, whether parallel (77.2 ± 63.5 µm/min vs. 5.9 ± 6.8 µm/min; mean ± S.D.) or perpendicular (52.9 ± 50.1 µm/min vs. 8.1 ± 9.1 µm/min; mean ± S.D.) to the direction of growth (Figures 4G and 4H; Movie S6). Together, these results indicate that leader cells experience greater mechanical tension than follower cells, associating high tension with endoreplication and hypertrophy.
Mechanical Tension Promotes Endoreplication
Transduction of mechanical tension through ECMs and the actin cytoskeleton to intracellular signaling programs can regulate cell proliferation, migration, and differentiation (Happe and Engler, 2016; LeGoff and Lecuit, 2015; Chanet and Martin, 2014; Heisenberg and Bellaiche, 2013; Lecuit et al., 2011; Provenzano and Keely, 2011; Hadjipanayi et al., 2009; Wozniak and Chen, 2009; Wang and Riechmann, 2007). High tension can increase cell proliferation in several contexts, whereas relaxation of tension reduces proliferation (Provenzano and Keely, 2011; Ulrich et al., 2009; Liu et al., 2007; Wang and Riechmann, 2007; Peyton et al., 2006; Nelson et al., 2005). For instance, mammalian myoblasts response to cyclic physical stretch by increased proliferation (Kumar et al., 2004), and quiescent epithelial cells reenter the cell cycle following applied mechanical strain (Gudipaty et al., 2017; Benham-Pyle et al., 2015). On the other hand, fluid flow-induced shear force inhibits vascular endothelial cell proliferation and causes G2/M arrest in tumor cells (Chang et al., 2008; Akimoto et al., 2000). Although many experiments have linked mechanical tension to cell cycle behavior, it has not, to our knowledge, been implicated in endoreplication.
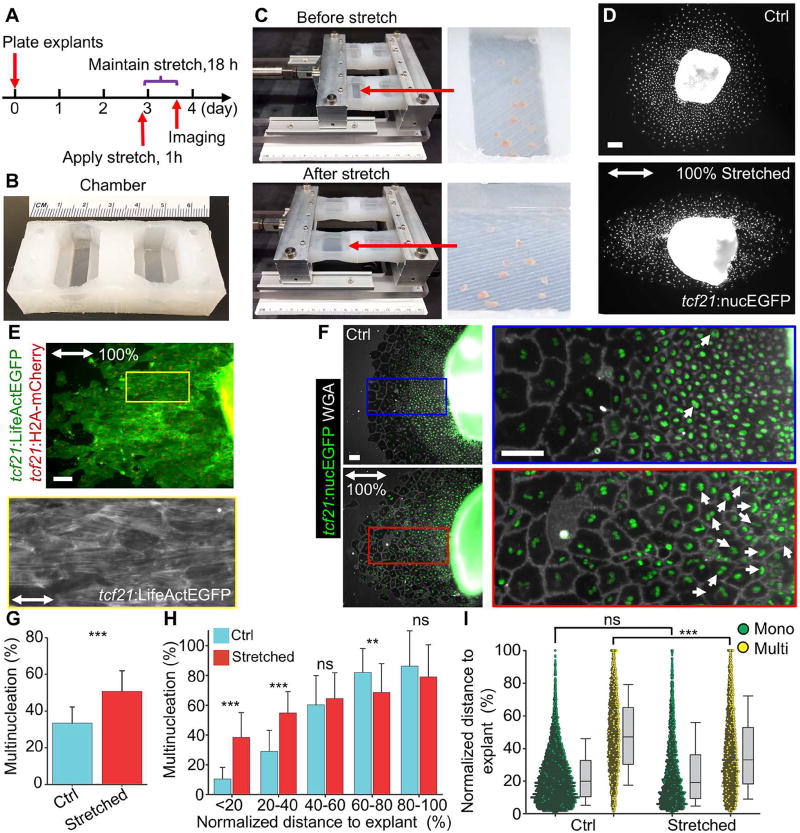
To directly test the effects of mechanical tension on cell cycle decisions, we plated epicardial explant cultures on elastic chambers (Figures 5A and 5B). Three days after plating, the chambers were mounted in a bioreactor and uniaxial stretch was applied to the culture surface to create tension anisotropy (Figure 5C). A constant elongation rate was applied over one hour to extend the chamber an additional 50%, 100% or 200% of the initial dimension, after which the tissue was held at the stretched length for 18 h (Figures 5A, 5C and S5). Stretching at 100% generated an elliptical shape and the most consistent effects, and this condition was chosen for further analysis (Figures 5D and S5). Cellular stress fibers were oriented in parallel with the stretch axis, indicating increased and anisotropic mechanical tension (Figure 5E).
Figure 5. Mechanical Tissue Stretching Promotes Epicardial Endoreplication.
(A) Schematic of experimental design in (C–F).
(B) An elastic chamber with culture surface areas of 7 × 23 mm2 (right) and 7 × 26 mm2 (left).
(C) Culture chambers before (top) and after stretch (bottom). Enlarged views of a chamber with heart explants and PBS are shown on the right.
(D) Explant culture from tcf21:nucEGFP animals with (bottom) or without (top) 100% stretch. The double arrow indicates stretch direction. nucEGFP is shown in white. Scale bars, 100 µm.
(E) Explant culture from tcf21:LifeAct-EGFP;tcf21:H2A-mCherry animals after 100% stretch. (Top) LifeActEGFP is shown in green and H2A-mCherry is shown in red. The framed region is enlarged to show LifeAct-EGFP (grayscale) below. The double arrow indicates stretch direction. Scale bars, 100 µm.
(F) Experiment as in (D) is shown, visualizing tcf21:nucEGFP tissue with cell shapes outlined using a WGA stain (grayscale). Top, unstretched control; bottom, 100% stretch. The double arrow indicates stretch direction. White arrows denote multinucleate cells close to the explant. Scale bars, 100 µm.
(G) Quantification of epicardial cell multinucleation in a quadrant of the cell sheet shown in (F). n = 28 (Ctrl) and 26 (Stretched) explants, respectively. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(H) Distribution plot showing the percentages of multinucleate cells at different regions of the epicardial cell sheet for both unstretched (Ctrl) and stretched cultures. Distances of cells to explant were normalized to the migration distances of the cell sheets and expressed as a percentage. n = 27 (Ctrl) and 31 (Stretched) explants respectively. *** P < 0.001; ** P < 0.01; ns, not significant; Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(I) Frequency plot showing the distances of cells to explants for mononucleate (Mono, green) and multinucleate (Multi, yellow) cells, respectively, for both unstretched (Ctrl) and stretched cultures. Distances of cells to explant were normalized to the migration distances of the cell sheets and expressed as a percentage. n = 7,183 (Ctrl, Mono), 3,390 (Ctrl, Multi), 3,959 (Stretched, Mono) and 3,994 (Stretched, Multi) cells, respectively. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate S.D.
See also Figure S5.
To determine the effects of stretching epicardial tissue sheets on endoreplication, we measured the distribution of mono- and multinucleate cells. We observed a ~51% higher overall percentage of multinucleate cells in stretched samples versus those without additional mechanical force (50.7% vs. 33.5%; Figures 5F and 5G). Furthermore, the distribution of multinucleate epicardial cells was heavily shifted toward the explant in stretched sheets compared to unstretched sheets (Figures 5H and 5I). Strikingly, tissue regions < 20% and 20–40% distant from the explant contained 239% and 101% higher frequencies of multinucleate cells, respectively, than the corresponding regions in unstretched samples (35.3% vs. 10.4% (region < 20%), and 54.2% vs. 27.0% (region 20–40%); Figure 5H). Thus, experimental increases in tension promote endoreplication in epicardial tissue sheets.
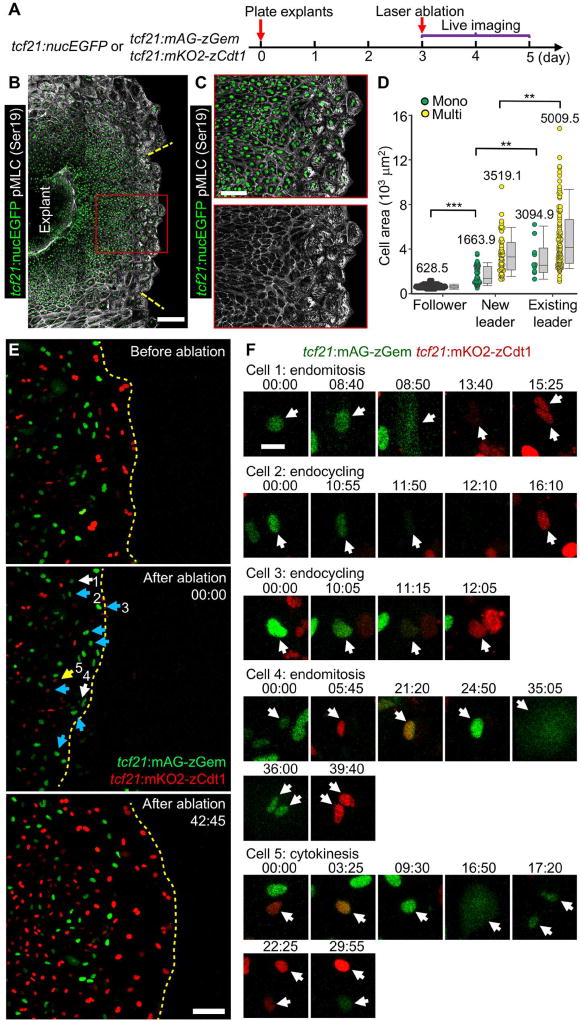
To attempt to visualize de novo leader cell creation and how position within the migrating tissue sheet can instruct epicardial cell phenotypes, we ablated leader cells in explant cultures using a laser (Figure 6A). Within 24 h of leader cell ablation, follower cells positioned at the new tissue front displayed stronger pMLC staining, resembling that of unperturbed leader cells (Figures 6B and 6C). Moreover, most of these follower cells (28 of 33 observed cell cycle events) underwent endoreplication to reconstruct a polyploid leader cell tissue front (Figures 6E and 6F; Movie S7). We found that 56% (49 of 88) of new leaders were multinucleate, with an average size of 3,519.1 µm2, with the remainder mononucleate with an average size of 1,663.9 µm2. Both values were greater than those of unperturbed follower cells (628.5 µm2, Figure 6D). Unperturbed leader cells had a higher frequency of multinucleation (89 of 99, or 90%) and were much larger (5,009.5 µm2 on average). These results suggest that the de novo leader cells transition to acquire leader cell features like multinucleation, hypertrophy, and high mechanical tension. In total, our experiments indicate that elevated mechanical tension at the contact-free front of the epicardial sheet creates a zone of endoreplication. This tension-regulated process results in regionalized populations of regenerating diploid and polyploid epicardial cells.
Figure 6. Follower Cells Undergo Endoreplication after Leader Cell Ablation.
(A) Schematic for experiments in (B–F).
(B, C) Ablation experiment using tcf21:nucEGFP explants (green). Twenty-four h post laser ablation, the explant culture was stained to detect pMLC (Ser19, grayscale). The framed regions in (B) are enlarged to show details in (C). The yellow dashed lines approximately mark the region that was ablated. Scale bars, 200 µm (B) or 100 µm (C).
(D) Quantification of nucleation and cell area of de novo leader cells and pre-existing leader and follower cells in (B). Leader cell region is defined by the strong staining of pMLC. Mono, mononucleate; Multi, multinucleate. n = 111 (follower), 39 (new leader, Mono), 49 (new leader, Multi), 10 (existing leader, Mono), and 89 (existing leader, Multi), respectively. *** P < 0.001; ** P < 0.01; Mann-Whitney Rank Sum Test. Bars indicate S.D.
(E) Video frames of a tcf21:FUCCI epicardial explant culture subjected to live imaging and laser ablation. The top panel is an image before ablation; the middle panel, immediately after ablation; the lower panel, a reconstructed leader cell region at 42 h, 45 min after ablation. The white, cyan and yellow arrows indicate nuclei that underwent endomitosis, endocycling and cytokinesis, respectively. Timing, hh:mm. Scale bar, 100 µm.
(F) Cropped video frames of the numbered cells indicated in the middle panel of (E). Timing, hh:mm. Scale bar, 20 µm.
See also Movie S7.
Autonomous but Distinct Regenerative Properties of Leader and Follower Cells
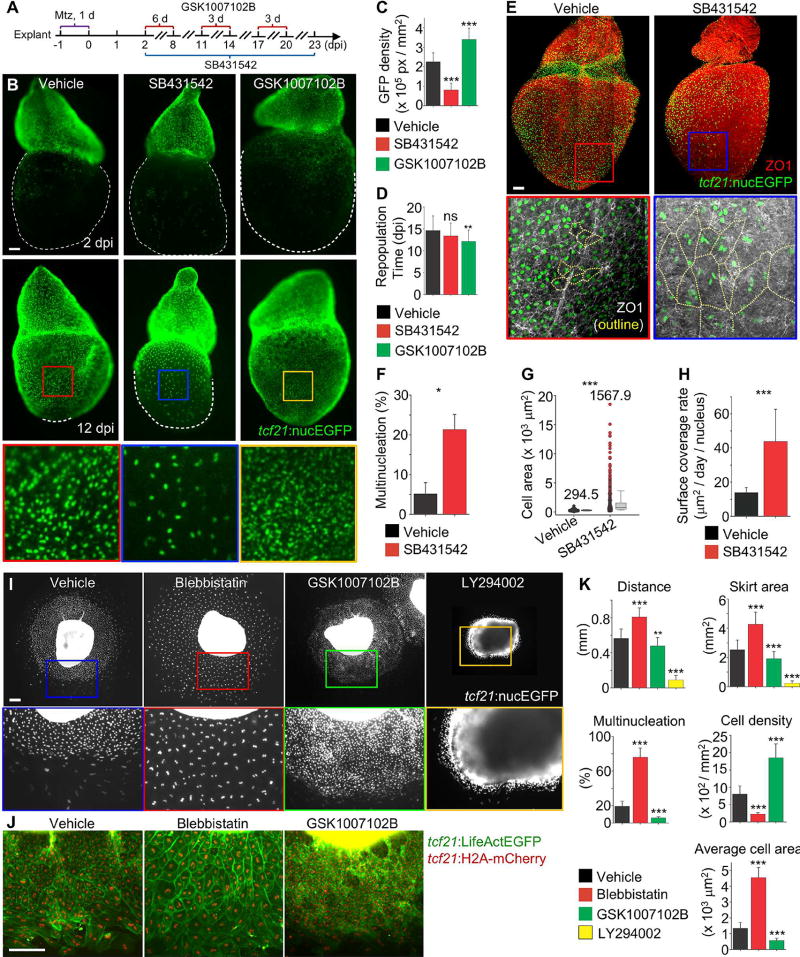
What is the function, if any, of creating a localized zone of endoreplicating cells during regeneration? To address this question, we screened ~340 known signaling pathway and kinase inhibitors for effects on the epicardial cell cycle (Elkins et al., 2016; Wang et al., 2015). One compound, GSK1007102B (at 0.5 µM), a potent inhibitor of Protein Kinase C and Akt (Rouse et al., 2009), generated an epicardium with ~75% higher cell density than vehicle-treated samples after 6 d treatment (Figures S6A and S6B). Conversely, treatment with the Tgf-β receptor inhibitor SB431542 (10 µM) inhibited the initial proliferative response of epicardial cells upon placement of whole hearts in explant culture (Figures S6C and S6D). We postulated that these small molecules could be used to deconstruct epicardial regeneration by promoting either the proliferative follower cell phenotype (GSK1007102B) or the non-proliferative leader cell phenotype (SB431542).
To test the effects of these compounds on epicardial regeneration, we ablated ventricular epicardial cells from whole hearts cultured ex vivo (Figures 7A and 7B). Hearts treated with GSK1007102B repopulated the ventricular surface with a ~51% higher estimated epicardial cellularity than vehicle-treated samples (Figure 7C), completing regeneration on average 2.5 d (or ~17%) faster (Figures 7B and 7D; Movie S8). Interestingly, SB431542 treatment enabled completion of regeneration at a similar rate as treatment with vehicle, albeit from an estimated 64% lower number of epicardial cells (Figures 7B–7D). SB431542-treated epicardial sheets were enriched throughout with cells possessing leader characteristics, with their cells ~432% larger on average than vehicle-treated cells and displaying a ~310% higher degree of multinucleation at 14 dpi (21.3% vs. 5.2%; Figures 7E–7G). Strikingly, this equates to ~215% greater surface coverage per nucleus (43.8 vs. 13.9 µm2/day/nucleus; Figure 7H), and an estimated ~264% greater surface coverage per cell (53.1 vs. 14.6 µm2/day/cell; calculated from the frequencies of multinucleate cells). This estimate of surface coverage on a per cell basis is conservative, as many multinucleated cells have more than two nuclei. These data, combined with assays revealing elevated migration velocities (Figure 4C), indicate that leader cells have enhanced regenerative potential. A third compound, LY294002 (at 50 µM), a potent inhibitor of phosphoinositide 3-kinase (PI3K), blocked epicardial regeneration, which resumed after drug washout (Figures S6E and S6F). This finding is consistent with known roles of PI3K in collective cell migration (Yamaguchi et al., 2015).
Figure 7. High Regenerative Capacity in Endoreplicated Cells.
(A) Schematic for experiments in (B–H). tcf21:NTR; tcf21:nucEGFP animals were used.
(B) Epicardial regeneration ex vivo in the presence of DMSO (Vehicle), 10 µm SB431542, or 0.5 µm GSK1007102B. White dashed lines outline the explants. Framed regions are enlarged to show details below. Scale bar, 100 µm.
(C) Quantification of GFP fluorescence density from images captured at 12 dpi. n = 18 (Vehicle), 18 (SB431542), and 13 (GSK1007102B), respectively. *** P < 0.001, Student’s t-test (compared with Vehicle). Bars indicate mean ± S.D.
(D) Quantification of timing to repopulate the ablated ventricular surface. n = 36 (Vehicle), 26 (SB431542), and 21 (GSK1007102B), respectively. ** P < 0.01; ns, not significant; Mann-Whitney Rank Sum Test (compared with Vehicle). Bars indicate mean ± S.D.
(E) Flattened images of whole-mounted hearts with ZO1 staining (red). Hearts were treated with DMSO (Vehicle) or SB431542 for 12 d (14 dpi). Framed regions are enlarged below, with nucEGFP shown in green, ZO1 in grayscale and outlined in yellow. Scale bar, 100 µm.
(F) Quantification of epicardial cell multinucleation in hearts treated with DMSO (Vehicle) or SB431542 for 12 d (14 dpi). n = 4 hearts for each. * P < 0.05, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(G) Quantification of epicardial cell area in hearts treated with DMSO (Vehicle) or SB431542 for 12 d (14 dpi). n = 822 (Vehicle) and 477 (SB431542) respectively. Numbers on the plot indicate mean values. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate S.D.
(H) Regenerative capacity on a per cell basis, with or without SB431542 treatment, measured by area covered per day by a digitalized nucleus. Area was measure at day 0 (2 dpi) and day 10 (12 dpi) of treatment, and cell density was measured at day 10 (12 dpi). One digitalized nucleus is defined as 30 GFP pixels. n = 18 explants for each group. *** P < 0.001, Mann-Whitney Rank Sum Test. Bars indicate mean ± S.D.
(I) Epicardial explant culture assays with Vehicle, Blebbistatin (10 µM), GSK1007102B (0.5 µM) or LY294002 (50 µM) treatment. tcf21:nucEGFP explants (shown in white) were plated for 5 d. Blebbistatin was added from day 3 to day 5, and GSK1007102B and LY294002 were added from day 0 to day 5. Framed regions are enlarged below to show details. Scale bar, 200 µm.
(J) Experiment as in (H) using tcf21:LifeAct-EGFP; tcf21:H2A-mCherry tissue. Scale bar, 200 µm.
(K) Quantification of epicardial growth. Skirt area is defined as the area covered by epicardial tissue growth from the explant. For migration distance and skirt area, n = 40 (Vehicle), 30 (Blebbistatin), 27 (GSK1007102B), and 19 (LY294002) explants, respectively. For nucleation, cell density, and average cell area, n = 10 explants for each treatment. *** P < 0.001; ** P < 0.01; Student’s t-test for migration distance and skirt area, Mann-Whitney Rank Sum Test for cell density, nucleation, and average cell area. All comparisons were versus Vehicle-treated group. Bars indicate mean ± S.D.
See also Figures S6 and S7, Movie S8.
To further examine leader and follower regenerative capacities, we inhibited cytokinesis by applying the myosin II inhibitor blebbistatin (Straight et al., 2003) at a low dose to epicardial explant cultures. Cultures were treated after 3 d of plating, a point when leader and follower features were established. This treatment sharply increased the frequency of multinucleation (~294% increase; Figures 7I–7K) and the average size (~238% increase; Figure 7K) of epicardial cells migrating from explants. Blebbistatin-treated epicardial cell sheets migrated ~45% farther and covered a ~69% larger surface area than vehicle-treated cell sheets, with a much lower (~72% lower) cell density (Figure 7K). Additional blebbistatin treatments with lower doses or for different durations did not affect pMLC staining and had similar effects (Figures S7A–S7D). Moreover, epicardial sheets treated with 10 µM SB431542 for 5 d covered the same distance and area as the vehicle-treated cell sheet, but with ~47% lower cell density and ~52% higher multinucleation (Figures S7E and S7F). We compared these effects with those of GSK1007102B on epicardial explant cultures (0.5 µM for 5 d). GSK1007102B–treated epicardial sheets had a ~131% higher cell density, generating cells that were ~58% smaller on average with a ~69% lower frequency of multinucleation than vehicle controls. These tissue sheets migrated ~15% shorter distances and covered ~24% less surface area than controls (Figures 7I–7K). Multinucleate cells were detectable at the leading edge with GSK1007102B treatment as in controls, whereas the leader cell region expanded during treatment with SB431542 (Figures S7G and S7H). Overall, our findings indicate that both leader and follower cell subpopulations have independent capacities to carry out regeneration. Yet, hypertrophic, polyploid cells generated through endoreplication possess an enhanced potential for surface coverage.
DISCUSSION
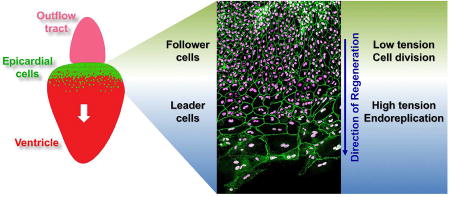
Here, we report a mechanism for tissue regeneration in which spatiotemporal control of the cell cycle partitions the cell source into subpopulations with distinct features. In the setting of extreme ablation, the epicardial sheet has an innate capacity to regenerate, whether on the beating myocardial surface or on artificial surfaces. We find that spared epicardial cells undergo endoreplication at the tissue front to form a hypertrophic, polyploid leader cell subpopulation. Our experiments indicate that increased tension on these cells based on their position is sufficient to induce endoreplication. Polyploid cells created by endoreplication support tissue regeneration through an enhanced capacity for surface coverage, whereas follower cells in the tissue sheet contribute through cytokinesis to increase cell number. In this way, source cells can be functionally specialized for features like migration and stress resistance, as well as cell renewal.
Leader cell existence is short-lived, as most multinucleate cells are normally eliminated after directing regeneration. Transience in this specific context leaves open the possibility that endoreplication may generally be more common during regeneration throughout animal phyla than appreciated. Indeed, in this study, longitudinal assessment using live cell cycle phase reporters in ex vivo models was necessary to reinforce what our in vivo observations suggested. Polyploidy has been associated with genome instability (Storchova and Pellman, 2004; Mayer and Aguilera, 1990), but in some settings is proposed to afford resistance to stress and cell death (Hassel et al., 2014; Zhang et al., 2014; Duncan, 2013; Mehrotra et al., 2008). The cause of apoptosis in polyploid epicardial cells described here is unclear; only one of the 23 dying polypoid cells we observed had just undergone nuclear division. After repopulation of the ventricular surface, new contacts were established between leader cells and cell density increased. It is possible that new contact inhibition, relaxation of mechanical tension, and genome instability each contribute to leader cell death. Ostensibly, the epicardium harnesses the high velocity and surface coverage of leader cells, but they are only temporary participants with reduced long-term fitness.
Although the locations and features of leader and follower cells are distinct, the cell phenotypes are in some ways interchangeable upon system perturbation. That is, blockade of epicardial cytokinesis and promotion of the leader cell phenotype does not disrupt regeneration, nor does promotion of the follower cell phenotype. Similarly, we saw that follower cells could reconstitute leader cell wavefronts that were experimentally ablated. This plasticity is reminiscent of findings describing murine liver regeneration, during which inhibiting polyploidization of hepatocytes after injury by cell-type-specific mutation of E2F7/8 does not affect liver regeneration, and blocking proliferation by telomere de-protection switches the mode of regeneration to endoreplication and hypertrophy (Chen et al., 2012; Pandit et al., 2012; Lazzerini Denchi et al., 2006). The capacity of epicardial cells to undergo either endoreplication or cytokinesis might facilitate specialization for functions like surface coverage or for cell amplification, and likely increases system robustness during regeneration. It is remarkable that epithelial cells, which have similar properties as epicardial cells, have not to our knowledge been reported to demonstrate the spatiotemporal regulation of phenotypes we have reported here.
Our evidence indicates that leader cell endoreplication is a product of high mechanical tension, which in the epicardial cell type impairs midbody formation and abscission of the cleavage furrow. How differences in mechanical tension manifest is unclear. It is possible that slowed follower cells increase cellular tension in leader cells by creating drag, and/or that customized ECM production by leader cells is a contributor. Additionally, the changing diameter of the ventricular chamber or growth path may generate tension in leading cells traversing the surface. Tension can increase cell division by exerting effects on cytoskeletal sensing proteins and their signal transduction to proteins like transcription factors (Benham-Pyle et al., 2015; Provenzano and Keely, 2011; Liu et al., 2007; Wang and Riechmann, 2007; Nelson et al., 2005; Kumar et al., 2004). For instance, cadherins transmit externally applied force to activate Hippo pathway transcription factors Yap/Taz, causing cell cycle entry (Benham-Pyle et al., 2015). Tension can also reduce cell division by impairing activities of proteins like cyclin-dependent kinases (Cdks) and increasing expression of the Cdk inhibitor p21 (Chang et al., 2008; Akimoto et al., 2000). The exact outcome of localized tension may be difficult to predict, and dependent on variables like a cell’s genetic program and mechanical properties, and the developmental stage of the tissue.
As mechanical force changes constitute a very early response to tissue injury, it is intriguing to speculate that injury-induced changes in tension are a generalizable mechanism to establish initial cell cycle dynamics during regeneration. Control of tension to strategically instruct cell cycle decisions thus has potential applications in tissue engineering and in directing therapeutic repair.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kenneth Poss (kenneth.poss@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
Zebrafish of the Ekkwill and Ekkwill/AB strains were maintained between 26–28.5°C with a lighting cycle of 14:10 h (light: dark) (Wang et al., 2011; Poss et al., 2002). Adults between 4 and 12 months of both sexes were used. Transgenic lines used in this study were Tg(tcf21:H2A-EGFP)pd251 (described below), Tg(tcf21:H2A-mCherry)pd252 (described below), Tg(tcf21:LifeAct-EGFP)pd253 (described below), Tg(tcf21:mAG-zGeminin)pd254 and Tg(tcf21: mKO2-zCdt1)pd255 (tcf21:FUCCI, described below), Tg(tcf21:mCherry-NTR)pd108 (tcf21:NTR) (Wang et al., 2015), Tg(tcf21:nucEGFP)pd41 (Kikuchi et al., 2011a), Tg(tcf21:DsRed2)pd37 (Kikuchi et al., 2011a). All transgenic strains were analyzed as hemizygotes. For epicardial ablation experiments in adults, animals were bathed for 24 hours in 10 mM Mtz (Sigma, M1547) as described and returned to water (Singh et al., 2012). Animal procedures were approved by Animal Care and Use Committee at Duke University.
Mouse
E11.5 embryos of the C57BL/6J mouse strain (Jackson Laboratories, cat#000664) were used for epicardial outgrowth assays. Sex could not be determined at this embryonic stage. Animal procedures were approved by Animal Care and Use Committee at University of Rochester.
METHOD DETAILS
Construction of tcf21:H2A-EGFP and tcf21:H2A-mCherry Zebrafish
The ORF sequence of zebrafish H2A F/Z (Pauls et al., 2001) was amplified using primers: CGGAATTCATGGCAGGTGGAAAAGC and CGGGATCCCCTGCGGTTTT CTGCTGG. The digested PCR fragment was inserted into the EcoR I-BamH I sites of pEGFP-N1 to make a pH2A-EGFP construct. To make a pH2A-mCherry construct, the EGFP cassette (Age I - Not I) in pH2A-EGFP was replaced with PCR amplified ORF of mCherry. To make BAC constructs, the translational start codon of tcf21 in the BAC clone DKEYP-79F12 was replaced with the H2A-EGFP (or mCherry)-SV40 ployA cassette by Red/ET recombineering technology (Gene Bridges) (Singh et al., 2012). The 5’ and 3’ homologous arms for recombination were a 50-base pair (bp) fragment upstream and downstream of the start codon, and were included in PCR primers to flank the H2A-EGFP (or mCherry)-SV40 polyA cassette. To avoid aberrant recombination between the H2A-EGFP (or mCherry)-SV40 polyA cassette and the endogenous loxP site in the BAC vector, we replaced the vector-derived loxP site with an I-Sce I site using the same technology. The final BAC was purified with Nucleobond BAC 100 kit (Clontech) and co-injected with I-Sce I into one-cell-stage zebrafish embryos. Stable transgenic lines with bright fluorescence but apparently normal mitosis and cell survival were selected.
Construction of tcf21:FUCCI Zebrafish
A construct containing mAG-zGeminin (1/100)-SV40 polyA or mKO2-zCdt1 (1/190)-SV40 polyA was used as a template to generate a PCR fragment with 50-bp homologous arms (Sugiyama et al., 2009). The tcf21:FUCCI (tcf21:mAG-zGem and tcf21:mKO2-zCdt1) BAC constructs were generated and injected as described above. Stable transgenic lines with moderate fluorescence were selected and tested for oscillation of red and green colors in explant cultures.
Construction of tcf21:LifeAct-EGFP Zebrafish
A 63-bp fragment (ATGGGTGTCGCAGATTTGATCAAGAAATTCGAAAGCATCTCAAAGGAAGAAGGCG GCAGCGGC) containing LifeAct probe sequence was fused to the 5’ end of the EGFP sequence followed by the SV40 polyA sequence, to make a LifeAct-EGFP-SV40 polyA cassette (Riedl et al., 2008). The tcf21:LifeAct-EGFP BAC construct was generated and injected as described above. Stable transgenic lines with bright fluorescence but apparently normal mitosis and cell survival were selected.
Cardiac Explants
Adult zebrafish hearts were collected and cultured as described previously (Cao et al., 2016; Cao and Poss, 2016; Wang et al., 2015). Briefly, zebrafish were euthanized using Tricaine (Sigma-Aldrich, A5040), and hearts were collected and rinsed several times with PBS. Rinsed hearts were transferred to dishes with DMEM medium (Thermo Fisher, cat#11965) plus 2 mM L-glutamine (Thermo Fisher, cat#25030), 10% fetal bovine serum (FBS, Thermo Fisher, SH30071), 1% MEM non-essential amino acids (Thermo Fisher, cat#11140), 100 U/ml penicillin, 100 µg/ml streptomycin (Thermo Fisher, cat#15140) and 50 µM 2-mercaptoethanol (Thermo Fisher, cat#21985) at 28°C and 5% CO2, while rotating at 150 r.p.m. For ex vivo epicardial ablation, dissected intact hearts were bathed for 24 h in 1 mM Mtz before washout, as described (Wang et al., 2015). For epicardial explant cultures, dishes were coated with 0.1% gelatin (Sigma, G1393) or 10 µg/ml fibronectin (Thermo Fisher, cat#33010018) overnight. Collagen (0.01%, VWR, cat#47743-656), poly-L-lysine (0.01%, Sigma-Aldrich, P4832) or fibrin gel (Kim et al., 2012) coatings were used for experiments examining different ECMs. Collected ventricular explants were cut into 3 pieces each and plated for 72 h in the same culture medium as for whole-mount explant culture, with or without chemicals. For EdU incorporation experiments, 25 µM EdU was added to culture medium for 1 h before fixation. For chemical screening, fresh tcf21:nucEGFP cardiac explants were treated with compounds in 24-well-plates, changing medium every other day over 6 d, with DMSO as a negative control. Images were acquired at day 6 to assess cell density. The GlaxoSmithKline Published Kinase Inhibitor Set (GSK-PKIS, 336 compounds) was used for screening, and only GSK1007102B potently increased proliferation (Elkins et al., 2016). We also tested several selected signaling inhibitors in addition to the library and identified SB431542 as a potent inhibitor of proliferation, LY294002 as a potent inhibitor of epicardial regeneration. SB431542 (Selleck Chemicals), LY294002 (Sigma), Blebbistatin (Sigma) and GSK1007102B (GSK) were dissolved in DMSO to make stock solutions with concentrations of 20 mM, 10 mM, 10 mM and 1 mM respectively.
Murine epicardial cells were plated by explant culture assays as previously described (Trembley et al., 2016). Briefly, pregnant C57BL/6J dams at 11.5 days post-coitum were anesthetized with ketamine via intraperitoneal injection and euthanized by cervical dislocation. The uterus was surgically removed, and the decidua carefully separated from the embryos in Hank’s Balanced Salt Solution (HBSS) pre-warmed to 37°C. The thoracic walls of embryos were removed us ing forceps to expose the chest cavity. Fetal hearts were then isolated by grasping the outflow tract and pulling the hearts away from the embryo body. Embryonic hearts were placed dorsal side down on collagen-coated chamber slides (BD Biosciences). After culturing overnight in M199 medium (HyClone) with 5% FBS at 37°C and 5% CO2, the hearts were removed, revealing a monolayer of epicardial cells attached to the collagen surface. Primary epicardial cells were cultured for 24 h in M199 with 1% FBS followed by 48 h in medium supplemented with TGF-β1 (10 ng/ml, R&D Systems, cat#100-B-001) to induce epithelial-to-mesenchymal transformation.
Histology and Microscopy
Freshly collected hearts and explants in culture were fixed with 4% paraformaldehyde (PFA) overnight at 4°C. After rinsing in PBS, hearts were blocked with 2% bovine serum albumin (BSA, VWR, cat#97061), 1% DMSO, 0.5% goat serum (Thermo Fisher, cat#16210) and 0.5% Triton X-100 in PBS for 1 h at room temperature (RT). Primary antibody, mouse anti-ZO1 antibody (Thermo Fisher, cat#33–9100) or rabbit anti-phospho-Histone H3 antibody (H3P, Cell Signaling Technology, cat#9701), was diluted in the blocking buffer and incubated with hearts overnight at 4°C. Hearts were then washed with PBS plus 0.5% Triton X-100 and incubated with the secondary antibody diluted in the blocking buffer for 1.5 h at RT. Hearts were stained with DAPI (Thermo Fisher, D3571) to visualize nuclei and mounted with Fluoromount G (Southern Biotechnology, cat#0100-01) between two coverslips, allowing imaging of both ventricular surfaces. Fluorescence images were captured using a Zeiss 700 confocal microscope (Wang et al., 2015; Poss et al., 2002).
Plated explants for immunostaining were cultured in glass bottom dishes (35 mm, MatTek). The tissue was fixed with 4% PFA for 10 min at RT, permeabilized with 0.5% Triton X-100 in PBS for 15 min, blocked with 1% BSA and 0.1% Tween-20 in PBS for 1 h at RT, incubated with the primary (2h at RT) and the secondary antibodies (1h at RT) in blocking buffer, and washed with 0.1% Tween-20 in PBS. An anti-Phospho-Myosin Light Chain 2 (Ser19) antibody (pMLC, Cell Signaling Technology, cat#3671) and an anti-β-catenin (Sigma, C2206) antibody were used. The secondary antibodies were Alexa Fluor 594 goat anti-mouse and goat anti-rabbit, and Alexa Fluor 633 goat anti-rabbit (Thermo Fisher). EdU was detected through a click reaction as described previously with fluorescent azide (Alexa Fluor 647, Thermo Fisher) (Salic and Mitchison, 2008). Fluorescent transgenes in floating explanted whole hearts were monitored using a Leica M205FA stereofluorescence microscope or a Zeiss Axiozoom microscope. Fluorescent transgenes in plated explant cultures were monitored using a Zeiss Axiozoom microscope. For time-lapse live imaging of plated explant cultures, cardiac explants were plated in glass bottom dishes (35 mm, MatTek). Confocal images were captured from day 3 or day 4 of culture at 5-min intervals, using a Zeiss 710 inverted confocal microscope equipped with a humidity chamber to preserve temperature (28°C) and CO2 (5%). For laser ablation, multinucleate leader cells were ablated by scanning 85 µm × 85 µm regions with a 405 nm laser at 30 mW power for 10 seconds. For high-magnification, time-lapse live imaging of explanted whole hearts, we used a Leica SP8 upright confocal microscope with a water immersion lens and INUG2F–UK lens incubator to preserve temperature and CO2. Explants were mounted in agarose and covered by medium. Images were taken at 10-min intervals for up to 5 days.
Murine epicardial cell cultures were stained as described previously (Trembley et al., 2016). Briefly, explant cultures were rinsed with Dulbecco’s phosphate-buffered saline (DPBS) and fixed with 4% PFA for 5 min at RT. Epicardial cells were then blocked with 3% BSA in PBS for 30 min at RT. Primary antibodies were diluted in the blocking buffer and added to explants for an overnight incubation at 4°C. Explants were then washed with DPBS and incubated with the secondary antibodies diluted in DPBS for 1 h at RT. Explants were mounted with mounting medium supplemented with DAPI (Vectashield) to visualize nuclei. Primary antibodies included mouse anti-WT1 (Thermo Fisher, MS-1837) and rabbit anti-ZO1 (Thermo Fisher, cat#402200). Secondary antibodies were Alexa Fluor 594 goat anti-mouse and Alexa Fluor 488 goat anti-rabbit (Thermo Fisher). Images were captured on an Olympus IX81 inverted scanning confocal microscope. Cell areas were quantified from 20× images using NIH ImageJ.
Laser Incision and Recoil
Cardiac explants carrying tcf21:LifeAct-EGFP; tcf21:H2A-mCherry reporters were plated in glass bottom dishes for 5 days before experiments. Laser incisions along a 12.8 µm line were applied using a Nd:YAG UV laser (355 nm; Continuum) interfaced on a Zeiss Axio Imager at a power range of 4–5.5 µJ (Peralta et al., 2007). Leader cell incisions were limited to the first two layers of the migrating front. Images were captured on the Zeiss Axio Imager with a 40 × / NA 1.2 water objective using Micro-Manager acquisition software. Live images were captured at 5 frames per second (fps) rate. The width of incision was measured in the middle of the cut every 0.6 second for 3 seconds following the end of laser cutting. The initial recoil velocity was linearly fitted from the increase of the cut width (Mayer et al., 2010; Peralta et al., 2007).
Mechanical Stretch Experiments
Elastic chambers with a culture surface area of 7 × 23 mm2 or 7 × 26 mm2 (Figure 5B) were generated by curing silicone rubber (Ecoflex 00–10, Smooth-On Inc.) in customized molds. The chambers were coated with 10 µg/ml fibronectin after sterilization with 70% ethanol and dried. Heart explants were plated as described above. Three days after plating, the culture chambers were mounted in a bioreactor (Figure 5C) with a linear actuator (UltraMotion) operated by LabVIEW software via a motor controller (Motion Mind 3, Solutions Cubed LLC). The culture surface was uniaxially stretched at a constant elongation rate for 1 h to extend 50%, 100% or 200% of the initial 7 mm dimension (10.5 mm, 50% stretch, 14 mm, 100% stretch or 21 mm, 200% stretch; Figures 5C and S5), and then held at the stretched length for 18 h. Stretching at 50% caused a milder increase in multinucleation than stretching at 100%. With 200% stretch, migrated epicardial cells lost connections with each other (Figure S5). Thus, only 100% stretched samples were chosen for further analysis. Fluorescent images were taken by using a Zeiss Axiozoom microscope. Alexa Fluor® 594-conjugated Wheat germ agglutinin (WGA, ThermoFisher, #W11262) was incubated with explant culture at 2.5 µg/ml for 5 min in PBS right before imaging.
Flow Cytometry
Ventricles from tcf21:NTR; tcf21:nucEGFP animals, either uninjured or at 7 or 14 dpi, were dissociated as described previously and stained with propidium iodide (PI) (Cao et al., 2016). Briefly, freshly collected ventricles ware rinsed with PBS, dissociated in 0.13U/ml Liberase DH (Roche, cat#05401054001), fixed with 2% PFA in PBS, and stained with 20 µg/ml PI (Sigma-Aldrich, P4170) with in the presence of 200 µg/ml RNase A (Sigma-Aldrich, R6513). EGFP+ cells were gated and assessed for PI fluorescence using a BD FACSCalibur system.
QUANTIFICATION AND STATISTICAL ANALYSIS
Clutchmates, or hearts collected from clutchmates, were randomized into different groups for each treatment. No animal or sample was excluded from the analysis unless the animal died during the procedure. All experiments were performed with at least 2 biological replicates. Sample sizes were chosen on the basis of previous publications and experiment types, and are indicated in each Figure legend. Quantification of cell area, density and nucleation were assessed by a person blinded to the treatments. For quantification of nucleation of whole-mounted samples, Z-stack images were used, and only nuclei of the outermost layer (Cao et al., 2016) were counted using the NIH ImageJ software (Figure S1A and S1B). The nuclei of each multinucleate cell tend to be close to each other and often are overlapping (Figure S1C and Movie S1). In fixed samples, these nuclei often presented as large nuclei with irregular shapes; thus, were defined as multinucleate cells (Figure S1D). When the signal of ZO1 staining traversed nuclei, the nuclei and the cells on each side of the ZO1 signal were discarded from quantification. When one cell was registered with no nucleus, that cell was discarded from quantification. All statistical values are displayed as Mean +/− Standard Deviation (S.D.). Sample sizes, statistical tests, and P values are indicated in the figure or figure legends. Student’s t-tests (two-tailed) were applied when normality and equal variance tests were passed. A Mann-Whitney Rank Sum test was used when these failed. Analysis of covariance (ANCOVA) was applied in regression analysis.
Supplementary Material
Live imaging of the apex of a cardiac explant. tcf21:NTR; tcf21:H2A-EGFP hearts were incubated with 1 mM Mtz for 24 h and cultured until 12 dpi. Then, the explant was subjected to live imaging for 61.5 h. The tcf21:H2A-EGFP reporter is shown in white. Timing, hh:mm:ss.
A 3-day culture of a tcf21:FUCCI explant subjected to live imaging for 13 h. Timing, hh:mm:ss.
High-magnification view of three cells in Movie S2, showing cytokinesis (left), endomitosis (middle) and endocycling (right). Timing, hh:mm:ss.
tcf21:LifeAct-EGFP; tcf21:H2A-mCherry epicardial explants were cultured for 4 days before live imaging for 15.5 h. Timing, hh:mm:ss.
High-magnification view of explanted tcf21:LifeAct-EGFP; tcf21:H2A-mCherry epicardial cells, showing normal cytokinesis (left) and cytokinesis failure (endomitosis; right). Cropped from Movie S4. Timing, hh:mm:ss.
A 5-d tcf21:LifeActEGFP epicardial explant culture was subjected to laser incision and assessed for recoil velocity. LifeActEGFP is shown in grayscale.
A 3-d tcf21:FUCCI epicardial explant culture was subjected to live imaging and laser ablation. Timing, hh:mm:ss.
Shown are explants imaged daily over a 11-d period following epicardial ablation (2–12 dpi) and chemical treatment (day 0 - day 10), visualized for tcf21:nucEGFP+ cells.
Highlights.
-
-
Regeneration of zebrafish epicardium involves both cell division and endoreplication
-
-
Endoreplication constructs a leading edge of large, multinucleate cells
-
-
Mechanical tension induces endoreplication in epicardial tissue sheets
-
-
Spatiotemporal control of cell cycle dynamics for tissue regeneration
Acknowledgments
We thank J. Burris, S. Miller, B. Thomas, and N. Mariano for fish care, W. Zuercher for providing the GSK-PKIS library, T.W. Pfeiler and G.C. Engelmayr for assistance with bioreactor design and construction, and D. Fox, J. Bear, M. Bagnat, and J. Kang for comments on the manuscript. This work was supported by postdoctoral fellowships (14POST20230023 and 16POST30230005) from American Heart Association (AHA) to J.C., a grant (15SDG25710444) from AHA to J.W., graduate research fellowships from National Science Foundation (NSF) to C.P.J. (2013126035) and B.D.C. (2014175655), a predoctoral training grant position (T32-GM007184) from National Institutes of Health (NIH) to A.H.C., a predoctoral training grant position (T32-HL066988-15) from NIH and predoctoral fellowship (16PRE30490009) from AHA to M.A.T., a network grant from Foundation Leducq to N.B. and K.D.P., and grants from NIH to S.D.T. (R00 HD074670), E.M.S. (R01 HL120919), D.P.K. (R01 GM033830), N.B. (R01 HL132389), and K.D.P. (R01 HL131319 and R01 HL081674).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, J.C. and K.D.P.; Methodology, J.C, K.D.P., C.P.J., N.B., D.P.K. and E.M.S.; Formal Analysis, J.C., B.D.C, A.D.S. and S.D.T.; Investigation, J.C., J.W., C.P.J., A.H.C., M.A.T., J.J.B., and A.L.D.; Resources, D.P.K. and N.B.; Writing – Original Draft, J.C.; Writing – Review & Editing, J.C., E.M.S., D.P.K., N.B. and K.D.P.; Funding Acquisition, K.D.P.
References
- Abmayr SM, Balagopalan L, Galletta BJ, Hong SJ. Cell and molecular biology of myoblast fusion. Int. Rev. Cytol. 2003;225:33–89. doi: 10.1016/s0074-7696(05)25002-7. [DOI] [PubMed] [Google Scholar]
- Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–56. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto S, Mitsumata M, Sasaguri T, Yoshida Y. Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1) Circ. Res. 2000;86:185–90. doi: 10.1161/01.res.86.2.185. [DOI] [PubMed] [Google Scholar]
- Benham-Pyle BW, Pruitt BL, Nelson WJ. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science. 2015;348:1024–7. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Navis A, Cox BD, Dickson AL, Gemberling M, Karra R, Bagnat M, Poss KD. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–43. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Poss KD. Explant culture of adult zebrafish hearts for epicardial regeneration studies. Nat. Protoc. 2016;11:872–81. doi: 10.1038/nprot.2016.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S, Desdouets C. Polyploidization of liver cells. Adv. Exp. Med. Biol. 2010;676:123–35. doi: 10.1007/978-1-4419-6199-0_8. [DOI] [PubMed] [Google Scholar]
- Chanet S, Martin AC. Mechanical force sensing in tissues. Prog. Mol. Biol. Transl. Sci. 2014;126:317–52. doi: 10.1016/B978-0-12-394624-9.00013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SF, Chang CA, Lee DY, Lee PL, Yeh YM, Yeh CR, Cheng CK, Chien S, Chiu JJ. Tumor cell cycle arrest induced by shear stress: Roles of integrins and Smad. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3927–32. doi: 10.1073/pnas.0712353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Ouseph MM, Li J, Pecot T, Chokshi V, Kent L, Bae S, Byrne M, Duran C, Comstock G, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 2012;14:1192–202. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 2005;171:153–64. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 2013;24:347–56. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Invest. 2012;122:3307–15. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J, Romanov S, Sepetov N, Huang XP, Roth BL, et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol. 2016;34:95–103. doi: 10.1038/nbt.3374. [DOI] [PubMed] [Google Scholar]
- Engel FB, Schebesta M, Keating MT. Anillin localization defect in cardiomyocyte binucleation. J. Mol. Cell Cardiol. 2006;41:601–12. doi: 10.1016/j.yjmcc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty SA, Lindblom J, Loftus PD, Redd MJ, Edes K, Davey CF, Krishnegowda V, Rosenblatt J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature. 2017 doi: 10.1038/nature21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjipanayi E, Mudera V, Brown RA. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil. Cytoskeleton. 2009;66:121–8. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- Happe CL, Engler AJ. Mechanical Forces Reshape Differentiation Cues That Guide Cardiomyogenesis. Circ. Res. 2016;118:296–310. doi: 10.1161/CIRCRESAHA.115.305139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassel C, Zhang B, Dixon M, Calvi BR. Induction of endocycles represses apoptosis independently of differentiation and predisposes cells to genome instability. Development. 2014;141:112–23. doi: 10.1242/dev.098871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–62. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC. Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol. Biol. Cell. 2012;23:1846–59. doi: 10.1091/mbc.E11-12-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kano Y, Amano M, Onishi H, Kaibuchi K, Fujiwara K. Rho-kinase--mediated contraction of isolated stress fibers. J. Cell Biol. 2001;153:569–84. doi: 10.1083/jcb.153.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15895–900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011a;138:2895–902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011b;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Rubin N, Huang Y, Tuan TL, Lien CL. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat. Protoc. 2012;7:247–55. doi: 10.1038/nprot.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Murphy R, Robinson P, Wei L, Boriek AM. Cyclic mechanical strain inhibits skeletal myogenesis through activation of focal adhesion kinase, Rac-1 GTPase, and NF-kappaB transcription factor. FASEB J. 2004;18:1524–35. doi: 10.1096/fj.04-2414com. [DOI] [PubMed] [Google Scholar]
- Lazzerini Denchi E, Celli G, de Lange T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 2006;20:2648–53. doi: 10.1101/gad.1453606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2011;27:157–84. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- LeGoff L, Lecuit T. Mechanical Forces and Growth in Animal Tissues. Cold Spring Harb. Perspect. Biol. 2015;8:a019232. doi: 10.1101/cshperspect.a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Liu WF, Nelson CM, Tan JL, Chen CS. Cadherins, RhoA, and Rac1 are differentially required for stretch-mediated proliferation in endothelial versus smooth muscle cells. Circ. Res. 2007;101:e44–52. doi: 10.1161/CIRCRESAHA.107.158329. [DOI] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr. Biol. 2013;23:2224–32. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen CD, Pedersen JT, Venning FA, Singh LB, Moeendarbary E, Charras G, Cox TR, Sahai E, Erler JT. Hypoxia and loss of PHD2 inactivate stromal fibroblasts to decrease tumour stiffness and metastasis. EMBO Rep. 2015;16:1394–408. doi: 10.15252/embr.201540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Depken M, Bois JS, Julicher F, Grill SW. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467:617–21. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- Mayer VW, Aguilera A. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat. Res. 1990;231:177–86. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 2008;22:3158–71. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 2012;22:1166–75. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11594–9. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noria S, Xu F, McCue S, Jones M, Gotlieb AI, Langille BL. Assembly and reorientation of stress fibers drives morphological changes to endothelial cells exposed to shear stress. Am. J. Pathol. 2004;164:1211–23. doi: 10.1016/S0002-9440(10)63209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand G, King RW. Understanding cytokinesis failure. Adv. Exp. Med. Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL. When bigger is better: the role of polyploidy in organogenesis. Trends Genet. 2015;31:307–15. doi: 10.1016/j.tig.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PC, Cornelissen PW, Toussaint MJ, Lamers WH, de Bruin A. E2F8 is essential for polyploidization in mammalian cells. Nat. Cell Biol. 2012;14:1181–91. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- Pauls S, Geldmacher-Voss B, Campos-Ortega JA. A zebrafish histone variant H2A.F/Z and a transgenic H2A.F/Z:GFP fusion protein for in vivo studies of embryonic development. Dev. Genes Evol. 2001;211:603–10. doi: 10.1007/s00427-001-0196-x. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. Actin stress fibres. J Cell Sci. 2007;120:3491–9. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- Peralta XG, Toyama Y, Hutson MS, Montague R, Venakides S, Kiehart DP, Edwards GS. Upregulation of forces and morphogenic asymmetries in dorsal closure during Drosophila development. Biophys. J. 2007;92:2583–96. doi: 10.1529/biophysj.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials. 2006;27:4881–93. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Provenzano PP, Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J. Cell Sci. 2011;124:1195–205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid K, Lu J, Zimmet JM, Jones MR. Roads to polyploidy: the megakaryocyte example. J. Cell Physiol. 2002;190:7–20. doi: 10.1002/jcp.10035. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–7. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley PR. An epicardial floor plan for building and rebuilding the mammalian heart. Curr. Top. Dev. Biol. 2012;100:233–51. doi: 10.1016/B978-0-12-387786-4.00007-5. [DOI] [PubMed] [Google Scholar]
- Roland AB, Ricobaraza A, Carrel D, Jordan BM, Rico F, Simon A, Humbert-Claude M, Ferrier J, McFadden MH, Scheuring S, et al. Cannabinoid-induced actomyosin contractility shapes neuronal morphology and growth. Elife. 2014;3:e03159. doi: 10.7554/eLife.03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse MB, Seefeld MA, Leber JD, McNulty KC, Sun L, Miller WH, Zhang S, Minthorn EA, Concha NO, Choudhry AE, et al. Aminofurazans as potent inhibitors of AKT kinase. Bioorg. Med. Chem. Lett. 2009;19:1508–11. doi: 10.1016/j.bmcl.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415–20. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell. 2012;22:879–86. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–7. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20812–7. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamori Y, Deng WM. Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev. Cell. 2013;25:350–63. doi: 10.1016/j.devcel.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley MA, Velasquez LS, Small EM. Epicardial Outgrowth Culture Assay and Ex Vivo Assessment of Epicardial-derived Cell Migration. J. Vis. Exp. 2016 doi: 10.3791/53750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzahor E, Poss KD. Cardiac regeneration strategies: Staying young at heart. Science. 2017;356:1035–1039. doi: 10.1126/science.aam5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–74. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. 2015;522:226–30. doi: 10.1038/nature14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Riechmann V. The role of the actomyosin cytoskeleton in coordination of tissue growth during Drosophila oogenesis. Curr. Biol. 2007;17:1349–55. doi: 10.1016/j.cub.2007.06.067. [DOI] [PubMed] [Google Scholar]
- Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, Zhu W, Fajardo G, Noseda M, Nakamura K, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525:479–85. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Mizutani T, Kawabata K, Haga H. Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin beta1 and PI3K. Sci. Rep. 2015;5:7656. doi: 10.1038/srep07656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Mehrotra S, Ng WL, Calvi BR. Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS Genet. 2014;10:e1004581. doi: 10.1371/journal.pgen.1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 2011;121:1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live imaging of the apex of a cardiac explant. tcf21:NTR; tcf21:H2A-EGFP hearts were incubated with 1 mM Mtz for 24 h and cultured until 12 dpi. Then, the explant was subjected to live imaging for 61.5 h. The tcf21:H2A-EGFP reporter is shown in white. Timing, hh:mm:ss.
A 3-day culture of a tcf21:FUCCI explant subjected to live imaging for 13 h. Timing, hh:mm:ss.
High-magnification view of three cells in Movie S2, showing cytokinesis (left), endomitosis (middle) and endocycling (right). Timing, hh:mm:ss.
tcf21:LifeAct-EGFP; tcf21:H2A-mCherry epicardial explants were cultured for 4 days before live imaging for 15.5 h. Timing, hh:mm:ss.
High-magnification view of explanted tcf21:LifeAct-EGFP; tcf21:H2A-mCherry epicardial cells, showing normal cytokinesis (left) and cytokinesis failure (endomitosis; right). Cropped from Movie S4. Timing, hh:mm:ss.
A 5-d tcf21:LifeActEGFP epicardial explant culture was subjected to laser incision and assessed for recoil velocity. LifeActEGFP is shown in grayscale.
A 3-d tcf21:FUCCI epicardial explant culture was subjected to live imaging and laser ablation. Timing, hh:mm:ss.
Shown are explants imaged daily over a 11-d period following epicardial ablation (2–12 dpi) and chemical treatment (day 0 - day 10), visualized for tcf21:nucEGFP+ cells.