Abstract
Aberrant glycosylation is a hallmark of various disease states, including cancer, and effective detection and discrimination between healthy and diseased cells is an important challenge for the diagnosis and treatment of many diseases. Here, we describe the use of boronic acid functionalized synthetic lectins (SLs) in an array format for the differentiation of structurally similar cancer associated glycans and cancer cell lines; discrimination is based on subtle variations in glycosylation patterns. We further demonstrate the utility of our SLs in recognizing glycoproteins with up to 50-fold selectivity, even in 95% human serum. Given their robust and selective nature, these SLs were able to effectively distinguish (a) five structurally similar glycans with 94% accuracy; (b) seven normal, cancerous and metastatic colon cancer cell lines, including three isogenic cell lines, with 92% accuracy; and (c) these same seven cell lines using a guided statistical analysis to improve our analysis to 97% accuracy. In total, these data suggest that an SL-based array will be useful for the diagnosis of cancer.
Introduction
The intracellular and extracellular biomarkers displayed by healthy and diseased cells provide unique signatures by which these cells can be distinguished. For example, in healthy cells, post-translational glycosylation of proteins plays a critical role in cell–cell interactions and in cell signaling.1 However, aberrant protein glycosylation is a hallmark of numerous diseases including inflammation and cancer, thus providing a means for the detection and classification of healthy and diseased states. Related to cancer, distinguishing between healthy and cancer cells that possess either low or high metastatic potentials typically relies on detecting subtle variations in the types and levels of specific biomarkers (e.g., DNA, RNA, and proteins) using high-affinity, target-selective sensors, e.g. antibodies. Regardless of the analyte, these approaches all require prior knowledge of the markers targeted and no specific biomarker or combination of biomarkers has been identified to sufficiently differentiate between healthy, cancerous/non-metastatic and cancerous/metastatic cell types. An alternative to this “lock-and-key” approach2–6 would be to use cross-reactive recognition elements as part of a sensor array.
Cross-reactive sensor arrays incorporate multiple receptors with different affinities such that each component has a selective and unique interaction with the targeted analyte(s). As a result, the response from the entire array produces a fingerprint pattern characteristic of the analyte to which it is responding. That is to say that classification is not based on the response from a single receptor, but rather it is the composite response from the entire array that allows for identification and classification of the analyte. This practice has often been referred to as the “electronic nose” approach,7–13 though, in this case, used for solution-based analysis.
While natural lectins (sugar binding proteins) display cross-reactivity, and lectin arrays can often offer an effective approach to cancer diagnostics, the methodology is often complex and the constituents are of inherently low stability and high cost.14–17 Here we describe an alternate approach based on the covalent yet reversible binding between boronic acid functionalized synthetic lectins (SLs) and cancer associated glycans and glycoproteins. This design does not require previous knowledge of the biomarkers targeted; rather it is focused on identifying changes in glycosylation patterns, a factor that is known to play a significant role in oncogenesis and metastasis.
In cancerous cells, the expression of specific glycan structures can be increased, decreased, or even newly expressed. These changes often co-opt cellular signaling pathways to promote growth, division and metastasis.1 For example, sialyl Lewis X (sLex) and sialyl Lewis A (sLea) (Fig. 1A) are overexpressed in breast, colon and pancreatic cancers,1 and the increased expression of sLex is known to enhance tumor metastasis.18–21 Tests to detect specific aberrant glycosylation events are used for both initial disease diagnosis and monitoring disease progression yet suffer from limitations including a high number of false positives and a reliance on inherently unstable and costly antibodies or natural lectins.14–17 For example, elevated levels of CEA (carcinoembryonic antigen), an aberrantly glycosylated glycoprotein, are associated with an increased risk of colon cancer relapse and metastasis.15 However, the test for CEA is only effective in 4% and 25% of Stage I and II cancers, respectively, which is problematic for a cancer diagnostic because it is during these early stages when the disease is most effectively treated.22
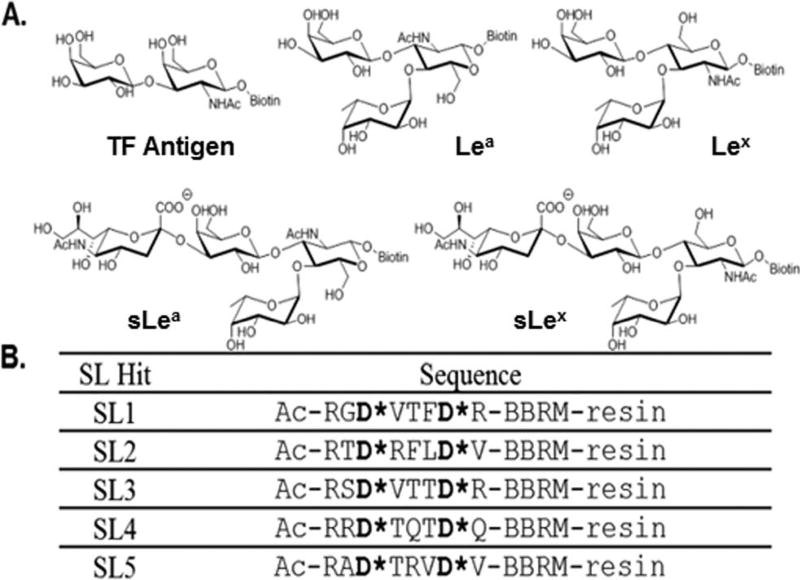
Fig. 1.
(A) The structures of biotinylated cancer associated glycans used in this study. (B) The sequences of the SLs used for validation studies and in the array assessments.
The development and use of boronic acid functionalized synthetic lectins (SLs) for saccharide detection and cancer diagnosis is a rapidly growing field.23–36 Boronic acids are incorporated into the SLs to enhance glycan binding via their ability to form covalent yet reversible bonds to the 1,2- and 1,3-diols present on many saccharides. These small molecule SLs generally show enhanced stability compared to antibodies and natural lectins, and it has been shown that incorporation of synthetic lectins into an array format allowed for the recognition and discrimination between simple monosaccharides and oligosaccharides in neutral aqueous media as well as real-world beverage samples, i.e. sweet tea with added Splenda.37 Further advances using cross-reactive nanoparticle-conjugated polymer based arrays have shown utility in differentiating normal, cancerous and metastatic cell types.38
We previously described the design, synthesis and utility of boronic acid functionalized peptide-based SLs in binding to glycoproteins36 and highlighted efforts in library design optimization and peptide sequencing.35 SLs, that were both cross-reactive and up to 5-fold selective for a particular glycoprotein, were identified.
Herein, we report the identification and characterization of three additional SLs that bind to proof-of-concept glycoproteins with up to 50-fold selectivity, even in complex matrices (i.e., human serum). Additionally, a four-component SL array was used to detect and differentiate five structurally similar cancer associated glycans (Fig. 1), as well as one ‘healthy’ and six cancer cell lines with high classification accuracy. By combining selective and cross-reactive SLs within the array, the selectivity of an individual SL need not be high as each sensor need only be incrementally different to create an array that maximizes variation in the array response to different analytes.39,40 Further analyses using directed partitioning, based on similarities in metastatic potential, was used to enhance the classification accuracy. Our results demonstrate the utility of using SL arrays for the diagnosis of cancer. Furthermore, since the analyte for which each SL was selected is not found on any of the cancer-associated cells studied, our array displays inherent adaptability.39,40 That is to say that this relatively small array was able to “learn” and accurately classify never before seen analytes.39,40
Results and discussion
Employing the same approach used to identify SL1 and SL2,35,36 SL3, SL4 and SL5 (Fig. 1B) were identified by screening our bead-based fixed position library with fluorescein isothiocyanate (FITC)-tagged versions of ovalbumin (OVA) and porcine stomach mucin (PSM). These SLs were subsequently re-synthesized and their selectivity and cross-reactivity evaluated using OVA, PSM, BSM (bovine submaxillary mucin) and BSA (bovine serum albumin). OVA, PSM and BSM are all glycoproteins, and it is noteworthy that the two mucins contain the same type of glycans but to differing extents and displayed in different environments. BSA, which is not glycosylated, was used as a control for non-specific protein binding.
SL selectivity studies
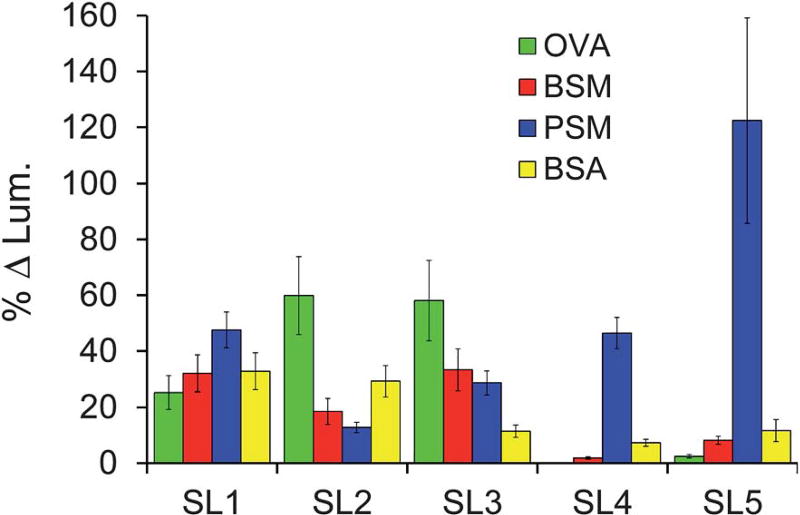
To control for differences in the extent of labeling or glycosylation, the fluorescence intensity of a similarly sized set of the SL library was used as a reference. The fluorescence intensity of the library was subtracted from the fluorescence intensity of the re-synthesized SL incubated with the same FITC-tagged glycoprotein (Fig. S1, ESI†), providing a change in fluorescence intensity upon binding. A percent change in binding was obtained by dividing this difference by the fluorescence intensity of the library (Fig. 2). To compare the ability of each SL to differentially bind to glycoproteins, a selectivity factor was obtained by dividing the percent increase for each analyte by the percent increase of the weakest binder for that SL (Table 1). The library was chosen as the reference because it provides a control containing all of the potential cross-reactive elements that could interfere with our assessment of binding selectivity. Outliers from the control were removed using the studentized t-test at the second quartile to give an accurate average for standardization purposes.
Fig. 2.
Percent change in luminosity of each identified SL towards four different analytes (analyte identification indicated in the legends above). Error bars represent the standard error of the percent change relative to the control as this propagated uncertainty is based on the variance between replicate measurements for the sample and control reference.
Table 1.
Selectivity factors for each SL screened against four different glycoproteinsa
| OVA | BSM | PSM | BSA | |
|---|---|---|---|---|
| SL1 | 1.0 | 1.3 | 1.9 | 1.3 |
| SL2 | 4.7 | 1.4 | 1.0 | 2.3 |
| SL3 | 5.1 | 2.9 | 2.5 | 1.0 |
| SL4 | 0.1b | 1.0 | 24.8 | 3.9 |
| SL5 | 1.0 | 3.4 | 49.9 | 4.8 |
The fold selectivity of an SL for one glycoprotein over another can be obtained by dividing their respective selectivity factors. SL1 and SL2 data from Bicker et al.35
The fold selectivity for SL4 was determined using BSM as the reference. OVA was not used as the reference because the %Δ luminosity was practically zero and dividing by such a small number resulted in fold selectivities that were quite meaningless.
The data for SL1 and SL2 have been previously described35 and are included in Fig. 2 and Table 1 for comparison. Here we see that SL1 is completely cross-reactive, binding with no more than 2-fold selectivity for any one analyte. In contrast, SL2 shows modest selectivity for binding OVA. The 3- and nearly 5-fold selectivity SL2 shows over BSM and PSM, respectively, demonstrated the ability of this approach to distinguish between similar analytes. However the 2-fold selectivity of SL2 for OVA over BSA suggests high non-specific, background binding for this SL, thereby decreasing its potential utility in a diagnostic array.
The newly reported SL3 was selected from screening the library against OVA, and showed only 2-fold selectivity towards OVA over BSM and PSM, while exhibiting relatively low background binding, as indicated by the 5-fold selectivity over BSA. SL4 and SL5 were identified from screening the library for PSM binders. Although SL4 displays an impressive 25-fold selectivity for PSM over BSM, it exhibits only ~6-fold selectivity for PSM over BSA. Thus, while exhibiting some degree of selectivity and showing a particular preference for binding certain analytes (i.e., PSM vs. BSM), this SL can also be considered cross-reactive with respect to PSM vs. BSA. As such, this SL is an ideal candidate for inclusion in a sensor array because it possesses differential analyte binding. Note that SL4 shows virtually no affinity for OVA and as such the percent change in luminosity relative to the library control is very small (0.15%). Thus, for the discussion of selectivity, presented in Table 1, BSM was used as the weakest binder because it was not reasonable to use OVA and divide by such a small number (e.g. PSM selectivity vs. OVA is 250).
Similar to SL4, SL5 displayed exquisite selectivity, exhibiting 50-fold selectivity for PSM over OVA and ~15-fold selectivity over BSM. The excellent selectivity of SL4 and SL5 for PSM over BSM (~25- and ~15-fold selectivity, respectively) is particularly impressive because these two glycoproteins possess identical types of glycans, though to a different extent and differentially displayed.41–43 These results suggest that these SLs not only bind to the saccharide, but also the protein. Nevertheless, it is important to recognize that we have previously shown that glycans are significant for the SL–glycoprotein interaction.36
The robustness of the SL-glycoprotein interaction was assessed using SL2 and SL5 with differing percentages of human serum (0, 25, 50 and 95%) in screening buffer. Both SLs retained excellent selectivity for the respective glycoproteins in all concentrations of serum (Fig. S2, ESI†). Control experiments confirmed that no serum components caused any changes in the assay response (Fig. S3, ESI†). To examine the contribution of valency, dissociation constants (Kd) were determined for both the bead-based polyvalent SL5 and a monovalent SL5. The dynamic nature of the beads44 (i.e., being a gel resin) allows for multiple interactions between bead-based SLs and the many glycans expressed on PSM. Therefore, incubating polyvalent, bead-based SL5 with varying concentrations of fluorescently labeled PSM (having a polyvalent display of glycans) yielded a Kd of 2.5 ± 0.29 µM (Fig. S4, ESI†).45 A fluorescence polarization (FP) assay was used to measure the affinity of the fluorescently-labeled, monovalent SL5 (FITC-SL5) for PSM.46 However, saturation of the FP signal was not observed because of limited glycoprotein solubility (Fig. S5, ESI†), thus Kd values could not be determined. Nevertheless, the observed response validated the assay and suggested that the Kd for the monovalent SL5-PSM interaction is significantly higher than 10 µM, the highest concentration tested. These results indicate that the polyvalent nature of the beads is critical for high affinity binding and suggest that multiple SLs on a single bead interact with each glycoprotein.
Glycan competition studies
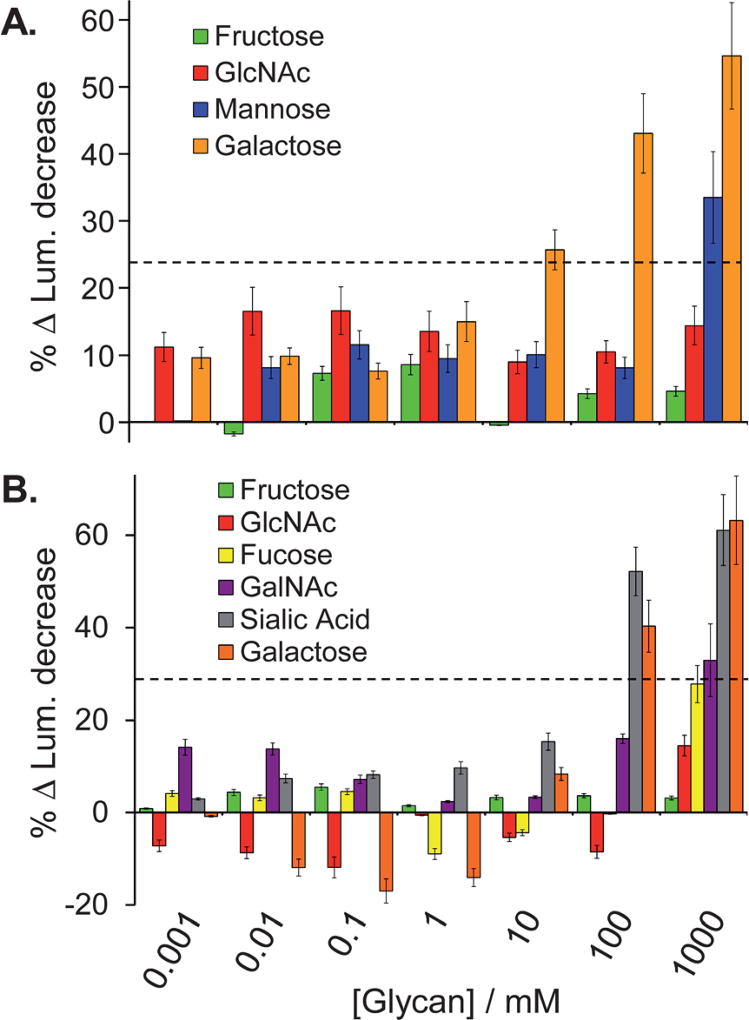
Glycan competition assays were used to identify the glycan structure(s) that were responsible for SL2-OVA and SL5-PSM binding. For these studies, SL2 was selected over SL3 because of the higher selectivity shown for OVA over BSM and PSM, while SL5 was chosen over SL4 because of the larger signal intensity response. In this study, varying concentrations of different monosaccharides were independently incubated with equal portions of resin-bound SLs and a constant concentration of the FITC-glycoprotein (0.1 mg mL−1) that the SL preferentially binds. The glycans used in the study of SL2 were those found on OVA, namely galactose, mannose and N-acetylglucosamine (GlcNAc).36,47 For SL5, galactose, GlcNac, sialic acid, fucose and N-acetylgalactosamine (GalNAc),36,48 which are all found on PSM, were used. Fructose was used to probe non-specific saccharide binding between the SLs and glycoproteins because it is one of the strongest known 1 : 1 boronic acid binders.49,50 It was expected that effective competition between a monosaccharide and a FITC-glycoprotein, for binding to the resin-bound SL, would result in a decrease in luminosity. Such a decrease in the binding signal would suggest that a particular monosaccharide was important for glycoprotein binding to the SL. Note that the response values in Fig. 3 have been mathematically defined such that increasing bar height corresponds with more effective competition (intensity = (initial − final)/initial) to more clearly show the competition trends.
Fig. 3.
Percent change in luminosity for the glycan competition studies used to explore the SL2-OVA (A) and SL5-PSM(B) binding interactions (analyte identification indicated in the legends above). Error bars represent the standard error of the percent change relative to the control as this propagated uncertainty is based on the variance between replicate measurements for the sample and control reference. The dashed lines in each panel indicate a competition threshold, based on three standard deviations above the noise. Signal response above this threshold indicates significant competition.
It is noteworthy that effective competition was only observed at high concentrations of the monovalent saccharides being studied. This result is likely due to the fact that these monosaccharide guests poorly compete with the multivalent display of saccharides found on the glycoproteins for binding to the multivalent display of SLs on the bead, as multivalent interactions are nearly always stronger than the sum of the monovalent interactions.51 Also note that reducing glycosides and non-reducing monosaccharides (as found on the glycoproteins) were both used for these competition experiments, and that both classes of compounds showed similar trends in the data. The results from the competition studies with the reducing sugars are shown in Fig. 3 and the non-reducing sugar competition study data are summarized in the supporting information (Fig. S6, ESI†). Given that reducing monosaccharides can isomerize to the furanose form to provide a diol that more effectively binds to boronic acids in a 1 : 1 manner,52–56 these monosaccharides provide a more stringent test of ligand binding than the non-reducing saccharides because they provide a “dual-competition” pathway. Namely via 1 : 1 furanose–boronic acid binding as well as the proposed pyranose–SL binding predicted for the saccharides found on the glycoproteins. Note that significant competition was defined as being three standard deviations above the noise (indicated by the dashed lines in Fig. 3). For this analysis the standard error for 1000 mM galactose was used because it displays the largest variance, thus for SL2 and SL5, the ‘cut-off’ percent change in luminosity was 23% and 29%, respectively.
For SL2, no appreciable decrease in luminosity was observed with N-acetylglucosamine (GlcNAc) even at concentrations as high as 1 M (Fig. 3A, red bars) indicating that N-acetylglucosamine does not interact with SL2, and thereby suggesting that this glycan is not critical for binding SL2 to OVA. In contrast, a significant decrease in luminosity was observed with both 1 M mannose and with as little as 10 mM galactose (Fig. 3A, blue and orange bars, respectively). These data indicate that SL2 is likely binding primarily with galactose, and to a lesser extent with mannose, both found on OVA. Competitive binding with non-reducing saccharides also showed significant competition with mannose (see ESI†). These results are particularly impressive because they suggest that SL2 interacts with both terminal (galactose) and core (mannose) glycan structures.47 Given that galactose is typically considered to be a weak boronic acid binder for simple 1 : 1 binding, the observed competition suggests that the binding site in this system is organized in a manner suitable for binding this sugar.31
Particularly small changes in luminosity corresponding to the addition of GlcNAc or fucose to SL5 (Fig. 3B, red and yellow bars, respectively) suggest that these glycans were not crucial for SL5 binding to PSM. Conversely, GalNAc competed for binding at high concentrations (Fig. 3B, purple bars), while both sialic acid and galactose displayed significant competition with PSM for binding to SL5 at concentrations above 100 mM (Fig. 3B, gray and orange bars, respectively), suggesting that SL5 is likely interacting with these terminal glycans. The data for the non-reducing sugars also demonstrates that sialic acid and GalNAc compete for binding to SL5.
The fructose competition studies are particularly impressive because neither SL2 nor SL5 showed any significant competition with up to 1 M saccharide, i.e., less than 10% observed decrease in the glycoprotein binding signal (Fig. 3, green bars). Since fructose is one of the strongest known 1 : 1 binders for boronic acids, the lack of competition with fructose provides further evidence that the SL–glycoprotein interactions are likely multivalent.
Discrimination of glycans
As an initial test of our approach towards binding biologically relevant targets, we used an array of SL1, SL3, SL4 and SL5 to distinguish between five structurally similar cancer associated glycans (TF antigen, Lea, Lex, sLea and sLex; Fig. 1A). These glycans were chosen because they represent some of the more common saccharide motifs overexpressed by cancerous cells as well as being composed of many of the same monosaccharides that were used in the above competitive binding assay with our SLs. SL2 was not included in the array to eliminate redundancy based on response similarities with SL3 and because of the high background binding to BSA as compared with SL3. It is worth noting that while SL1 has higher background binding to BSA than SL3; it was still included in the array due to its broad yet differential, cross-reactive response to all glycoproteins assessed.
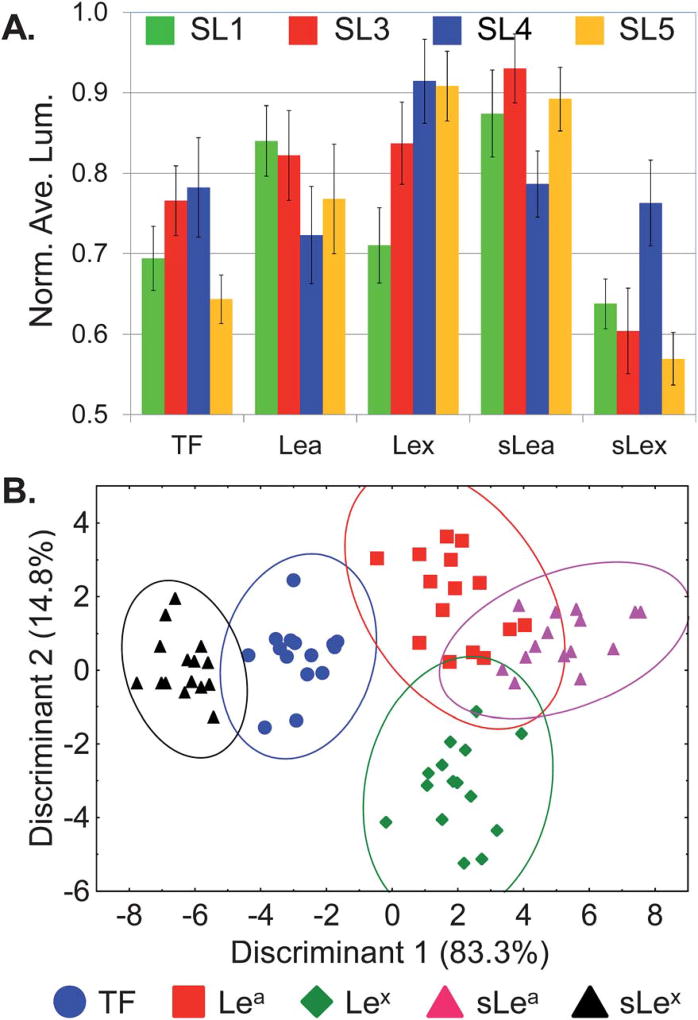
After screening each SL against a solution containing biotinylated glycan and fluorescently labeled streptavidin, luminosity values, from fluorescence microscope images, were analyzed (4 SLs by 5 glycans by 15 replicates). To account for differences in bead size and loading levels, luminosities were normalized against the highest luminosity within a given SL type (in this study the greatest degree of variability stems from bead-to-bead variations). The unique pattern generated for each different glycan based on the response of the four different SLs is shown in Fig. 4A. Note that the response for each glycan produces patterns that do not differ greatly between analytes, nevertheless the response is reproducible and the resulting patterns are unique and distinguishable within the limits of the associated error.
Fig. 4.
Differentiation of five glycans using a SL array. (A) Fingerprint pattern of the average normalized luminosity intensities from SL1, SL3, SL4 and SL5 responding to five different glycans (TF, Lea, Lex, sLea and sLex). (B) The two-dimensional LDA score plot derived from the patterns shown in (A) for 15 replicates. Ellipses indicate 95% confidence level, analyte identification indicated in the legends above.
Though these patterns are similar they are nonetheless unique, and therefore statistical analyses were used to identify the most significant features necessary for classification of the analytes. Specifically, linear discriminant analysis (LDA) was used.57 This analysis minimized variation within each glycan type while maximizing the differences between different glycans by creating linear combinations of each response pattern and transforming them into canonical discriminants. For this analysis, Discriminant 1 and Discriminant 2 contain 83.3% and 14.8% of the between group variation, respectively (Fig. 4B).58 Therefore, each point in the plot contains information for an explicit measurement from the four different SLs responding to a specific glycan. Note that the different glycans are clustered into five groups with an average standard deviation of ~6%. Furthermore, the Wilks’ lambda value for this analysis is 0.009 with a p-tail value of <0.000001, indicating that there is a statistically significant difference in the population means from this analysis at the 95% level of confidence.
While there is some overlap of the ellipses drawn in Fig. 4B, it is important to recognize that this plot only shows two dimensions out of the four dimensional data used for this analysis (displaying the data in three dimensions (four is not possible) does not visually enhance the ability of the plot to show discrimination).
Leave-one-out cross-validation was next used to assess the ability of the SL array to classify unknowns as the appropriate glycan.58 This procedure sequentially removes one sample point at a time and uses the remaining points as a new training set to create a model analogous to that shown in Fig. 4B. The classification accuracy was determined by whether or not the “left-out” data point was assigned to the correct glycan grouping. Using this method each analyte response can be used as an unknown and the classification accuracy determined for the entire data set. Based on this analysis, the SL array correctly classified 71 of the 75 measured samples (94.7% classification accuracy, with a chance accuracy of only 20%). Significantly, the Lewis antigens and their sialylated forms (Lea/Lex and sLea/sLex) were efficiently discriminated while only differing by the addition of a terminal sialic acid moiety. Additionally, this SL-array impressively distinguished between Lea and Lex, as well as between sLea and sLex, glycans where the only structural difference is the regiochemistry of the linkage to the core GlcNAc moiety (Fig. 1A). Of the four misclassified glycans (Table 2), Lea was twice identified as sLea, sLea was once classified as Lea, and Lex was once recognized as sLea.
Table 2.
Percent classification accuracies of glycans using an SL array as determined by different cross-validation techniques
| Jackknife | Boot-strapa | Training/test setb | |
|---|---|---|---|
| Lea | 86.6 | 85.8 | 88.2 |
| Lex | 93.3 | 95.3 | 96.0 |
| TF | 100 | 96.2 | 93.8 |
| sLea | 93.3 | 93.6 | 94.4 |
| sLex | 100 | 99.0 | 99.0 |
| Total | 94.6 | 94.2 | 93.9 |
Average values were calculated from 50 replicate analyses of independently randomized samples with N = 75.
Average values were calculated from 25 replicate analyses of independently randomized samples at approximately 50% exclusion (randomized test samples accounted for, on average, 37 samples (49.5%), ranging from 26–43 samples).
To further evaluate the validity of our SL array for discriminating between these five structurally similar glycans, and to circumnavigate the disadvantages associated with leave-one-out cross-validation (also referred to as delete-one jackknife) the more statistically robust “boot-strapping” approach was used.59 In the approach, multiple data sets (typically 20–10 000) are generated by randomly selecting points from the original data set. During this sampling, the probability that a data point will appear ‘n’ times is close to a Poisson distribution with mean unity.
The Mersenne–Twister random number generator60 was used for random selection of data points in Systat and data sets were created with 75 elements, the same number as the original data set. Fifty (50) separate and unique data sets were generated using this approach and were then evaluated for classification accuracy. Overall, this analysis yielded a 94.2 ± 2.0% classification accuracy for the array identifying these five glycans. This is consistent with the leave-one-out accuracy of 94.6%. Significantly, individual glycans were accurately classified from 86–99% (Table 2). As with the leave-one-out analysis, the three greatest misclassifications were due to Lea being misclassified as sLea (9.3%), sLea being misclassified as Lea (6.7%), and Lex being misclassified as sLea (4.7%).
Still further stressing the limits of this array for differentiating glycans, we chose to randomly split our data in half. Using one half as a training set, to create a statistical model, and the other half as a test set to assess the ability of this model to accurately identify these “unknowns.” Training and test sets were chosen at random from the Normal distribution.61 To minimize systematic error, random set generation and subsequent analyses were carried out 25 times to create replicates. The data in Table 2 represents the averages obtained for these replicate runs. Consistent with the previously described analyses, the overall classification accuracy of this approach was 93.9% ± 2.8%. This is by far the most stringent method used to assay the validity of the models generated from our SL array and still exhibits exceptional classification accuracy. The consistency displayed across the three methods further testifies to the strength of the outlined SL array design for discriminating structurally similar cancer associated glycans.
As indicated above, it is possible that the SLs interact, not only with the glycan, but also with the protein portion of glycoproteins. In this analysis the protein component, FITC-streptavidin, is the same for each glycan being analyzed. As such, any observed difference in the response from the array must be attributed to the glycan constituent. Given the structural similarities between these glycans, it is remarkable that there were not more misclassifications. In total, these results validate our ability to differentiate structurally similar cancer associated glycans with high accuracy using a small, cross-reactive SL array.
Discrimination of cancer cell lines
To further probe the utility of this four-component SL-array, we targeted an important goal in cancer diagnostics: to distinguish between healthy, cancerous/non-metastatic and cancerous/metastatic cell types. Specifically, we used our SL-array to discriminate between seven different cell types including: three colorectal carcinoma non-metastatic cell lines (HCT116, CT-26, HT-29), three colorectal carcinoma metastatic cell lines (CT-26-F1, CT-26-FL3, LoVo), and one murine fibroblast cell type (NIH/3T3) to serve as a “healthy” control cell line. Note that CT-26-F1 and FL3 cell lines were derived from the parental CT-26 cell line by in vivo education selection through serial passage in Balb/c mice and represent a series of highly similar isogenic cell lines that only differ in their metastatic potential (CT-26 <10% metastatic, CT-26-F1 ~50% metastatic and CT-26-FL3 ~95% metastatic).
Unlike the identification of discrete, structurally similar glycans, we predicted that cell type discrimination would result from a general response to the distinctive membrane protein composition of each cell type, thus affording a unique cellular signature, as previously demonstrated by Bunz and Rotello.38 For this study, cell membrane proteins and glycoproteins were isolated62 and fluorescently labeled to detect binding to the SL-array. While we note that this labeling approach is less than ideal for the development of a diagnostic, it does suffice to demonstrate the utility of using an SL array towards discriminating between cell lines. To account for differences in the extent of fluorescent labeling and protein concentration between each cell extract, luminosities were normalized against the highest luminosity within a given cell type (in this study the greatest degree of variability stems from cell line-to-cell line variations).63 Note that replicates obtained for the LoVo, HCT116, NIH/3T3 and HT-29 cells were derived from multiple sample preparations of cell cultures grown by different researchers over the course of several months.
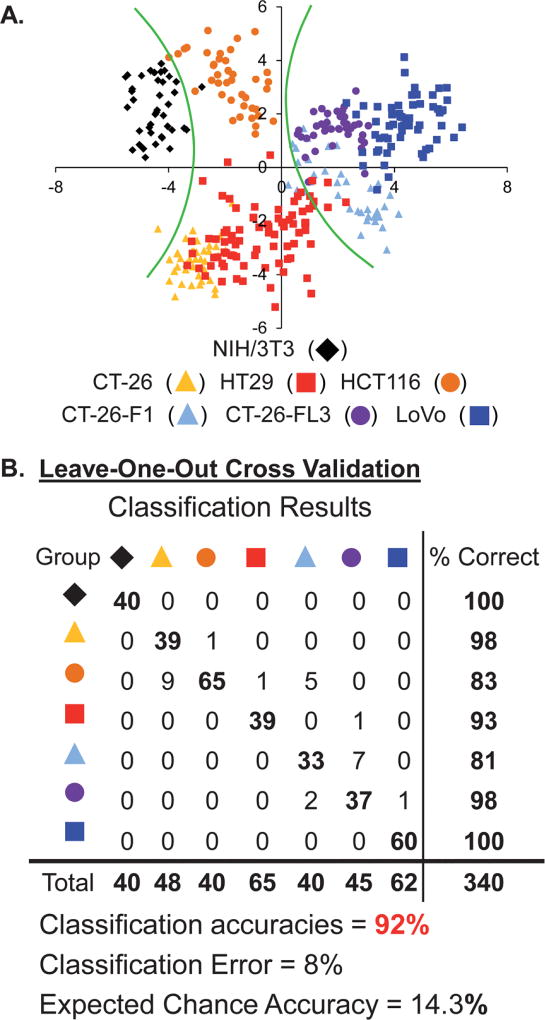
Fig. 5A shows the two-dimensional projection of the LDA results (4 SLs by 7 cell lines by 40 replicates each for NIH/3T3, CT-26, HT-29, CT-26-F1 and CT-26-FL3; 60 replicates for LoVo; and 80 replicates for HCT116). It is important to note that if all of the variance is captured in one discriminant then the statistical analysis is not really necessary; however successive discriminants containing large portions of the variance supports the validity of and the need for the statistical analysis. In this analysis Discriminant 1 contains 54% and Discriminant 2 contains 31% of the total variance, while the remaining 15% is partitioned between Discriminants 3 (11%) and 4 (4%) (i.e., this is four-dimensional data). This distribution of variance suggests that each of the SLs in the array is important for discriminating between cell lines.
Fig. 5.
(A.) The two-dimensional LDA score plot of the response of the SL array for discriminating seven cell types. Green curves indicate boundaries between healthy, cancerous/non-metastatic and cancerous/metastatic cell types. For clarity, the Discriminant 1 vs. Discriminant 2 data was rotated 20° about the z-axis (analyte identification indicated in the legends above). (B.) Leave-one-out cross validation classification matrix for the SL-array based assay.
Note that each of the same colored points cluster together indicating the ability of the statistical model to define similarity between replicates of a specific analyte. However, some of these different clusters are closely packed and some groups overlap suggesting that there are strong similarities between some of the analytes, as would be expected. Nevertheless, it important to recognize that the data is in fact four dimensional; therefore the overlap between groups shown in this two dimensional figure (Fig. 5A) is not necessarily indicative of poor classification.
To quantitatively evaluate the accuracy of this approach, leave-one-out cross-validation was used and demonstrated that this statistical model exhibited 92.1% accuracy, correctly identifying 313 out of 340 measured samples. Fig. 5B presents the LDA classification results matrix for the assay. The cross-diagonal of the matrix corresponds to the number of accurately identified samples (set in bold). Any numbers that fall off this diagonal represent the number of misclassifications for that cell type and correspond to the misclassified cell type identity. The column on the right of the matrix provides the classification accuracy for each cell type. While the overall classification accuracy for the array is 92.1%, the accuracy for each individual cell type varies between 81–100%.
Given the diversity of protein and glycan structures present on the cell membrane for each of these different cell types, it is difficult to speculate on the specific glycans that are recognized by the SLs and that contribute to the discrimination of these different cell lines. Still, there are clear trends in the statistical output that support the validity of this analysis. As one moves from left to right along the x-axis in Fig. 5A the metastatic potential of the cell lines increases. Specifically, the green curves in Fig. 5A provide boundaries between the “healthy” 3T3 cells (black) at the far left of this plot; the cancerous/non-metastatic cell lines (HCT116, CT-26 and HT-29 – orange, yellow, red, respectively) in the middle and the cancerous/metastatic cell lines (CT-26-F1, CT-26-FL3 and LoVo – light blue, purple, blue, respectively) to the right. This clustering of cell types with similar metastatic potential suggests that the basis upon which the first two discriminants are derived correlate highly with this attribute. Additionally, the Wilks’ lambda value for this analysis is 0.003 with a p-tail value of <0.000001, thus indicating that there is a statistical difference in the population means from this analysis at the 95% level of confidence. Further MANOVA treatment of the data provided a Wilks’ lambda value of 0.004 with a p-value of <0.000001 and sequential univariate F-Tests for each variable provided p-values of <0.000001 for each.
To further validate this approach, boot-strapping and training/test set analyses (at a 50% exclusion split) were carried out. The results provided in Table 3 indicate that these more rigorous validation methods provide classification accuracies consistent with those obtained for the leave-one-out cross-validation, 92.1 ± 1.1% and 92.7 ± 1.8%, respectively. As seen for the glycan analysis above, cell-line misclassifications were consistent across all three validation methods. Furthermore, the misclassified cell-lines were not random but often had a structural basis behind the result. For example, in the boot-strap analysis, CT-26-F1 displayed the lowest classification accuracy at 80.5%; and all of the misclassifications were as CT-26-FL3, an isogenic, highly metastatic cell line. Similarly, from the training/test set analysis, CT-26-FL3 has one of the lower classification accuracies (87.4%); here all of the misclassifications in this analysis were attributed to CT-26-F1 (85%) and LoVo (15%). Recall that both CT-26-FL3 and LoVo are highly metastatic and that CT-26-FL3 is isogenic with CT-26-F1. Finally, while the classification error for HCT116 is relatively large across the validation methods (classification accuracies from 81.3–89.2%), the majority of misclassifications are CT-26 and HT-29 cells. Since all three cell lines are cancerous non-metastatic, these misclassification are not unexpected because classification accuracy, in this model, correlates with metastatic potential.
Table 3.
Percent classification accuracies of cell lines using an SL array as determined by different cross-validation techniques
| Jackknife | Boot-strapa | Training/test setb | |
|---|---|---|---|
| 3T3/NIH | 100 | 100 | 100 |
| CT-26 | 97.5 | 97.0 | 96.7 |
| CT-26-F1 | 82.5 | 80.5 | 82.3 |
| CT-26-FL3 | 92.5 | 92.7 | 87.4 |
| HCT116 | 81.3 | 83.6 | 89.2 |
| HT-29 | 97.5 | 96.8 | 96.7 |
| LoVo | 100 | 99.9 | 100 |
| Total | 92.1 | 92.1 | 92.7 |
Average values were calculated from 100 replicate analyses of independently randomized samples with N = 340.
Average values were calculated from 25 replicate analyses of independently randomized samples at approximately 50% exclusion (randomized test samples accounted for, on average, 173 samples (50.9%), ranging from 153–191 samples).
Directed partitioning for enhanced cancer cell discrimination
With the advancement of cross-reactive sensor arrays, numerous statistical and non-statistical approaches have become available to evaluate the array responses; however, many do not scale well with increasing numbers of analyte classes. For analysis of these multi-class systems, the most common statistical approaches rely on multivariant analysis, such as feature selection algorithms. Alternatively, the analysis can be reduced to a series of multiple binary classification problems run in parallel, such as one-from-n (one-against-rest), pairwise (one-against-one) or hierarchical (decision trees) processes.
We have previously presented a hybrid approach, for the identification and discrimination of biogenic amines,64 where the multi-class system is simplified in a manner analogous to the binary classification routines. However, this class reduction did not rely on statistical methods; instead, we used insight into the chemical nature of the analytes to group these compounds into structurally related categories.
In training the array using this directed partitioning technique, previous knowledge about the nature of the samples is required, for example whether the cell lines are cancerous or not. However, as described above, no specific information about the exact identity of the analytes is necessary, for example the glycan being bound. This method is in direct contradiction with traditional routines that rely solely on statistical models. The quality of the results from this approach is often enhanced because logical reasoning, based on the inherent nature of the samples, is involved as part of the partitioning. Once classified into groups, these subsets could be further categorized as the individual components using a hierarchical, group–ungroup, multi-layered analysis approach to achieve enhanced classification. Therefore, directed partitioning was used to reduce classification error and the data were grouped according to their metastatic potential, i.e. healthy, cancerous/non-metastatic and cancerous/metastatic.
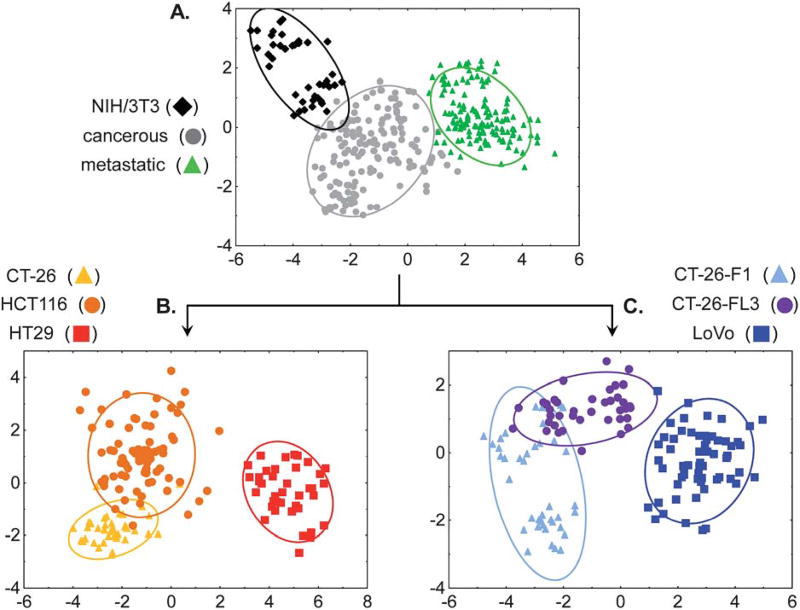
When the analysis was performed using these new groups, classification accuracies, based on leave-one-out cross-validation, improved to 97.1%, correctly identifying 330 out of 340 samples (Fig. 6A). The classification accuracy is unchanged using the training/test set analysis at 50% exclusion (97.3 ± 1.5%). From a diagnostic perspective, this is perhaps the most important classification; to determine whether the cancer is present or not. Of the 10 misclassified samples, 8 were cancerous/non-metastatic that were identified as cancerous/metastatic and the remaining 2 were cancerous/non-metastatic that were considered healthy, thus producing a 0.6% “false negative” rate. Additionally, note that the data for the 3T3 cells seems “bimodal,” showing two distinct clusters within the category. This separation results from combining data acquired by different experimentalists from different culture broths. Most significantly, while this separation is noticeable, the overall clustering is still quite tight and the 3T3 classification is 100% in the leave-one-out analysis. Based on the training/test set analysis, the within group misclassification is 6.7%, resulting in an overall 0.8% “false-positive” rate. These results clearly support the validity of this approach to identify cancerous from noncancerous cell lines. Furthermore, the low false negative rate compares quite favorably with current diagnostic tests such as the CEA test, where the false negative rate is 16%.22
Fig. 6.
(A) The 2-D LDA score plot of the response of the SL array for discriminating grouped healthy, grouped cancerous/non-metastatic and grouped cancerous/metastatic cell types. (B) 2-D LDA score plot of the array response to ungrouping the cancerous/non-metastatic cells: HCT116, CT-26 and HT-29. (C) 2D LDA score plot of the array response to ungrouping the cancerous/metastatic cells: CT-26-F1, CT-26-FL3 and LoVo. Ellipses indicate 95% confidence level, analyte identification indicated in the legends above.
By successively ungrouping each subset, a multi-layered analysis could be carried out to identify the individual cell type. The two-dimensional projections of the four-dimensional LDA results for these subset categorizations are shown in Fig. 6B–C. In Fig. 6B cancerous/non-metastatic cell lines were accurately discriminated in 150 out of 160 samples or 94%; an improvement from 89% in the single-layer analysis. Specifically, HT-29 cells were classified with 100% accuracy; CT-26 cells achieved 98% classification accuracy and HCT116 were classified with 89% accuracy. For the 10 misclassified analytes, 9 of the HCT116 samples were identified as CT-26 while one CT-26 was classified as HCT116. Given that all three of these cell lines are cancerous non-metastatic, these misclassification are not extraordinary because classification accuracy, in this model, correlates with metastatic potential.
Similarly, the cancerous/metastatic cell lines were separated into the individual components with 92% classification accuracy (129 out of 140 samples, Fig. 6C). In this analysis, 91% of the misclassifications resulted from mis-assignments between CT-26-F1 and CT-26-FL3. It is important to recall that these are highly similar isogenic cell lines, derived from the parental CT-26 cell line, and differ only in their metastatic potential. The impressive 88% classification accuracy, between the highly metastatic cell lines CT-26-F1 and CT-26-FL3, as well as 92% classification accuracy between the parent CT-26, and metastatic CT-26-F1 and CT-26-FL3 cell lines further validates our approach while indicating that there are distinct glycosylation patterns associated with metastatic potential. These results highlight the adaptability of this array-based approach for classifying cell types based on complex mixtures rather than a specific analyte, thereby mimicking the mammalian senses of taste and smell.39,40
Conclusions
In summary, selective and cross-reactive SLs have been identified by screening a resin-based SL library binding to glycoproteins. Selectivities as high as ~50-fold, for one glycoprotein over another, have been observed. The selectivity of the SL-glycoprotein interactions are maintained in 95% human serum, demonstrating their robustness. Significantly, SLs were assembled into an array format to distinguish between five structurally similar cancer associated glycans with 94% accuracy. Additionally, the same array was used to discriminate seven cell types, including three colorectal carcinoma non-metastatic cell lines, three colorectal carcinoma metastatic cell lines, and one healthy control cell line with high accuracy. Two statistical methods were employed for this analysis. In a single layered approach, analysis of all seven analytes at once provided overall classification accuracy above 92%. Using directed partitioning afforded 97% accuracy for distinguishing between cancerous non-metastatic, cancerous metastatic and healthy cells. By sequentially ungrouping these subsets the overall accuracy of the analysis was improved compared with the single-layer analysis. Current work is focused on identifying SLs for specific cancer associated targets to enhance detection sensitivity and discrimination ability, as well as expanding the array to discriminate between other glycans and cell types. Finally, we note that SLs themselves may possess therapeutic utility as targeting agents and metastatic inhibitors, as has been shown with natural lectins.65,66
Supplementary Material
Acknowledgments
We thank Dr J. E. Jones and Dr O. Obianyo for their help with fluorescence polarization. This work was supported by funds provided from NIH COBRE grant P20RR17698.
Footnotes
Electronic supplementary information (ESI) available: Complete methods including: labeling, membrane extraction and screening protocols, Supplementary Figures S1–S5 and LDA classification data. See DOI: 10.1039/c2sc00790h
Notes and references
- 1.Dube DH, Bertozzi CR. Nat. Rev. Drug Discovery. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 2.Harmat V, Naray-Szabo G. Croat. Chim. Acta. 2009;82:277–282. [Google Scholar]
- 3.Lavigne JJ, Anslyn EV. Angew. Chem., Int. Ed. 2001;40:3118–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Schmidtchen FP. Chem. Soc. Rev. 2010;39:3916–3935. doi: 10.1039/c0cs00038h. [DOI] [PubMed] [Google Scholar]
- 5.Umali AP, Anslyn EV. Curr. Opin. Chem. Biol. 2010;14:685–692. doi: 10.1016/j.cbpa.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behr J-P. The Lock and Key Principle: The State of the Art 100 Years on. Perspect. Supramol. Chem. 1994;1 [Google Scholar]
- 7.Brattoli M, de Gennaro G, de Pinto V, Loiotile AD, Lovascio S, Penza M. Sensors. 2011;11:5290–5322. doi: 10.3390/s110505290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole M, Covington JA, Gardner JW. Sens. Actuators, B. 2011;156:832–839. [Google Scholar]
- 9.Paolesse R, Monti D, Dini F, Di Natale C. Top. Curr. Chem. 300:139–174. doi: 10.1007/128_2010_97. [DOI] [PubMed] [Google Scholar]
- 10.Ranjan RK, Prasad K. Anal. Chem–Indian J. 2008;7:739–742. [Google Scholar]
- 11.Roeck F, Barsan N, Weimar U. Chem. Rev. 2008;108:705–725. doi: 10.1021/cr068121q. [DOI] [PubMed] [Google Scholar]
- 12.Wilson AD, Baietto M. Sensors. 2009;9:5099–5148. doi: 10.3390/s90705099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yinon J. Anal. Chem. 2003;75:98A–105A. [Google Scholar]
- 14.Hollingsworth MA, Swanson BJ. Nat. Rev. Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 15.Nakagoe T, Sawai T, Tsuji T, Jibiki MA, Nanashima A, Yamaguchi H, Yasutake T, Ayabe H, Arisawa K. Hepatogastroenterology. 2003;50:696–699. [PubMed] [Google Scholar]
- 16.Wang WS, Lin JK, Lin TC, Chiou TJ, Liu JH, Yen CC, Chen WS, Jiang JK, Yang SH, Wang HS, Chen PM. Hepatogastroenterology. 2002;49:388–392. [PubMed] [Google Scholar]
- 17.Magnani JL. Arch. Biochem. Biophys. 2004;426:122–131. doi: 10.1016/j.abb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Baldus SE, Zirbes TK, Monig SP, Engel S, Monaca E, Rafiqpoor K, Hanisch FG, Hanski C, Thiele J, Pichlmaier H, Dienes HP. Tumor Biol. 1998;19:445–453. doi: 10.1159/000030036. [DOI] [PubMed] [Google Scholar]
- 19.Fuster MM, Brown JR, Wang L, Esko JD. Cancer Res. 2003;63:2775–2781. [PubMed] [Google Scholar]
- 20.Ogawa J-i, Sano A, Koide S, Shohtsu A. J. Thorac. Cardiovasc. Surg. 1994;108:329–336. [PubMed] [Google Scholar]
- 21.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, Izumi Y, Irimura T. Dis. Colon Rectum. 1997;40:420–431. doi: 10.1007/BF02258386. [DOI] [PubMed] [Google Scholar]
- 22.Fakih MG, Aruna P. Oncology. 2006;20:579–587. [PubMed] [Google Scholar]
- 23.Walker D, Joshi G, Davis A. Cell. Mol. Life Sci. 2009;66:3177–3191. doi: 10.1007/s00018-009-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin S, Cheng Y, Reid S, Li M, Wang B. Med. Res. Rev. 2010;30:171–257. doi: 10.1002/med.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James TD, Samankumara KRAS, Shinkai S. Angew. Chem., Int. Ed. 1997;35:1911–1922. [Google Scholar]
- 26.James TD, Shinkai S. Top. Curr. Chem. 2002;218:159–200. [Google Scholar]
- 27.Lavigne JJ, Anslyn EV. Angew. Chem., Int. Ed. 1999;38:3666–3669. [PubMed] [Google Scholar]
- 28.Li M, Lin N, Huang Z, Du L, Altier C, Fang H, Wang B. J. Am. Chem. Soc. 2008;130:12636–12638. doi: 10.1021/ja801510d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto, Takeuchi MM, Shinkai S. Tetrahedron. 1998;54:3125–3140. [Google Scholar]
- 30.Yang W, Fan H, Gao X, Gao S, Karnati VVR, Ni W, Hooks WB, Carson J, Weston B, Wang B. Chem. Biol. 2004;11:439–448. doi: 10.1016/j.chembiol.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 31.James TD, Phillips MD, Shinkai S. Boronic acids in saccharide recognition. Royal Society of Chemistry; Cambridge, UK: 2006. [Google Scholar]
- 32.Duggan PJ, Offermann DA. Tetrahedron. 2009;65:109–114. [Google Scholar]
- 33.Pal A, Bérubé M, Hall DG. Angew. Chem., Int. Ed. 2010;49:1492–1495. doi: 10.1002/anie.200906620. [DOI] [PubMed] [Google Scholar]
- 34.James TD, Shinmori H, Shinkai S. Chem. Commun. 1997:71–72. [Google Scholar]
- 35.Bicker KL, Sun J, Lavigne JJ, Thompson PR. ACS Comb. Sci. 2011;13:232–243. doi: 10.1021/co100054e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou Y, Broughton DL, Bicker KL, Thompson PR, Lavigne JJ. ChemBioChem. 2007;8:2048–2051. doi: 10.1002/cbic.200700221. [DOI] [PubMed] [Google Scholar]
- 37.Edwards NY, Sager TW, McDevitt JT, Anslyn E. J. Am. Chem. Soc. 2007;129:13575–13583. doi: 10.1021/ja073939u. [DOI] [PubMed] [Google Scholar]
- 38.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. S10912/10911–S10912/10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavigne JJ, Anslyn EV. Angew. Chem., Int. Ed. 2001;40:3118–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Wright AT, Anslyn EV. Chem. Soc. Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 41.Karlsson NG, Nordman H, Karlsson H, Calstedt I, Hansson GC. Biochem. J. 1997;326:911–917. doi: 10.1042/bj3260911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Arcy SM, Donoghue CM, Koeleman CA, Eijnden DHVd, Savage AV. Biochem. J. 1989;260:389–393. doi: 10.1042/bj2600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martensson S, Levery SB, Fang TT, Bendiak B. Eur. J. Biochem. 1998;258:603–622. doi: 10.1046/j.1432-1327.1998.2580603.x. [DOI] [PubMed] [Google Scholar]
- 44.Combinatorial Chemistry Catalog and Solid Phase Organic Chemistry (SPOC) Handbook. Novabiochem; Laufelfingen: 1996. [Google Scholar]
- 45.GraFit, Erithacus Software Limited, Version 5.0.11 edn, 2004. Note that when the data were fit to a two-site binding model no significant differences in the calculated Kd values were apparent, Kd1 = 0.47 ± 40.51 µM and Kd2 = 43.47 ± 41.40 µM. However, the errors are quite large for this later analysis while the single site model afforded a significantly better fit to the data.
- 46.Moerke NJ. Curr. Protoc. Chem. Biol. 2009;1:1–15. doi: 10.1002/9780470559277.ch090102. [DOI] [PubMed] [Google Scholar]
- 47.Harvey DJ, Wing DR, Kuster B, Wilson IB. J. Am. Soc. Mass Spectrom. 2000;11:564–571. doi: 10.1016/S1044-0305(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 48.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. ChemBioChem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 49.Springsteen G, Wang B. Tetrahedron. 2002;58:5291–5300. [Google Scholar]
- 50.Jin S, Zhu C, Cheng Y, Li M, Wang B. Bioorg. Med. Chem. 2010;18:1449–1455. doi: 10.1016/j.bmc.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mammen M, Choi S-K, Whitesides GM. Angew. Chem., Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 52.Bielecki M, Eggert H, Norrild JC. J. Chem. Soc., Perkin Trans. 1999;2:449–456. [Google Scholar]
- 53.Draffin SP, Duggan PJ, Duggan SAM, Norrild JC. Tetrahedron. 2003;59:9075–9082. [Google Scholar]
- 54.Eggert H, Frederiksen J, Morin C, Norrild JC. J. Org. Chem. 1999;64:3846–3852. [Google Scholar]
- 55.Norrild JC, Eggert H. J. Am. Chem. Soc. 1995;117:1479–1484. [Google Scholar]
- 56.Norrild JC, Eggert H. J. Chem. Soc., Perkin Trans. 1996;2:2583–2588. [Google Scholar]
- 57.Systat, Version 11.00.01. Systat Software, Inc; 2004. [Google Scholar]
- 58.Beebe KR, Pell RJ, Seasholtz MB. Chemometrics. A Practical Guide. John Wiley & Sons, Inc; New York: 1998. [Google Scholar]
- 59.Wu CFJ. Ann. Stat. 1986;14:1261–1295. [Google Scholar]
- 60.Matsumoto M, Nishimura T. ACM Transactions on Modeling and Computer Simulation. 1998;8:3–30. [Google Scholar]
- 61.Casella G, Berger RL. Statistical Inference. Thomas Learning; Pacific Grove, CA: 2002. [Google Scholar]
- 62.Nakamura T, Hayashi T, Nishimura-Nasu Y, Sakaue F, Morishita Y, Okabe T, Ohwada S, Matsuura K, Akiyama T. Genes Dev. 2008;22:1244–1256. doi: 10.1101/gad.1632308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Control studies were carried out to assure that the array response was independent of concentration or extent of glycoprotein labeling with the fluorophore.
- 64.Nelson TL, Tran I, Ingaliinera TG, Maynor MS, Lavigne JJ. Analyst. 2007;132:1024–1030. doi: 10.1039/b708583d. [DOI] [PubMed] [Google Scholar]
- 65.Mannori G, Santoro D, Carter L, Corless C, Nelson RM, Bevilacqua MP. Am. J. Pathol. 1997;151:233–242. [PMC free article] [PubMed] [Google Scholar]
- 66.Minko T. Adv. Drug Delivery Rev. 2004;56:491–509. doi: 10.1016/j.addr.2003.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.