Abstract
The covalent coupling of fatty acids to proteins provides an important mechanism of regulation in cells. In eukaryotes, cysteine fatty acylation (S-fatty acylation) has been shown to be critical for protein function in a variety of cellular pathways as well as microbial pathogenesis. While methods developed over the past decade have improved the detection and profiling of S-fatty acylation, they are hampered in their ability to characterize endogenous protein S-fatty acylation levels under physiological conditions. Furthermore, understanding the contribution of specific sites and levels of S-fatty acylation remains a major challenge. To evaluate S-fatty acylation of endogenous proteins as well as determine the number of S-fatty acylation events, we developed the acyl-PEG exchange (APE) that utilizes cysteine-specific chemistry to exchange S-fatty acylation sites with mass-tags of defined size, which can be readily observed by western blot. APE provides a readily accessible approach to investigate endogenous S-fatty acylation from any sample source, with high sensitivity and broad applicability that compliments the current toolbox of methods for thioester-based post-translational modifications.
Keywords: S-fatty-acylation, PEGylation, Mass-shift, Post-translational modification quantification
Introduction
The covalent modification of cysteines with long-chain fatty acids provides a selective means of altering the hydrophobicity of proteins and regulating their trafficking, stability, and activity (Chamberlain & Shipston, 2015; Peng, Thinon, & Hang, 2016). In eukaryotes cysteines are predominantly acylated with 16-carbon palmitic acid (S-palmitoylation), as well as longer (stearic acid) and unsaturated (oleic acid) fatty acids (Muszbek, Haramura, Cluette-Brown, Van Cott, & Laposata, 1999; Thinon, Percher, & Hang, 2016), collectively referred to as S-fatty acylation. S-palmitoylation is a reversible modification, coupled and removed by DHHC-family acyltransferases (Fukata & Fukata, 2010; Linder & Deschenes, 2007) and palmitoyl-protein thioesterases (Lin & Conibear, 2015a, 2015b), respectively. The role of S-fatty acylation in the physiology and regulation of numerous classes of proteins (Chamberlain & Shipston, 2015) drives our interest in how this critical and potentially dynamic modification is regulated and quantitatively influences specific cellular pathways.
Current efforts in the investigation of S-fatty acylation have improved the selection of available methods to detect and enrich fatty acylated proteins, as well as our understanding of their regulatory mechanisms (Brett et al., 2014; Liang et al., 2001). Alkyne-modified fatty acid chemical reporters have been extensively used to metabolically label S-fatty acylated proteins for detection by fluorescence, or enrichment for mass spectrometry analysis using bioorthogonal ligation methods (Charron et al., 2009; Hang, Wilson, & Charron, 2011). Exploiting hydroxylamine (NH2OH)-mediated cleavage of thioesters has allowed the selective capture of S-fatty acylated proteins by acyl-biotin exchange (ABE) (Kang et al., 2008; Wan, Roth, Bailey, & Davis, 2007) or acyl-resin capture (acyl-RAC)(Forrester et al., 2011), enabling the detection of S-fatty acylation of proteins under physiological conditions, including tissue samples which are inaccessible to chemical probes.
These methods have been crucial to profile novel S-fatty acylation targets in various cell lines(Caballero et al., 2016; Chesarino et al., 2014; Peng et al., 2016) and tissues(Kang et al., 2008), as well as the validation of S-fatty acylation sites. However, these methods have inherent limitations to quantifying S-fatty acylation, including the S-fatty acylated fraction, and the number of S-fatty acylation sites present in endogenous proteins. By quantitatively measuring levels of S-fatty acylation, the effect of the modification on protein localization, function, and cellular phenotypes can be better understood.
To assess the levels of S-fatty acylation of endogenous proteins, we have developed the acyl-PEG exchange (APE), which selectively replaces S-fatty acids with maleimide-functionalized polyethylene glycol (mPEG-mal) reagents of defined mass. These ‘mass tags’ induce a mobility shift when the tagged protein sample is analyzed by gel electrophoresis, proportional to the number of S-fatty acylated cysteines. The APE utilizes the thioester specificity of NH2OH to cleave cysteine-coupled fatty acid groups, followed by selective cysteine coupling of the mPEG-mal. This enables facile detection by western blot of the multiple S-fatty acylation populations of endogenous proteins, without the interference of exogenous chemical reporters or affinity enrichment of proteins. In summary, The APE is an accessible method to monitor S-fatty acylation levels and dynamics(Percher et al., 2016) of endogenous proteins. Complementing current tools, the APE provides a strong example for the applicability of mass-tag labeling for the investigation of thioester-based post-translational modifications.
Basic Protocol: S-fatty acylation mass-tag labeling using maleimide-PEG
The Acyl-PEG Exchange (APE) method utilizes cysteine-specific chemistry to label S-fatty acylated proteins with defined mass tags for detection by western blot. This protocol enables the analysis of the whole-cell lysate without need for enrichment or the addition of an excess of chemical probes, providing a semi-quantitative analysis of S-fatty acylation levels.
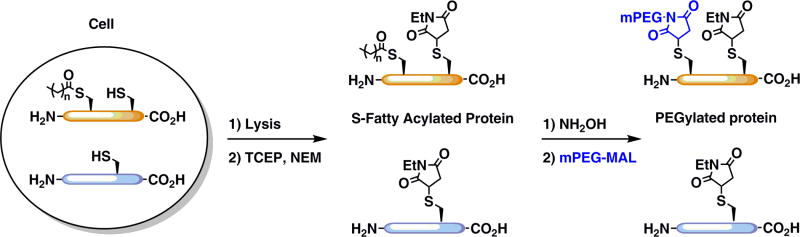
Starting with a protein sample obtained from tissue, cell culture, or recombinant protein, non-modified cysteines are capped with NEM to prevent further reaction with a methoxy-PEG-maleimide (mPEG-mal) tag. After removal of excess NEM by methanol-chloroform-H2O precipitation (MCHP), fatty acids coupled to cysteine via a thioester bond are cleaved with hydroxylamine (NH2OH). Excess NH2OH is removed and exposed cysteines are labeled with mPEG-mal at stoichiometries and levels reflective of the original number of S-fatty acylation events (Fig 1). The samples are then separated by SDS-PAGE and analyzed by western blot with appropriate antibodies.
Figure 1.
Mass-tag analysis of S-fatty acylation. With APE, cell lysates are reduced with TCEP, and free cysteine residues are capped with NEM. S-fatty acid groups are removed by NH2OH, and the exposed cysteines are reacted with mPEG-Mal. Proteins are separated by SDS/PAGE and analyzed by western blot, enabling the detection of both unmodified and S-fatty acylated proteins.
Materials list
0.25% trypsin-EDTA
PBS sterile
Sodium Dodecyl Sulfate (SDS)
(tris(2-carboxyethyl)phosphine) (TCEP)
Phenylmethanesulfonyl fluoride (PMSF)
Benzonase
Bicinchoninic acid assay kit (BCA).
N-ethyl maleimide (NEM)
Hydroxylamine Hydrochloride (NH2OH)
5 and 10 kDa Methoxypolyethylene glycol maleimide (mPEG-mal, Sigma)
4× Laemmli Buffer
Beta mercaptoethanol
Microfuge/Eppendorf tubes (1.5 ml)
Nitrocellulose membrane (0.2 µm)
Speed Vacuum (e.g. Centrivap Concentrator, Labconco)
Sonicator (e.g. Ultrasonic Cleaner, VWR)
Steps
Prepare cell lysate
-
1
Trypsinize cells, transfer the cell suspension to an Eppendorf tube (1.5 mL) and spin down for 5 min at 300 × g at 4°C.
We recommend 200 µg of proteins as starting material for one experiment. Two confluent wells of a 6-well plate are typically sufficient.
-
2
Wash cells with PBS three times to remove fetal bovine serum proteins from the medium.
The cell pellet can immediately be used for lysis, or frozen in liquid nitrogen and stored at −80°C for future use.
-
3
Lyse cell samples by resuspending spun down cell pellet in chilled lysis buffer (see recipe).
We typically lyse two confluent wells of a 6-well plate (962 mm2) in 150 µL buffer. Brief sonication facilitates complete lysis, assessed by loss of viscosity. It is essential that all buffers used throughout this protocol are within the pH range of 6.5–7.5 to ensure cysteine selectivity of maleimide
When a clear solution is obtained, add EDTA to a final concentration of 5 mM (Stock concentration of 0.5 M, pH 8, Sigma). Measure the protein concentration using BCA protein assay (unit 3.4; Olson and Markwell, 2007) or other equivalent method, and adjust protein concentration to 2 mg/mL with lysis buffer. Aliquot 92.5 µL (200 µg) of total protein to a separate Eppendorf tube.
EDTA inhibits benzonase if added directly to the lysis buffer. Benzonase hydrolyzes nucleic acids and is essential to reduce the viscosity of the sample.
Capping of exposed cysteines with NEM
-
4
Treat each sample with 5 µL of 200 mM neutralized TCEP (1/20, final concentration 10 mM) for 30 minutes at room temperature with nutation.
This first step is used to reduce any disulfide bonds in the samples. In contrast to the commonly used reducing agents dithiothreitol (DTT) or β-mercaptoethanol (BME), TCEP does not contain thiols which will quench NEM and will not hydrolyze the thioester bonds(Ji et al., 2013). We have used as little as 50 µg of total protein for our experiments, though 100–200 µg is recommended. To facilitate future steps in the protocol, we recommend starting volumes of 140 µL max protein lysate, otherwise during later steps the volume will exceed that of a standard microcentrifuge tube.
-
5
Cap the reduced cysteines by adding 2.5 µL of NEM from a freshly prepared 1 M stock in ethanol (1/40, final concentration 25 mM). Incubate for 2 hours at room temperature with nutation.
To reduce evaporation of ethanol from the NEM stock solution while pipetting, keep chilled on ice.
-
6
Terminate the NEM-capping reaction by methanol-chloroform-H2O precipitation (4:1.5:3, relative to sample volume): (A) sequentially add methanol (400 µL), chloroform (150 µL) and distilled H2O (300 µL) (all prechilled on ice). Mix by inversion several times and centrifuge at 20,000× g for 5 min at 4°C. Two separate phases should be observed with a protein pellet between the two. (B) Carefully remove the aqueous (top) layer, being careful not to disturb the protein pellet, and add 1 mL of prechilled methanol. Gently invert several times (the pellet should sink to the bottom of the tube), and centrifuge again at 20,000 × g for 3 min at 4°C. (C) Decant the supernatant and wash the protein pellet once more with 800 µL of prechilled methanol. Centrifuge again, decant the methanol, and speed-vacuum the protein pellet for 10 minutes.
Extra care should be taken while precipitating low protein amounts (<50 µg) with methanol-chloroform-H2O. During the methanol washes, vortexing should be avoided as it could break the protein pellet and could result in a loss of sample. For small protein volumes, it is recommended to use longer (10 minutes) spins to ensure complete collection of the pellet fragments.
-
7
To ensure complete removal of NEM from the protein pellets, repeat step 6 two times by resuspending the dried protein pellet in TEA buffer with 4% SDS (wt/vol).
We have routinely stored dried protein pellets at −80 °C during the 3× chloroform-methanol precipitation, to continue work the next day. We recommend minimizing the number of freeze–thaw cycles.
NH2OH mediated thioester cleavage, and mPEG-maleimide alkylation
-
8
After the third methanol-chloroform-H2O precipitation, resuspend the protein pellet in 60 µL TEA buffer containing 4% SDS, 4 mM EDTA and split in two for the +/− NH2OH conditions. For the + NH2OH sample, add 90 µL 1 M neutralized NH2OH dissolved in TEA buffer 0.2% Triton X-100 to obtain a final concentration of 0.75 M NH2OH. For the negative (− NH2OH) control add 90 µL TEA buffer 0.2% Triton X-100. Incubate the samples at room temperature for 1 hour with nutation.
EDTA is essential for efficient cleavage of the thioester, while the protease inhibitor mixture or PMSF should be omitted, as these reagents can interfere with the NH2OH reactivity
-
9
Remove the NH2OH using the previously described methanol-chloroform-H2O precipitation
-
10
Resuspend the dried pellet in 30 µL TEA buffer containing 4% SDS and 4 mM EDTA. Sonicate briefly (~5 sec) to ensure complete resolubilization of the pellet, spin down, and dilute the sample with 90 µL TEA buffer with 0.2% Triton X-100 and 1.33 mM mPEG-Mal (5 or 10 kDa) for a final concentration of 1 mM mPEG-Mal. Incubate samples for 2 hours at room temperature with nutation before a final methanol-chloroform-H2O precipitation.
Analyze by SDS-PAGE and western blot
-
11
Resuspend the pellet in 50 µL 1× Laemmli buffer and then heat for 5 min at 95 °C. Typically, 15 µL of the sample is loaded in 4–20% Criterion-TGX Stain Free polyacrylamide gels (Bio-Rad), separated by SDS/PAGE, and analyzed by western blot.
For every mPEG-mal coupled to the protein, we observe a mass-shift corresponding to twice the mass of the tag used (see Troubleshooting).
We typically use 4–20% acrylamide gels, though non-gradient gels are also suitable. For higher molecular proteins (>100 kDa), a lower percent acrylamide gel is recommended to ensure the sufficient separation to observe a 10 kDa mass shift.
A good control to assess the completion of the APE is to blot against calnexin (1:2,000 ab22595; Abcam; HRP-conjugated goat anti-Rabbit, 1:5000, DC03L, Calbiochem))
Reagents and Solutions
Use deionized water (dH2O), Milli-Q-purified water or equivalent in all recipes and protocol steps.
TEA Buffer 10× (50 mM Triethanolamine, 150 mM NaCl, pH7.3)
372.98 mg of triethanolamine
438.3 mg of sodium chloride
Add 40 mL of dH2O
Adjust pH to 7.3 with concentrated HCl
Add water to 50 mL
Store up to 1 year at room temperature
TEA Buffer 4% SDS
5 mL TEA Buffer 10×
2 g SDS
45 mL dH2O
Store up to 1 year at room temperature.
TEA Buffer 0.2% Triton X-100
5 mL TEA Buffer 10×
100 µL Triton X-100
44.9 mL dH2O
Store up to 1 year at room temperature.
To ensure accurate addition of Triton X -100, we usually measure the needed volume with a pipette and drop the whole tip into the bottle.
TEA buffer with 0.2% Triton X -100 should be prepared the night before to ensure the detergent has completely gone into solution.
Lysis Buffer
Transfer the needed volume of TEA 4% SDS buffer into a new tube and add 1× protease inhibitor cocktail (Roche, EDTA free), 1,500 units/mL benzonase, 5 mM PMSF (Sigma) just before use.
Always prepare fresh before use.
1× Laemmli Buffer
50 µL 4× Laemmli buffer
5 µL beta mercaptoethanol
150 µL TEA buffer 4%SDS
Prepare fresh before use.
Hydroxylamine (NH2OH)
347.4 mg of hydroxylamine hydrochloride
Add 4 mL of TEA buffer 0.2% Triton X-100
Neutralize to pH 7.3 with NaOH 10N
Adjust volume to 5 mL with TEA buffer 0.2% Triton X-100
Always prepare fresh before use.
NEM: 184.96 g/mole (Cysteine selective at PH 6.5 – 7.5)
25.03 mg of NEM
Add 200 µL of Ethanol
Always prepare fresh before use.
PEG-MAL 5 kDa (1.33 mM)
6.66 mg of 5 kDa PEG-maleimide (Sigma)
Add 1 mL of TEA Buffer 0.2% Triton X-100
Always prepare fresh before use.
PEG-MAL 10 kDa (1.33 mM)
13.33 mg of 5 kDa PEG-maleimide (Sigma)
Add 1 mL of TEA Buffer 0.2% Triton X-100
Always prepare fresh before use.
200 mM TCEP solution
286.6mg TCEP-HCl
Add 4 mL of dH2O
Neutralize to pH 7.3 with NaOH 10N
Add water to 5 mL
Always prepare fresh before use.
200 mM phenylmethylsulfonyl fluoride (PMSF)
34.84 mg of PMSF
1 mL of ethanol
COMMENTARY
Background Information
Further discussion of APE, ABE and metabolic labeling with chemical probes
Though the use of ABE and chemical probes to detect and profile S-fatty acylation are essential tools for the field, their experimental designs have inherent limitations that might lead to erroneous interpretations of results when quantifying S-fatty acylation.
Alkyne chemical probes for metabolic labeling
The use of alkyne-containing fatty-acid probes to metabolically label proteins requires the incubation of the cell culture with the alkyne-fatty acid (usually in the µM range for several hours), followed by lysis, immunoprecipitation and copper-catalyzed azide-alkyne cycloaddition (CuAAC). Despite the versatility of probe design and means of detection, there are several caveats that should considered during the interpretation of results:
Chemical probes do not allow for the quantification of S-fatty acylation levels. The measurement of S-fatty acylation is by fluorescence, which cannot measure the total amount of non-S-fatty acylated protein. This prevents the quantification of the S-fatty acylated fraction.
Incubation of the cell culture with exogenous lipids might influence intracellular lipid levels, altering physiological concentrations of free fatty acid.
Exogenous alkyne-fatty acid competes with endogenous fatty acids for the metabolic labeling of proteins, and both can be incorporated. Additionally, proteins with low S-fatty acylation turnover will be poorly labeled with chemical probes, interfering with detection by fluorescent signal.
Chemical probes can be metabolized to different chain lengths and degrees of saturation, potentially leading to false positive results.
Acyl Biotin Exchange
The ABE and acyl-RAC methods both bypass the need for exogenous chemical probes to metabolically label S-fatty acylated proteins. S-fatty acylation can be measured under physiological conditions, and from samples inaccessible to metabolic labeling (tissues). These methods can quantify what fraction of the protein is S-fatty acylated, but encounter some limitations:
The ABE and acyl-RAC can observe quantitative difference between non-modified (apo), and S-fatty-acylated fractions, assuming complete enrichment of the S-fatty-acylated sample. This cannot distinguish between proteins with different numbers of S-fatty acylation sites, and can only determine what fraction of the protein is modified at least once.
For comparing proteins with multiple S-fatty acylation sites, as long as there is at least one S-fatty acylated protein, no additional cysteine S-fatty acylation can be detected.
The ABE exploits the NH2OH-selective cleavage of thioesters to detect S-fatty-acylated proteins. Similar to the APE, this method cannot confirm the structure and chain length of the S-fatty acid and care should be given to confirm through alternative methods the characteristics of the modification.
Theoretical limitations of APE
Though the APE enables the simultaneous detection of multiple acylation levels, the total signal intensity of the +NH2OH sample is consistently greater than that of the −NH2OH control, suggesting APE might not be completely quantitative. We speculate this might be due to differences in transfer to the western blot membrane, or PEG-mediated differences in exposure of the protein to antibodies or HRP. While the signal differences challenge the accuracy of the S-fatty acylation ratios, the APE still provides a reliable method to determine the qualitative measurement of S-fatty acylation states (Roberts et al., 2016), as well as a semi-quantitative comparative assay between different conditions (Nathan P. Westcott, In Press; Yokoi et al., 2016).
The APE provides a sensitive assay to detect the presence of multiple S-fatty acylation events of endogenous proteins. While we have applied the APE to numerous peripheral and integral membrane proteins, we have noticed that our most robust results are observed for small-medium range proteins. Larger proteins 90 kDa+ have been shown to be palmitoylated using chemical probes, but have exhibited no mass shift under our conditions, even with the use of larger (10 kDa) mPEG-mal mass tags. Other papers since have published similar mass-shift based assays, with varied conditions (see Assay Variations).
Assay Variations
Since the publication of our method in April, 2016, a similar mass-tag based approach was reported (Yokoi et al., 2016). Briefly, samples were lysed in buffer containing 4% SDS, with and without 8 M urea, reduced with 25 mM TCEP at 55° C, and capped with 50 mM NEM for 3 h at room temperature. After treatment with 1 M NH2OH, the sample was PEGylated with 20 mM mPEG-Mal.
Critical Parameters points to consider before beginning
When preparing samples for the APE, several aspects in the experimental design that are not detailed in the protocol merit prior consideration. Starting volumes of >50 µg per condition ensure an easier precipitation and are recommended for all conditions, including negative controls. To minimize sample handling and errors, we routinely conduct the NEM capping on samples of 100 µg, then split the sample in two for +/− NH2OH incubation, and further labeling with mPEG-mal. Additionally, it is recommended that the western blot of different proteins are conducted in sequence or in separate blots if it is unclear the number of S-fatty acylation events of the protein, as this might lead to band overlap.
Troubleshooting suggestions for overcoming or avoiding commonly encountered problems
Pellet Breaking Apart During Methanol-Chloroform-H2O Precipitation (MCHP)
During MCHP, we observe pellet fragmentation for starting sample sizes of <50 µg. It is possible to complete the experiment with longer centrifugation (10 min per spin) and careful decanting, though fragmentation can be avoided by increasing the sample size to >100 µg.
Dried pellet won’t completely resuspend in 4% SDS
After incubating the dried protein pellet in 4% SDS for 10–15 minutes, sonicate briefly (5 seconds) to disrupt the remaining insoluble pellet.
Mass Shift in (−) NH2OH Control
A band shift in the − NH2OH control could be due to incomplete cysteine-capping with NEM, or background labeling of non-cysteine nucleophiles with the mPEG-mal.
-
-
A time course assay is recommended for mPEG-mal labeling to ensure that a complete mass shift has occurred for the protein of interest, without background bands appearing in the − NH2OH control.
-
-
As maleimide–cysteine selectivity occurs at a pH range of 6.5–7.5, all buffers should be tested to ensure they are within the correct pH range.
Incomplete mass shift
An incomplete reaction of the sample with mPEG-mal can be observed using a positive control. Common causes we have encountered include:
-
-
Incomplete labeling of the exposed cysteines with mPEG-mal, either due to insufficient incubation times, or an interfering reagent in the reaction conditions such as high concentrations of TCEP(Henkel, Rockendorf, & Frey, 2016; Kantner & Watts, 2016) interacting with the mPEG-mal. Residual NEM present during NH2OH cleavage would also result in alkylation of the newly revealed cysteine.
-
-
Incomplete cleavage of thioesters with NH2OH. The presence of EDTA is critical during NH2OH thioester cleavage. It is also recommended that a time course for NH2OH incubation is conducted for the protein of interest.
Absence of mass shift
A positive control is recommended to ensure that proper conditions were present throughout the experiment. Blotting against endogenous calnexin has proven a robust, reliable control in our hands in a variety of cell lines.
Larger mass shift than expected
Under our conditions we routinely observe a mass shift double the size of the mass-tag labeling the protein (e.g. a 10 kDa increase in protein migration, when labeled with a single 5 kDa mPEG-mal). The PEG tag used to label S-fatty acylated proteins has different chemical properties than the denatured protein backbone. These likely result in a different charge/mass ratio, altering its migration in the SDS-PAGE.
A larger mass shift than expected, equivalent to multiple mPEG-mal couplings, might suggest either background labeling (check − NH2OH control), or additional S-fatty acylation sites on the protein.
Anticipated Results
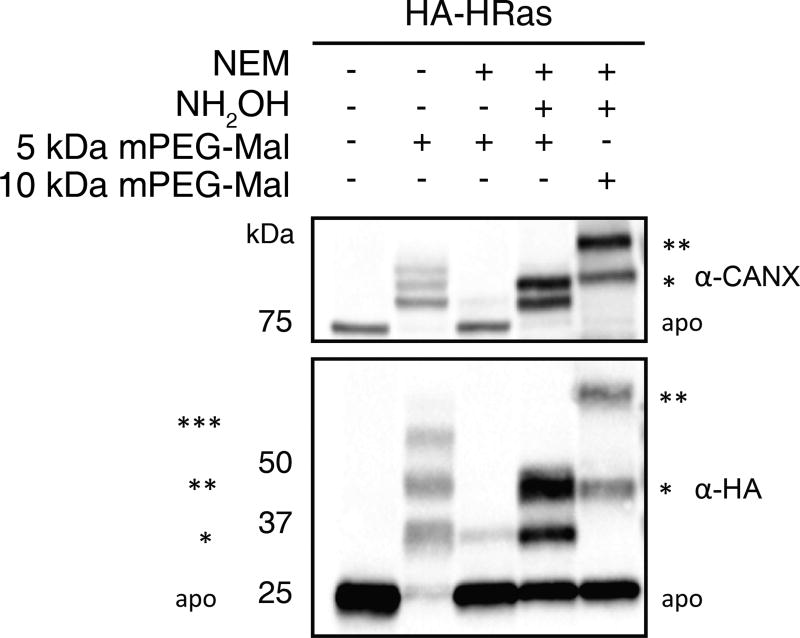
An example of a standard APE western blot (Fig. 2) demonstrates an observed mass shift, as well as the expected mass shift of the positive control endogenous calnexin.
Figure 2.
APE enables mass-shift based detection of protein S-fatty acylation levels. Experimental replicate of Fig.2 from Percher et al. (2016)(Percher et al., 2016). HEK293T transfected with HA-HRas were lysed and total cell lysates were subjected to APE with NEM (25 mM), NH2OH (0.75 M), and mPEG-Mal (1 mM), and compared with negative controls. Samples were analyzed by western blot using anti-HA and anti-CANX antibodies. The number of PEGylation events are indicated by asterisks (*). Apo refers to non-PEGylated protein.
Time Considerations
Prepare cell lysate
Harvesting, lysing, and measuring protein concentration of tissue culture samples (1–4 different conditions) typically takes 1–2 hours.
Capping of exposed cysteines with NEM
Reducing and capping the protein samples requires 2.5 hours of incubation time. Three consecutive methanol-chloroform-H2O precipitations usually require 2–3 hours. The samples can be frozen after any methanol-chloroform-H2O precipitation and stored at −80 °C to be continued the next day.
NH2OH mediated thioester cleavage and mPEG-maleimide alkylation
These steps usually require 5 hours to complete.
Analysis by SDS-PAGE and western blot
Running the gel should take 1–2 hours. Depending on the apparatus used for western blot, the transfer will take between 30 minutes to several hours, followed by 4–5 hours of blocking, incubation with primary and secondary antibodies, and washing.
Acknowledgments
This work was supported in part by a National Science Foundation graduate fellowship (A.P.); a Marie Sklodowska-Curie actions for a postdoctoral fellowship (E.T.); and National Institutes of Health-National Institute of General Medical Sciences Grant R01GM087544 (H.C.H.).
Citations
- Brett K, Kordyukova LV, Serebryakova MV, Mintaev RR, Alexeevski AV, Veit M. Site-specific S-acylation of influenza virus hemagglutinin: the location of the acylation site relative to the membrane border is the decisive factor for attachment of stearate. J Biol Chem. 2014;289(50):34978–34989. doi: 10.1074/jbc.M114.586180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero MC, Alonso AM, Deng B, Attias M, de Souza W, Corvi MM. Identification of new palmitoylated proteins in Toxoplasma gondii. Biochim Biophys Acta. 2016;1864(4):400–408. doi: 10.1016/j.bbapap.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. 2015;95(2):341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131(13):4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- Chesarino NM, Hach JC, Chen JL, Zaro BW, Rajaram MV, Turner J, Yount JS. Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 2014;12:91. doi: 10.1186/s12915-014-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52(2):393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. 2010;11(3):161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- Hang HC, Wilson JP, Charron G. Bioorthogonal chemical reporters for analyzing protein lipidation and lipid trafficking. Acc Chem Res. 2011;44(9):699–708. doi: 10.1021/ar200063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel M, Rockendorf N, Frey A. Selective and Efficient Cysteine Conjugation by Maleimides in the Presence of Phosphine Reductants. Bioconjug Chem. 2016;27(10):2260–2265. doi: 10.1021/acs.bioconjchem.6b00371. [DOI] [PubMed] [Google Scholar]
- Ji Y, Leymarie N, Haeussler DJ, Bachschmid MM, Costello CE, Lin C. Direct detection of S-palmitoylation by mass spectrometry. Anal Chem. 2013;85(24):11952–11959. doi: 10.1021/ac402850s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456(7224):904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantner T, Watts AG. Characterization of Reactions between Water-Soluble Trialkylphosphines and Thiol Alkylating Reagents: Implications for Protein-Conjugation Reactions. Bioconjug Chem. 2016;27(10):2400–2406. doi: 10.1021/acs.bioconjchem.6b00375. [DOI] [PubMed] [Google Scholar]
- Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276(33):30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife. 2015a;4:e11306. doi: 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DT, Conibear E. Enzymatic protein depalmitoylation by acyl protein thioesterases. Biochem Soc Trans. 2015b;43(2):193–198. doi: 10.1042/BST20140235. [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8(1):74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Muszbek L, Haramura G, Cluette-Brown JE, Van Cott EM, Laposata M. The pool of fatty acids covalently bound to platelet proteins by thioester linkages can be altered by exogenously supplied fatty acids. Lipids. 1999;34(Suppl):S331–337. doi: 10.1007/BF02562334. [DOI] [PubMed] [Google Scholar]
- Nathan P, Westcott JPF, Molina Henrik, Hang Howard C. Chemical proteomics reveals ADP-ribosylation of small GTPases during oxidative stress. Nature Chem Bio. doi: 10.1038/nchembio.2280. (In Press) (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Thinon E, Hang HC. Proteomic analysis of fatty-acylated proteins. Curr Opin Chem Biol. 2016;30:77–86. doi: 10.1016/j.cbpa.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc Natl Acad Sci U S A. 2016;113(16):4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BJ, Svoboda RA, Overmiller AM, Lewis JD, Kowalczyk AP, Mahoney MG, Wahl JK., 3rd Palmitoylation of Desmoglein 2 Is a Regulator of Assembly Dynamics and Protein Turnover. J Biol Chem. 2016;291(48):24857–24865. doi: 10.1074/jbc.M116.739458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinon E, Percher A, Hang HC. Bioorthogonal Chemical Reporters for Monitoring Unsaturated Fatty-Acylated Proteins. Chembiochem. 2016;17(19):1800–1803. doi: 10.1002/cbic.201600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2(7):1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 Depalmitoylating Enzymes. J Neurosci. 2016;36(24):6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]