SUMMARY
The postsynaptic density (PSD) contains a collection of scaffold proteins used for the assembly of synaptic signaling complexes. However, it is not known how the core-scaffold machinery associates in protein-interaction networks and how proteins coding for genes involved in complex brain disorders are distributed through spatio-temporal protein complexes. Here, using immunopurification, proteomics, and bioinformatics, we isolated 2876 proteins across 41 in-vivo interactomes and determined their protein domain composition, correlation to gene expression levels, and developmental integration to the PSD. We defined clusters for enrichment of schizophrenia (SCZ), autism spectrum disorders (ASD), developmental delay (DD), and intellectual disability (ID) risk factors at embryonic day 14 and the adult PSD. Mutations in highly-connected nodes alter protein-protein interactions modulating macromolecular complexes enriched in disease risk candidates. These results were integrated into a software platform: Synaptic Protein/Pathways Resource (SyPPRes), enabling the prioritization of disease risk factors and their placement within synaptic protein interaction networks.
INTRODUCTION
The postsynaptic density (PSD) is a morphological and functional specialization of the postsynaptic membrane. This specialized signaling machinery is composed of 1,500~2,000 proteins1,2 arranged in protein interactions that can be modulated by post-translation modifications (PTMs)3–5. A common regulatory mechanism to ensure that signaling components encounter their intracellular partners in the right place and time is the association of components in protein complexes6,7. These interactions use protein scaffolds with specialized protein-interaction modules (protein domains) as a key mechanism to achieve specificity. A core feature of the PSD is the presence of a variety of scaffold molecules containing protein domains specialized in protein-protein interactions8,9. A schematic view of PSD-scaffolds can describe three layers of scaffold proteins: a top layer composed of Membrane Associated Guanylate Kinases (MAGUKs) family members that connect to glutamate receptors, a bottom layer of SHANK family scaffolds, and a middle connecting layer composed of Disk Large Associated Guanylate Associated Proteins (DLGAPs) proteins. These three scaffold families may be considered as fundamental organizers of the core protein-protein interaction machinery of the postsynaptic site10–13. A number of components of the PSD have been linked to a plethora of brain diseases1,14. However, there is a lack of information on how these components associate in protein interaction networks, and how these molecules might change their protein interactions through development and within different cellular contexts, where they might represent different functional pathways and not only a single PSD component.
Here, we investigated the organization of the core-scaffold machinery of the PSD by immunoisolation of MAGUKs, DLGAPs and SHANKs at embryonic day 14 (e14), postnatal day 7 (p7), p14, and adult mouse prefrontal cortex (PFC) and determined their association with in-vivo protein interactors in physical complexes. We show that core scaffolding components of the PSD have both shared and distinct interactors throughout development, which define a unique interactome prior to PSD maturation. We then immuno-isolated and determined protein complexes for interactors of PSD scaffolds, which enabled the total identification of 41 interactomes and 2876 individual interactors. Using de novo coding mutations on exome sequenced parent-proband trio cohorts from a variety of brain and psychiatric disorders, we describe where mutations implicated in contributing to autism spectrum disorders (ASD), developmental delay (DD), intellectual disability (ID), and schizophrenia (SCZ) were distributed within protein interactomes. These results were integrated into a software platform that allows the visualization of disease risk factors within protein networks, and we provide examples of how highly connected components of the PSD signaling network disrupt protein-protein interaction associated with psychiatric and other complex brain disorders.
RESULTS
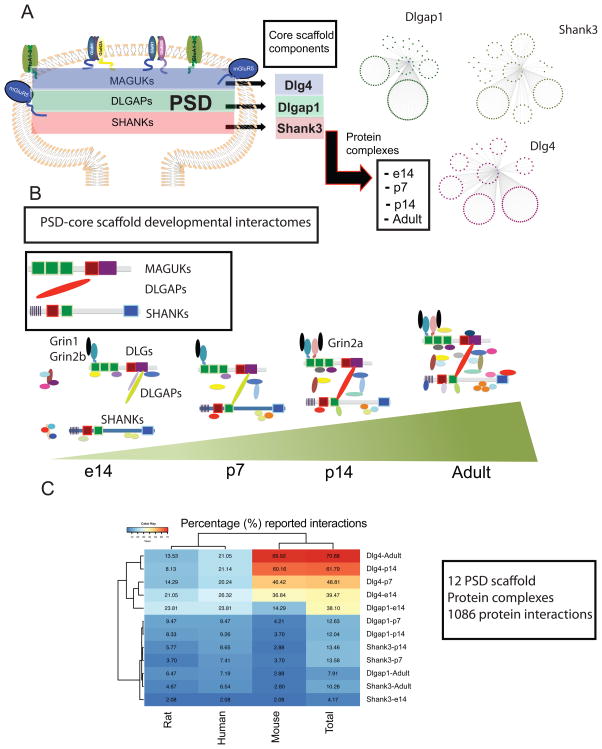
First, we determined the developmental profile of PSD-scaffold protein complexes. We isolated Dlg4, Dlgap1 and Shank3 in-vivo protein interactomes at embryonic day 14 (e14), postnatal day 7(p7), p14 and adult mouse pre-frontal cortex (PFC). All of the immuno-purified protein complexes were obtained by in-vivo immuno-isolation of the target protein using knock-out, knock-in mouse lines, anti-GST antibodies and reciprocate immuno-precipitations as controls (See Methods). Protein complexes were fractionated by SDS-PAGE, and analyzed by nano-LC MS/MS (Methods) (Fig. 1A–C, Supplementary table 1). This developmental interactome of PSD-scaffolds resulted in 1085 in-vivo protein interactors from 12 protein complexes in partially overlapping protein-interaction networks (Fig. 1A, B, Supplementary table 1). Although the roles of PSD scaffolds in the organization of protein interactions have been widely recognized, only a relative small number of protein interactions have been reported. We extracted and manually curated protein interactions reported for Dlg4, Dlgap1 and Shank3, using the BioGRID15 and Mentha16 protein-protein interaction databases. We extracted protein interactions identified in the human, mouse, and rat species and compared them to our protein-protein interaction data considering the individual species in which protein interactions were reported in or a combination of all three species. With respect to protein complexes identified at different time points in development, we compared each developmental stage-specific complex or the combination of all time points for a total set of non-redundant interactions. For a majority of the reported protein complexes approximately 90% of the non-redundant interactions reported are novel, with the least percentage of novel interactions in any one data-set being 70% in non-redundant and 56% for redundant interactions in the case of Dlg4 (Fig. 1D, Supplementary table 1, 2, Supplementary fig. 1) Moreover, databases include a collection of protein interactions reported with a variety of methods, including direct and indirect assays. While direct methods show binary protein-protein interactions, indirect methods can also contain associations of proteins in larger, non-binary interactions where the reported proteins might not be directly interacting. Due to this, we analyzed the experimental methods used to characterize each of the previously reported interactions (Supplementary fig. 2). This was able to show that a majority of reported interactions were determined via indirect assays while a lower proportion were shown using direct methods such as yeast two hybrid (Y2H), co-crystallization studies or both.
Figure 1.
Developmental PSD-scaffold interactomes
A. Cartoon of distribution of core PSD-scaffold families. B. Protein interactomes of core PSD-scaffolds Dlg4, Dlgap1 and Shank3 at embryonic day 14 (e14), postnatal day 7 (p7), postnatal day 14 (p4) and Adult mouse prefrontal cortex (PFC). Interactomes are derived from three independent experiments. C. Cartoon representation of protein interactions from core PSD-scaffolds. Figure represents a developmental change in core scaffolds through development with DLG scaffolds binding to NMDAR (NR1/NR2B) and DLGAPs in e14 protein complexes but not to SHANKs scaffolds. SHANKs protein complexes associate to downstream protein interactors at e14, without linking to DLGs and DLGAPs. Upstream components are first associated at p7. NR2A receptors at p14 and full scaffold interactions are observed in adult protein complexes. D. Percentage of scaffold-interactions reported in databases (Biogrid/Mentha) considering human, mouse and rat samples in in-vivo and in-vitro assays.
The relationship between protein interactions and gene expression levels have been widely studied17–19. It has been proposed that tissue specific proteins can interact with core cellular components and that universally expressed proteins (housekeeping proteins) can also interact with tissue-specific proteins19. This scenario has also been proposed for a number of PSD-specific proteins19. Therefore, it has been suggested that the biology of particular tissues might be better interpreted by the individual protein interactions rather than the specific proteins expressed within a tissue19. However, these analyses compare the gene expression of protein interactors through different tissues rather within the tissue where the protein interaction occurs. Therefore, it is still not known if the expression levels of protein interactors within protein complexes are better correlated when compared to the background of proteins expressed within the same tissue. Thus, we used the Brainspan database to obtain a developmental gene expression profile for the human orthologues of each protein present in scaffold complexes and compare their expression profiles at their correspondent human developmental stage e14: pregnancy second trimester, p7: late-fetal/Neonatal, p14: Early-Childhood, Adult: Young Adult/Adult.
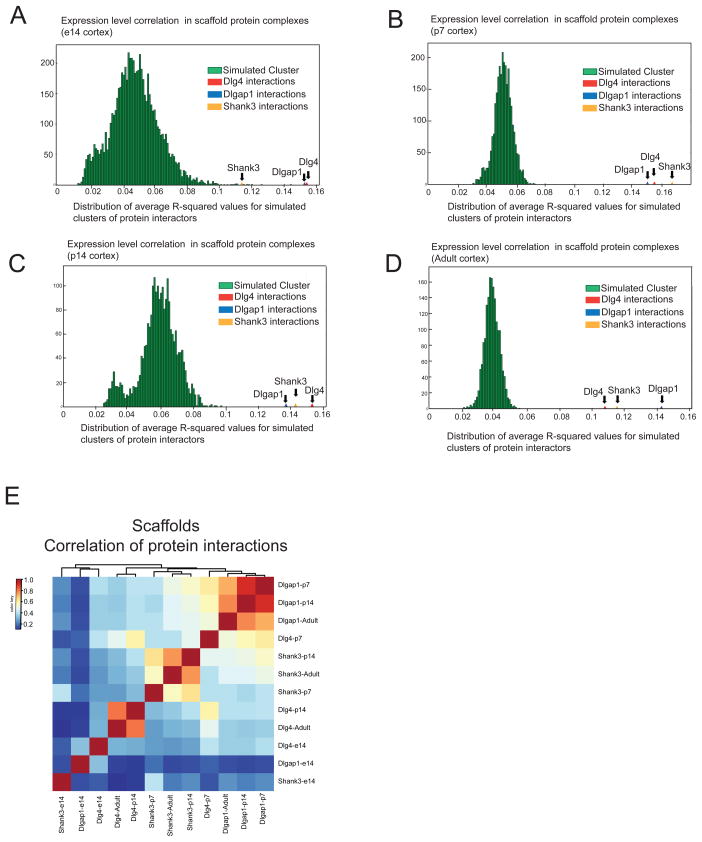
For each protein-complex, we considered three parameters including the number of proteins in the complex, the developmental period from which the complex was derived, and the corresponding gene expression levels of those proteins involved in the complex. We generated pseudo-complexes by randomly sampling an equal number of proteins for each complex from a tissue– and development time period-specific background and simulations were performed to obtain an average sample correlation (ASC) across each pseudo-complex, thus providing an empirical p-value. Pseudo-complexes failed to produce an ASC greater than observed in the protein complexes of interest (Figure 2A–D), suggesting a significant correlation between protein complex composition and tissue expression. Moreover, no differences were observed along different developmental stages with similar correlation levels being observed in scaffold complexes at e14, p7, p14 and adult stages (Fig. 2A–D). Therefore, within brain genes with similar expression levels, protein interactions grouped in scaffold complexes are significantly more correlated (Fig. 2A–D), suggesting a tighter correlation and regulation of expression levels, within tissue, for protein interacting partners.
Figure 2.
Human developmental transcriptome correlation in scaffold protein complexes
A–D: Shows the correlation of mRNA expression levels for components of Dlg4, Dlgap1 and Shank3 protein complexes using Brainspan database. Average pairwise squared sample correlation (ACS) across all genes in Dlg4, Dlgap1 and Shank3 protein complexes are compared to pseudo-complexes (green) created by randomly sampling genes with similar expression values (n = 10,000 trials). The average sample correlation across each pseudo-complex shows that protein interactions grouped in scaffold complexes are significantly more correlated at each developmental stage. E. Correlation analysis illustrating the degree of similarity between scaffold complexes through development.
We then determined the degree of similarity between scaffold complexes through development. We constructed a matrix plot and hierarchical clustering analysis of protein interactors for each individual protein complex and analyzed their shared interactors. While there is a general trend towards increasing similarity between complexes of a particular scaffold protein through development, the differences for any cluster at different developmental stages are not statistically significant. Moreover, while embryonic complexes (e-14) cluster separately from other stages; postnatal and adult protein complexes for any given scaffold show a greater overlap with each other than to complexes of a different scaffold protein at any other stage. This suggests a differential contribution of individual components of the scaffold machinery of the PSD in the clustering of protein interactions through mouse development (Fig. 2E, Supplementary table 1).
Developmental organization of core PSD-scaffolds interactions
The clustering of PSD scaffolding complexes through development (Fig. 2E) indicates a differential association of Dlg4 and Dlgap1 complexes compared to Shank3 interactions (Fig. 2E). Analysis of individual protein complexes shows that embryonic PFC contain a pre-assembly of the core scaffold machinery of the PSD with DLGs associated to NMDAR receptor subunits and DLGAP scaffolds. These top and middle layers of scaffolds are still not associated to the lower layer composed by SHANK proteins (Supplementary table 1, Fig. 1C). We observed that Dlg4 and Dlgap1 protein complexes contained NMDAR subunits Grin1, Grin2b and Grin2d, together with top (Dlg1, Dlg2, Dlg3) and middle (Dlgap2, Dlgap3, Dlgap4 and Dlgap5) scaffolds. However, none of these protein complexes contained any member of the SHANK family of protein scaffolds nor did Shank3 complexes contain any DLG or DLGAP family member (Supplementary table 1). Shank3 complexes show an early developmental association to a different set of adaptor and scaffold proteins that are usually described in PSD fractions such as Baiap2, Lrrc7, Shank1-2 and Homer1-3, that are not present in DLG or DLGAP protein complexes (Supplementary table 1). Overall, this suggests a differential building of the core scaffold machinery of the PSD through development, where the top and middle layers of the PSD connect to glutamate receptors as early as mouse e14 stage, while the SHANK layer still not a part of the core scaffold machinery. However, at e14 this layer associates to a number of PSD signaling components which will be incorporated to the scaffold core at later stages, beginning at p7.
The protein domain landscape of scaffold interactomes
Protein domains are considered to be basic units of biological function6,20. Therefore, the protein domain composition and the molecular organization of protein domains may indicate the range of cellular functions within protein complexes. To analyze the set of protein functions that are integrated to the PSD through development we determined the distribution of protein domains within protein complexes and their combinatorial interactions.
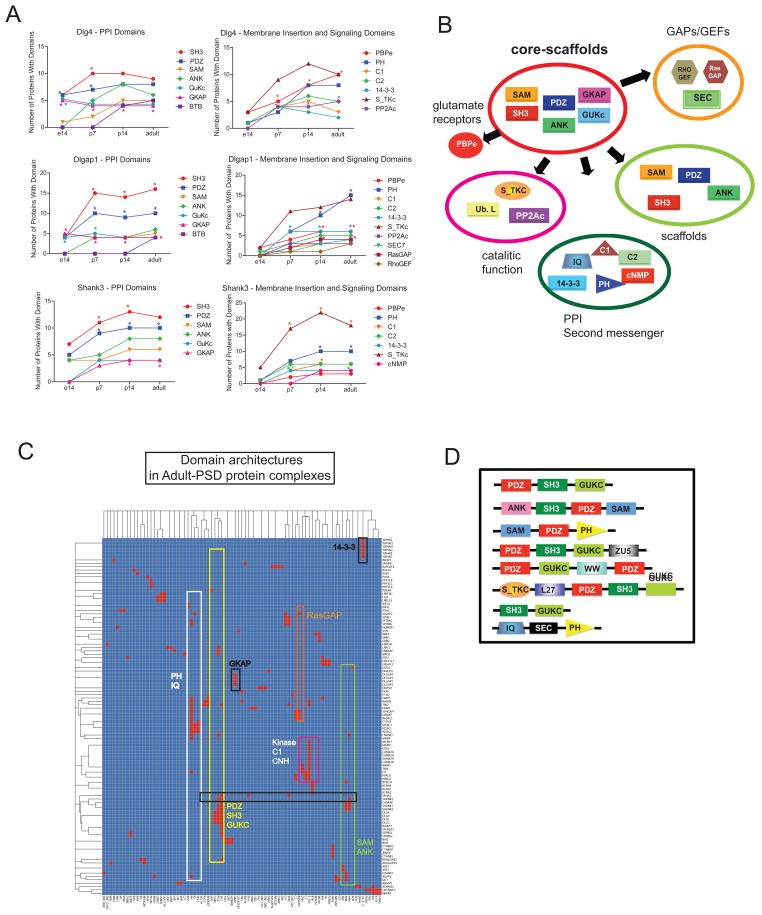
We first extracted and manually curated protein-domains using the SMART21 and Pfam22 databases (Methods) and determined a) the protein domain composition, b) protein domain enrichment, and c) protein domain architectures in each protein complex (Fig. 3A–D, Supplementary table 3). It has been previously described that adult PSDs are enriched in a number of protein interaction domains2. In adult mouse PFC this include SH3, PDZ, GuKc, and PDZ domains (Supplementary table 3). However, we observed a differential contribution of DLG, DLGAP and SHANK scaffolds to the protein domain composition of the PSD through development. Starting at e14 stage, Dlg4 and Dlgap1 complexes begin to incorporate GKAP, GuKc, and PDZ domains to glutamate receptor containing complexes (PBPe, SMART: SM00079) (Figure 3A, Supplementary table 4), while Shank3 complexes provide a higher number of poly-proline binding domains such as SH3 and WH1 (Figure 3A, Supplementary table 4). Moreover, while the top layer of scaffolds represented by Dlg4 incorporate an increasing number of glutamate receptors through development, the middle and lower scaffolds (Dlgap1/Shank3) provide a variety of protein domains involved in second messenger signaling, cytoskeletal binding and enzymatic function (Figure 3A, Supplementary table 4). In particular, there is an early enrichment in serine/threonine kinase domains in Shank3 complexes starting at p7. This indicates a developmental building of PSD functions organized by different members of PSD scaffolds using core protein-protein interaction domains to recruit functional modules to the PSD.
Figure 3.
Protein domain profiling of PSD-scaffold interactomes
A. Plots show developmental change and enrichment of protein domains present in Dlg4, Dlgap1 and Shank3 protein complexes. Domains were manually curated from SMART and Pfam databases and considered for analysis when present in at least 4 proteins in the complex. *P < 0.05, hypergeometric test followed by Bonferroni correction. B. Cartoon representing protein domain-domain interactions of core-scaffold domains (SAM, PDZ, GKAP, GUKc, ANK, SH3) through development. C–D. Chart and cartoon of domain architectures present in adult-PSD scaffold complexes. Chart shows the distribution of domain (x-axis) architectures present in individual proteins (y-axis) in complexes.
Moreover, the analysis of the organization of protein domains (domain architectures) shows that the core protein interaction domains (SAM, PDZ, SH3, GuKc and ANK), are grouped in protein families containing multi-domain architectures (Figure 3C, Supplementary dataset 1). These multi-domain architectures serve as core-building components to associate a variety of domain combinations, including those involved in signaling mechanisms and incorporate a larger number of scaffolds to the PSD (Figure 3B, C, Supplementary dataset 1). The core-scaffolds provide the essential backbone to expand the PSD functional capacity that results in the acquisition of an enhanced signaling machinery with the capacity to modulate postsynaptic function through development.
Scaffold-protein interactors
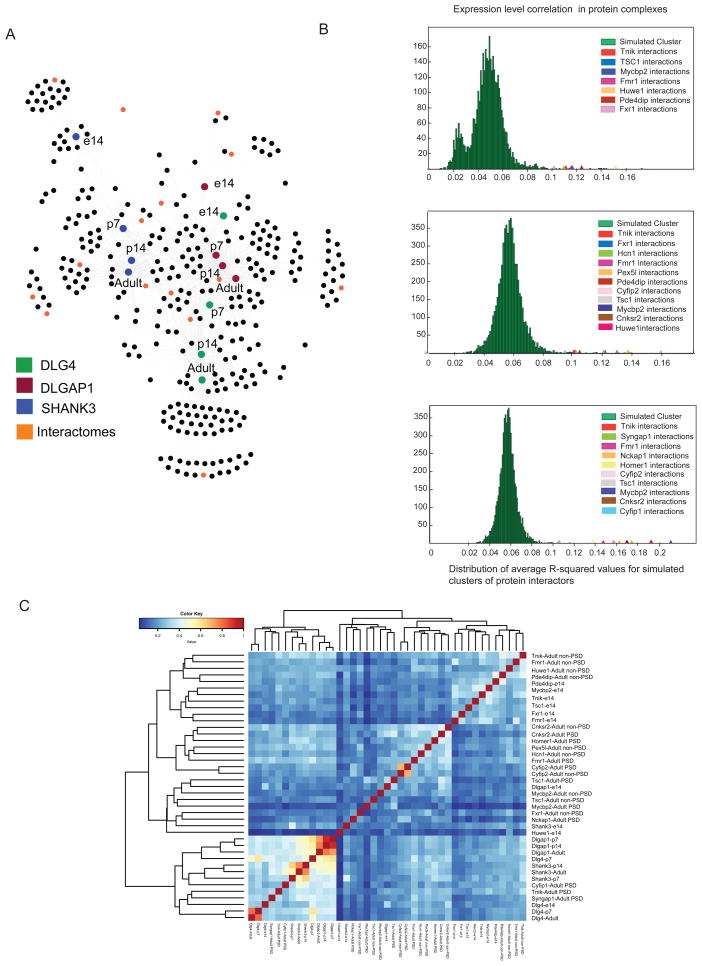
To expand the PSD protein interaction network, we first wanted to determine protein interactomes representing different regions of network. Thus, we first clustered the developmental Dlg4/Dlgap1/Shank3 network (Figure 4A, Supplementary table 5), and screened for specific antibodies directed against interactors from different functional groups and numbers of links (degree 1 to 9) within the scaffold network (Figure 4A), and determined their respective interactomes. Protein interactions were then probed at embryonic day 14 and adult PFC. While Dlg4/Dlgap1/Shank3 proteins are preferentially localized at the PSD, many of their interactors can also localize in different subcellular fractions (non-PSD). However, information about protein interactions corresponding to PSD or other subcellular fractions is lacking. Due to this, protein interactomes for this set of proteins were also screened in PSD and in non-PSD fractions. This latter fraction corresponded to PFC triton-soluble fractions3 (Methods) and allowed us to discriminate between PSD specific and non-specific (non-PSD) interactions. We thus expanded the PSD protein interaction network with 15 scaffold interactors, to 28 protein interactomes (Table 1), and 1,776 protein interactions (Supplementary table 1). As observed for protein scaffolds, these sets of protein interactions were largely unreported in literature. A majority of the interactions that were previously reported were discovered using non-stoichiometrical systems which do not consider developmental stage or localization of the protein interactions (Supplementary fig. 3, Supplementary table 2).
Figure 4.
Developmental PSD scaffold network
A. Developmental Dlg4, Dlgap1 and Shank3 protein interactions network. Color boxes show Dlg4, Dlgap1 and Shank3 developmental nodes (e14/p7/p14/Adult). Orange nodes indicate scaffold interactors used in interactome analysis. B. Brainspan mRNA expression correlation analysis of protein complexes of scaffold-interactors compared to pseudo-complexes (n = 10,000 trials). ACS shows significant correlation for protein interactions irrespective of developmental stage or cellular localization. D. Correlation analysis of protein interactions within protein complexes identifies major clustering components within components of adult PSD protein complexes associated to scaffolds and e14 stage.
Table 1.
Nodes immunoisolated from PSD networks
Table shows properties of nodes used in immunoisolation HPLC-MS/MS assays. Table shows developmental stages and biochemical fractions where nodes where found, node function, node degree within scaffold-network, and associations to Dlg4, Dlgap1, Shank3 scaffolds.
| Scaffold Interactor | Stage | Localization | Function | Scaffold | Degree Network |
|---|---|---|---|---|---|
|
| |||||
| Tnik | e14/Adult | PSD/non-PSD | S/T kinase | Dlgap1/Shank3 | 8 |
| Pde4dip | e14/Adult | non-PSD | Scaffold | Shank3 | 1 |
| Syngap1 | Adult | PSD | RasGAP | Dlg4/Dlgap1/Shank3 | 8 |
| Cnksr2 | Adult | PSD/non-PSD | Scaffold | Dlgap1 | 1 |
| Cyfip1 | Adult | PSD | Adaptor | Dlg4/Dlgap1/Shank3 | 9 |
| Fmr1 | e14/Adult | PSD/non-PSD | RNA binding | Dlg4/Dlgap1 | 2 |
| Fxr1 | e14/Adult | non-PSD | RNA binding | Dlgap1/Shank3 | 2 |
| Nckap1 | Adult | PSD | Cytoskeleton binding | Dlg4/Dlgap1/Shank3 | 9 |
| Pex5l | Adult | non-PSD | Channel binding | Shank3 | 1 |
| Huwe1 | e14/Adult | non-PSD | Ubiquitin ligase | Dlgap1 | 1 |
| Cyfip2 | Adult | PSD/non-PSD | Adaptor | Dlg4/Dlgap1/Shank3 | 9 |
| Hcn1 | Adult | non-PSD | Channel | Shank3 | 3 |
| Tsc1 | e14/Adult | PSD/non-PSD | Adaptor | Shank3 | 1 |
| Homer1 | Adult | PSD | Adaptor | Dlgap1/Shank3 | 7 |
| Mycbp1 | e14/Adult | PSD/non-PSD | Ubiquitin ligase | Dlg4 | 1 |
While we observed that the core-PSD scaffolds were highly correlated according to gene expression levels, they corresponded to a defined cellular localization and a defined PSD function. Therefore, we asked if this newly derived set of interactomes, which are functional heterogenous, were also correlated according to gene expression with respect to the transcriptome background.
Analysis of mRNA levels using the Brainspan developmental transcriptome showed a higher correlation between protein interactors than to proteins expressed in brain PFC (Figure 4B), irrespective of localization, developmental stage or protein function. Therefore, this suggests a general characteristic and correlation between protein interactions/mRNA expression levels rather than a specific property of the PSD scaffolds.
We then analyzed and clustered protein interactomes by their similarity in protein composition. While Dlg4/Dlgap1/Shank were tightly correlated, scaffold-interactors showed a lower degree of clustering and were correlated in accordance with the interactome localization and developmental stage (Figure 4C). The largest correlation was observed in a number of adult-PSD interactomes corresponding to adaptor and enzymatic functions such as Cyfip1, Tnik and Syngap1. These interactomes show a larger number of shared protein associations to Dlg4/Dlgap1/Shank3 indicating a role for these molecules in the modulation of core-PSD scaffolds interactions.
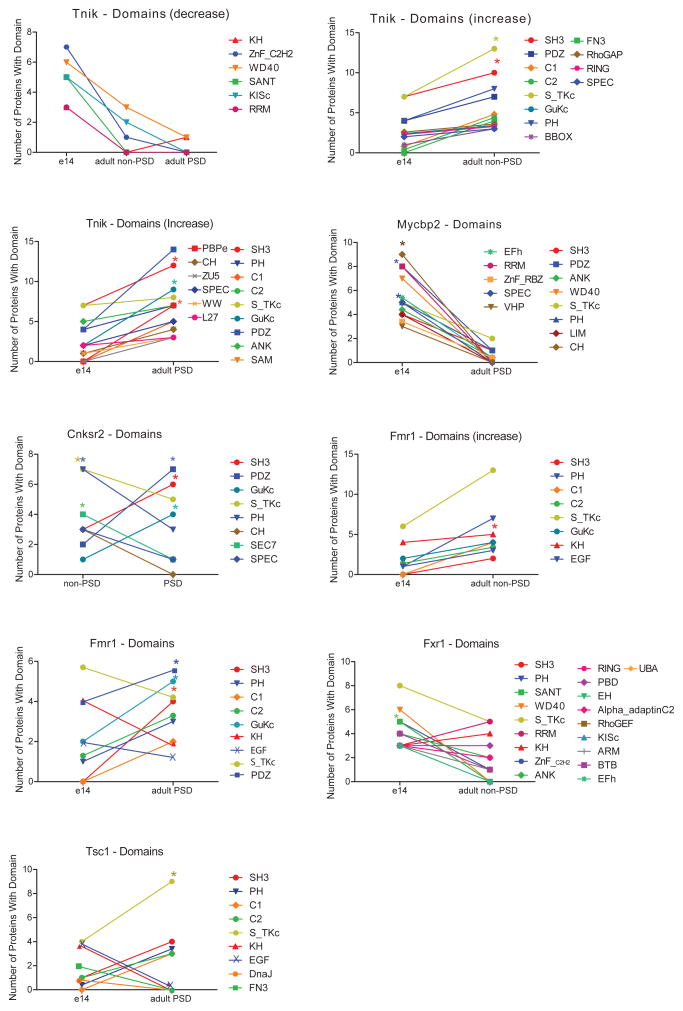
We then determined the protein domain profile for individual protein complexes (Figure 5, Supplementary table 6), considering protein domains that were present in at least 4 protein interactors in each protein complex, resulting in 355 domains from 966 proteins across protein complexes. Domain profiling confirms the pattern observed for scaffold complexes (Figure 3, 5), with adult PSD complexes showing an increase in protein interaction domains such as PDZ, SH3, PH, GuKC through development, reaching a peak at the PSD specialization. However, while scaffold complexes consistently presented a steady increase in interaction, signaling and glutamate receptor binding domains, PSD-scaffold interactors associated a number of protein domains involved in DNA/RNA binding: RING, ZNF_C2H2, and SANT. The abundance of these domains presented a steady decrease through development, and were not present in Adult-PSD complexes (Figure 5, Supplementary table 6). Moreover, contrary to PSD scaffold interactomes, analysis of protein domain architectures shows that scaffold interactors are not enriched in multimodular domain architectures (Supplementary dataset 2). Therefore, scaffold interactors might better represent a collection of pairs of direct interactions without forming large protein complexes. Thus, while PSD scaffolds serve as a template for the acquisition of functional domains, scaffold-interactors are engaged in more variable protein associations and therefore, they represent a more heterogeneous set of functions through development and cellular localization.
Figure 5.
Protein domain profiling of protein interactomes
Protein domain composition and enrichment in e14, adult, PSD and non-PSD fraction of protein complexes for scaffold interactors. Protein domains were manually curated from SMART and Pfam databases. Charts shows changes in the composition of protein domains between two different stages, PSD/non-PSD, e14/Adult. *P < 0.05, hypergeometric test followed by Bonferroni correction.
Developmental PSD interactomes in complex brain disorders
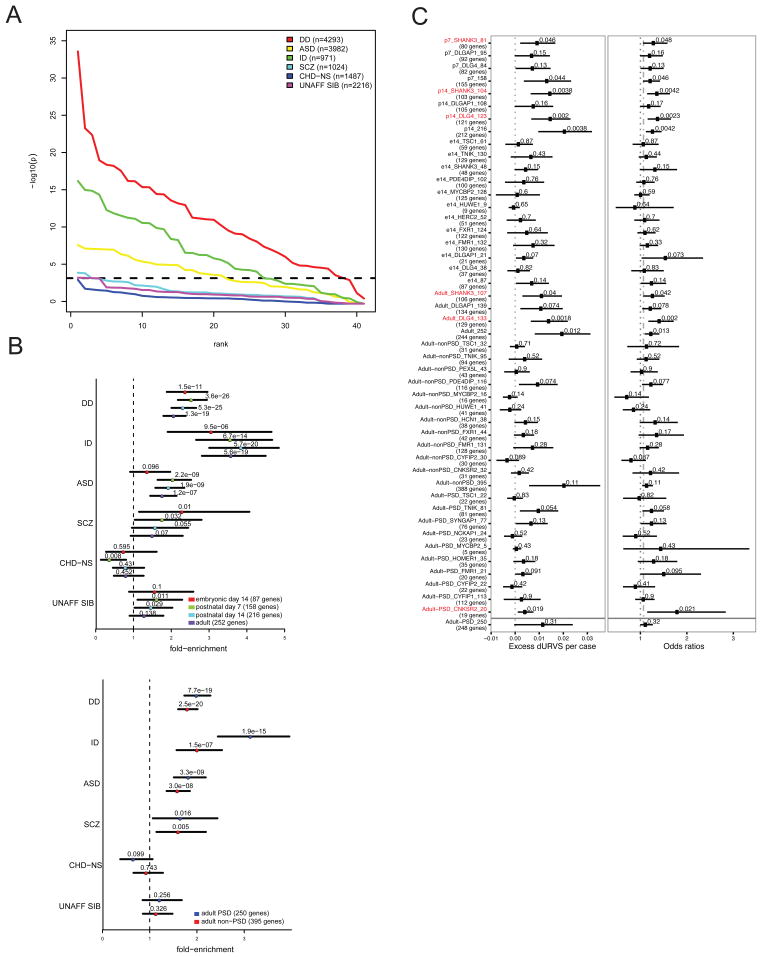
Several components of the PSD have been linked to a variety of brain diseases such as psychiatric disorders, intellectual disability and developmental delay23–31. In particular, human genetics studies have recurrently proposed a role of PSD components in psychiatric disease. However, the differential distribution of common, rare variants and syndromic genes in these diseases makes it difficult to interpret the contribution of PSD components. To show how components of the PSD and their protein interactions can help our understanding of brain disease more generally, we incorporated published de novo coding mutations on exome sequenced parent-proband trio cohorts from variety of brain and psychiatric disorders. Published lists of de novo mutations were collected from autism (ASD), developmental delay (DD), intellectual disability (ID), and schizophrenia (SCZ). Specifically, we tested for an enrichment of protein-truncating and missense mutations (collectively termed nonsynonymous) in protein interaction networks, using the mutational model described in Samocha et al. 2014 as our baseline expectation. We also examined de novo mutations from unaffected siblings of ASD and SCZ probands, as well as probands diagnosed with congenital heart disease, but without any associated neurodevelopmental or syndromic phenotype (CHD-NS) (Figure 6A, Supplementary table 7).
Figure 6.
Analyisis of synaptic interactomes in complex brain disorders
A. Enrichment of de novo coding mutations on exome sequenced parent-proband trio cohorts for autism (ASD), developmental delay (DD), intellectual disability (ID), and schizophrenia (SCZ) in protein interaction complexes. Chart shows enrichment of protein-truncating and missense mutations (nonsynonymous) in protein interaction networks with respect to the baseline, as described in Samocha et al 2014, which indicates the simulated multiple testing correction (alpha=0.05) level. Controls including de novo mutations from unaffected siblings (UNAFF-SIB) of ASD and SCZ probands, and probands diagnosed with congenital heart disease but without any associated neurodevelopmental or syndromic phenotype (CHD-NS) are included. Two-sided binomial exact test. B. Shows enrichment analysis of nonsynonymous mutations in developmental (e14, p7, p14, Adult) scaffold complexes and in same protein complexes from PSD and non-PSD interactomes. Color boxes indicate protein complexes cellular localization and developmental stage. Top plot shows Dlg4, Dlgap1 and Shank3 developmental complexes collapsed per developmental stage showing fold-enrichment over mutation model expectations. Bottom plot shows comparison of scaffold interactors with PSD and non-PSD interactomes, All PSD and all non-PSD interactios were collapsed and plot shows fold-enrichment over the mutation model expectation. Two-sided binomial exact test. C. Shows enrichment for ultra-rare protein-altering variants (URVs) in individuals with schizophrenia. Graphs shows enrichment for PSD scaffolds Dlg4, Shank3 at different developmental stages and for the GWAS hit, Cnksr2, protein complexes. Figure includes scaffolds and interactors protein complexes, together with all complexes at any developmental stage (e14, p7, p14, Adult). Number of genes considered for analysis are displayed between parenthesis (genes correspond to proteins present in interactomes). Enrichment analysis and P values were determined using a linear (left) and a logistic (right) regression model (right) using exome-wide dURV count as a covariant to correct for average exome-wide burden (dot-dashed line). Horizontal bars indicate 95% confidence interval. Enrichment and P values were computed as described in Genovese et al., 2016 (methods).
Across the spectrum of protein interaction networks that we generated, the enrichment in nonsynonymous de novo mutations is strongest in individuals diagnosed with DD or unresolved ID, where the majority of networks were significantly enriched in probands relative to model expectations, followed closely by individuals diagnosed with ASD. Within these disorders embryonic and early developmental protein complexes were more significant in DD and ID compared to ASD (Figure 6A–B, Supplementary table 7). Moreover, while the contribution to PSD and non-PSD complexes where equally significant for most disorders, PSD protein interactions were significantly more enriched in ID when compared to their non-PSD counterparts. These results suggest some differential contribution of protein interactions from PSD components along development and cellular localization in these disorders (Figure 6B).
However, SCZ presented a different scenario, with little to no enrichment across the protein interactor networks, with only a modest enrichment observed relative to non-syndromic CHD and unaffected siblings. In general, de novo protein truncating variants (PTVs) showed larger enrichment than missense, however the general patterns of significance across disease were consistent in both annotation types (Supplementary table 7). The null result in SCZ, however, is not that unexpected, as the burden of de novo mutations in SCZ is quite modest relative to other neuropsychiatric disorders, and the sample size much lower than DD and ASD. In fact, when collapsing networks by developmental stage and PSD/non-PSD membership, we see comparable enrichment with ASD, but with less power due owing to the smaller number of trios (Figure 6B). Therefore, these results might reflect the genetic architecture of the disease rather than a particular characteristic of PSD protein interaction networks. As an example comparison, 2 of the 41 protein interactor sets, Tnik in adult PSD (p = 2.8e-4) and Dlg4 in p7 (p = 3.3e-4), are more significantly enriched in nonsynonymous mutations than the activity regulated cytoskeleton (ARC) protein complex (p = 1.4e-3), a candidate gene set highlighted in both de novo SNV and CNV studies of SCZ23,26,32 (Supplementary table 5). Recently, an increased burden of ultra-rare protein-altering variants in individuals with schizophrenia32 has been shown. These ultra-rare variants (URVs) are concentrated in neuronal expressed genes. We then asked in particular protein interaction networks were enriched in SCZ-URVs. We found that 6 of the 41 protein complexes were enriched in SCZ-URVs (p<0.05) (Figure 6C), including different developmental stages of top PSD scaffold Dlg4, and lower PSD-Scaffold Shank3 (Figure 6C). Moreover, we also observed an enrichment in another Tnik, Shank3 and Dlg4 interactor: Cnksr2 (p = 1.9e-2). Cnksr2 complexes were enriched only at adult-PSD and not in non-PSD complexes. This PSD scaffold is incorporated at the PSD in late development and has a similar domain architecture to the Shank family of interactors (Supplementary Dataset 1, 2). Cnksr2 complexes are also enriched in nonsynonymous de novo mutations (p = 1.4e-3), at a similar level to ARC protein complexes. Moreover, CNKSR2 is candidate risk factor for SCZ as it is found within a single-gene, genome wide significant locus as determined by the Schizophrenia Working Group of the Psychiatric Genomics, Consortium33. This suggests that a small number of adult-PSD complexes with inter-connected protein interactions are associated with a variety of genetic signatures observed in SCZ.
Mutations in components of adult postsynaptic density complexes
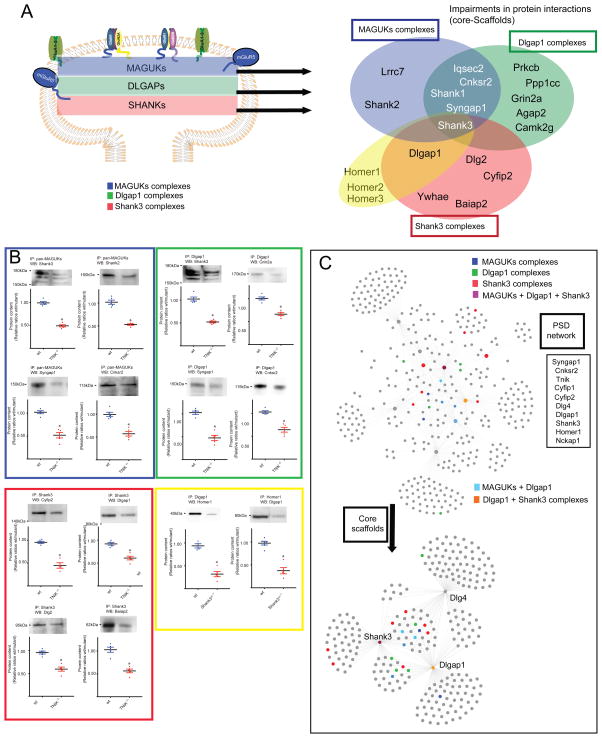
To answer if mutations in highly-connected components of PSD can affect the composition of PSD complexes enriched in brain disorders, we first clustered adult-PSD protein complexes and used two of the most highly connected genes Tnik and Shank3 (Figure 7C, Supplementary Table 5), with protein complexes enriched in multiple brain disorders (Figure 6A, C). To investigate how mutations in Tnik and Shank3 might dirsrupt protein interactions through the three layers of PSD complexes, we used a Tnik knockout mice (Tnik−/−), and a mouse line with a heterozygous deletion of Shank3 c-terminal region Shank3ΔC−/+. We then isolated MAGUKs (top), with a pan-MAGUK antibody, Dlgap1 (middle), and Shank3 (bottom) protein complexes as an approximation for the distribution of changes in protein complexes across the three main layers of the PSD (Figure 7A, C) (Methods). Protein interactions and ratios for wt/Tnik −/−; wt/Shank3ΔC−/+ were determined by nano-HPLC MS/MS. A normalized spectral abundance factor (NSAF) was calculated for each protein34 and was used for comparison between different samples. A ratio ≥ 2 or ≤ 0.5 in triplicate samples was considered as biological change, and confirmed by western blot (WB) (Figure 7A–C, Supplementary table 8, Supplementary fig. 4). To confirm that changes were a consequence of disruptions in protein interactions, we determined total protein levels in wt and mutant samples by WB and no changes were observed in ratios of wt/mutant sample (Figure 7C, Supplementary table 7, Supplementary fig. 3). We observed that mutations in highly connected components of the PSD protein-interaction network disrupted protein-protein interactions through the three scaffold-layers of the PSD (Figure 7A–C Supplementary table 8). This group of proteins were highly enriched in psychiatric risk factors with 45% described in GWAS-SCZ)33 and enriched in ASD, ID nonsynonymous de novo mutations and SCZ-URVs. Moreover, we found that proteins with impaired association to PSD complexes contained SH3, PH, SAM, ANK, PDZ, as the most common domain architectures (Figure 7B, Supplementary dataset 1, 2). This suggests that mutations in components of the adult-PSD common to many of the complexes studied can also alter the pattern of protein interactions and/or localization of multiple candidate genes involved in different brain disorders.
Figure 7.
Disruption of scaffold networks by mutations in high connected nodes
A. Venn diagram shows disruption in protein interactions in MAGUK (blue), Dlgap1 (green), Shank3 (red) protein complexes in and Homer1, Dlgap1 protein complexes in Tnik−/− and Shank3ΔC−/+ mutant mice. Protein interactions were determined by immunoisolation of scaffold complexes and quantitation by HPLC-MS/MS in adult wt and mutant mouse PFC. Ratios of mutant/wt that scored as <0.67 in three independent experiments were considered a biological change. C. Clustering of PSD complexes, showing Tnik, Shank3, Cnksr2, Syngap1, Cyfip1, Cyfip2, Homer1, Nckap1, Dlg4 and Dlgap1 protein interactions. Color-boxes indicates disrupted interactions within the PSD protein interaction network. Disrupted interactions correspond to highly connected nodes within the network. Inset shows most abundant (top) protein domains present in proteins with impaired associations in PSD complexes. C. Shows confirmation of impairments in protein complexes shown in Figure 7A, B (same color code) by WB analysis of protein complexes determined in wt and mutant mouse. Representative cropped images of WB from PFC samples n = 4 independent experiments or more: Tnik−/−, IP: Shank3, WB: Dlg2 (n = 4 independent experiments); Tnik−/−, IP: Dlgap1, WB: Cnksr2 (n = 4 independent experiments); wt, IP: Dlgap1, WB: Homer1 (n = 4 independent experiments); wt IP: Homer1, WB: Dlgap1 (n = 6 independent experiments). All data represented as mean ± s.e.m. *P < 0.05, two tailed unpaired t test.
Synaptic Protein/Pathways Resource
To prioritize the significance of disease risk factors within their protein interaction and cellular landscape, we developed the software platform Psychiatric Protein/Pathway Resource (SyPPRes) available at http://neurocomplex.usc.edu. The software tool was built upon the Phenolyzer tool35, which analyzes known gene-gene and gene-phenotype relationships from literature, then prioritizes candidate genes and generates interaction networks for each disease. By overlaying the molecular interaction information detected from our study to those inferred from existing biological knowledge, the SyPPRes tool uses these updated gene-gene relationships to prioritize genes with respect to different complex brain disorders, including autism spectrum disorders, schizophrenia, intellectual disability, epilepsy. For example, if a known ASD risk gene is located in the same protein complex with another gene previously not associated with ASD, then this new gene will be prioritized as potentially contributing to ASD. The SyPPRes tool also allows user to specify specific genomic regions (such has GWAS peaks) or gene list, to restrict the analysis to only a specific set of genes.
DISCUSSION
Starting with a developmental profile of the core-scaffold machinery of the PSD, we determined the molecular and cellular organization of PSD protein complexes and the context where PSD components associate to brain disorders. We show, that the core PSD scaffold-structure is already present at e14. This early structure starts to cluster components of the PSD and begins to define its future characteristics. However, while NMDAR subunits are already linked to MAGUKs and DLGAPs protein complexes, downstream components are kept in separate protein complexes organized by SHANKs. The core-NMDAR/MAGUK/DLGAP/SHANK three-layer-scaffold structure was first observed at p7, while p7–p14 interactions represent a transition from e14 to adult complexes. We observed a continuous incorporation and expansion of functional groups with increased associations to glutamate receptors at the top layers and proteins with enzymatic activities incorporated through the middle and lower layer of scaffolds.
Scaffold interactors form a more heterogeneous set of protein complexes when compared to core PSD scaffolds defining different sets of molecular complexes and cellular functions. Therefore, while the adult PSD protein interaction network is connected through a core-scaffold component using localization specific hubs, a number of PSD scaffold-interactors can also associate in protein networks enriched in “housekeeping” protein interactions at early developmental stages that are not exclusive to neurons.
PSD signaling and complex brain disorders
A role for components of the PSD has been recurrently proposed in a number of psychiatric disorders. However, the current lack of information on PSD protein interactomes and their developmental and spatial profile may generate an over-representation of individual adult protein complexes such as Arc and NMDAR23,26, this is particularly clear in SCZ genetics23,26,32,36. Thus, to answer which complex(es) best capture disease association will require the characterization of a much wider range of complexes at multiple developmental time-points and how they behave under different patterns of synaptic stimulation37. Our data supports a role for a relative small number of PSD protein complexes associated to the three main scaffold families (DLG/DLGAP/SHANK), with a core highly-connected component enriched in psychiatric and ID candidate genes. These sets of molecules are observed in several psychiatric and brain disorder datasets38–43, and belong to the major signaling component of the PSD. Therefore, highly connected molecules within PSD interaction networks might not be associated to an individual protein complex, but to the core protein-interaction machinery of the PSD. This structure allow the connection of different components of the network44 through their multi-domain composition. Therefore, risk factors affecting signaling hubs may disrupt the domain architecture of the synaptic signaling network. Since many of these connections are linked to psychiatric disease, it is expected that the network may be disrupted by mutations at different components. Therefore, kinases like Tnik and scaffolds such as Shank3 might represent two ends of the spectrum of adult PSD signaling complexes. While scaffolds can cluster risk factors through its interaction modules, kinases/phosphatases can dynamically regulate the interactions of these modules through protein phosphorylation.
As the list of psychiatric and brain disorder risk factors is likely to continue growing, it will be important to determine if the combination of risk factors in synaptic components indicate only a general deficit in PSD signaling, or if they integrate into specific signaling components of the PSD signaling machinery.
Beyond addressing molecular mechanisms at determined risk pathways, future studies will need to address how non-physically connected components of protein interaction networks, contribute to the susceptibility in brain disorders. It will be important to define if risk factors accumulate within one risk pathway, or if they need to be combined within different spatio-temporal networks. This raises the possibility that a combinatorial profile of risk factors, distributed along biological networks might contribute to the heterogeneity and overlap within disorders45. Thus, determining the cellular networks where risk factors are functionally active with tools such as SyPPRes, may help to define disease-risk scores, and develop strategies to stratify individuals for vulnerability to psychiatric and other brain disorders.
Methods
Animals
Animal welfare and all the procedures performed comply with the USC Institutional animal care and use committee (IACUC) and are registered under protocol number: 11782-CR001. Mice were housed in semi-natural light cycle of 12:12 hours and kept under USC IACUC guidelines. Mice used corresponded to e14, p7, p14 and adult males (3 – 4 months old). Cages were monitored daily and at least 3 times a week to detect and follow pregnant females for proper mice age recording. Mice were weaned at 21 days of age (4 mice/cage), moved to separate cages, and littermates were kept together. Mice were euthanized by carbon dioxide inhalation, using compressed CO2 gas in cylinders. Mice were introduced without pre-charging the chamber, followed by 100% carbon dioxide, with a fill rate of 10% to 30% of the chamber volume per minute. To reduce animals distress while tailing for genotyping, a local anesthetic (Ethyl chloride spray) were always applied to the tip of tail before tailing. All wild type mice (C57BL/6, C57LB/6N, and C57BL/6J) were purchased from Charles Rivers laboratories.
Biochemistry
PSD-enriched fractions were prepared using a 3-step protocol as previously described37. Tissue was homogenized in 10mM Hepes buffer pH 7.4, containing 2mM EDTA, 5mM Sodium Orthovanadate, 30mM NaF, 20mM β-glycerol phosphate and Roche Complete as protease inhibitor cocktail. Homogenates were spun, for 4 min at 500g supernatant collected, and centrifuged at 10,000g, and the membrane fraction solubilized in Hepes 50mM pH 7.4, containing 2mM EGTA, 2mM EDTA, 30mM NaF, 5mM Sodium Orthovanadate, 20mM β-glycerol phosphate, Roche Complete and 1% Triton X-100. Solubilized membranes were centrifuged at 30,0000 RPM in a Beckman Optima Max rotor MLA-130 for 40 minutes, pellet was collected and solubilized in 50mM Tris pH 9, 30mM NaF, 5mM Sodium Orthovanadate, 20mM β-glycerol phosphate, ZnCl2 20uM, Roche Complete and 1% Sodium Deoxycholate (DOC). Enrichment and quality of PSD fractions was monitored through western blotting against a number of protein controls including, glutamate receptor subunits, presynaptic markers, cytoplasmatic proteins, and core PSD scaffolds1.
We determined protein complexes at embryonic day 14 (e14), postnatal days 7 and 14 (p7, p14), and adult (12–16 weeks old). Adult samples were determined at both PSD and non-PSD fractions. 1.5 mg of total protein was used for immuno-isolation in all of the fractions tested. Antibodies were screened for specificity and recovery of total levels of a target protein by determining the right concentration of antibody to use by quantitation of the target protein in immunoprecipitation and flow-through fractions. Concentrations for antibodies ranged: 0.4–0.8 ug/ul, immunoprecipitations were performed as described37 with minor modifications. Antibodies were screened with Tnik−/−, Fmr1−/−, Shank3B, Cnksr2−/Y, Dlgap1−/−, Tsc1−/−, Syngap1−/+ mutant mice, and anti-GST as negative controls. To remove RNA binding proteins associated to RNA strands, e14 samples and Fmr1/Fxr1 IPs were pre-incubated with RNAse T1/H. A list of antibodies used for immune-isolation and western blotting is provided in supplementary table 11.
For western blotting, samples were loaded onto NuPAGE® Novex® 4–12% Bis-Tris Gels and separated at 135V. Proteins were then transferred onto PVDF membranes using a Bio-Rad Trans-Blot Turbo Transfer System. Membranes were then blocked for 1 hour in blocking buffer (5% Bovine Serum Albumin/0.05% TBST (TBS-Tween 20)) at room temperature and then incubated with the appropriate primary antibody overnight in 1% blocking buffer at 4 degrees Celcius. The next day, the membrane was washed 4 times for 10 minutes each with TBST followed by a 1 hour incubation with the respective secondary antibody in 1% blocking buffer for 1 hour at room temperature. Membranes were then washed 4 times for 10 minutes each with TBST. To characterize antibodies specificities, all target proteins were immunoisolated and fractionated in 12 bands by in-gel digestion. After Mass spectrometry analysis of each band, the appropriate molecular weight of target protein was identified. For western blot analysis, blots were cropped at the identified molecular weight for each target and developed by chemiluminescence assays. Membranes were probed using ECL western blotting substrate (ThermoFisher) and imaged using a Carestream Image Station 4000MM Pro.
For mass spectrometry sample preparation, samples were separated by NuPAGE® Novex® 4–12% Bis-Tris Gels and subsequently fixed and then stained with Instant blue®, for qualitative QC. Lanes were cut and placed into 96-well plates for de-staining, and digested by trypsin at 37°C for 1 hour. Peptides were then extracted with acetonitrile. Peptide desalting and reverse phase separation of peptides was performed using the Nano/Capillary Liquid Chromatography (LC) System Ultimate 3000 (Thermo/Dionex).
Mass spectrometry data processing
MS data were processed using Proteome Discoverer 1.4 (PD, Thermo Scientific) and searched by both Sequest and Mascot V2.4 (Matrix science) against a modified mouse database which was downloaded from Uniprot and was combined with its decoy database. The mass tolerance used for searching was set as 10 ppm for precursor ions and 0.8 Da for fragment ions. No more than two missed cleavage sites were allowed. Static modification was set as cysteine carboxyamidation and dynamic modification was set as methionine oxidation. False discovery rates (FDR) were automatically calculated by the Percolator node of PD based on decoy database hits. A peptide FDR of 0.01 were used for cut-offs. Peptides with high confidence were considered as true hits and proteins with at least two different peptides present in triplicate assays and absent in controls were considered positive. In addition to the mouse KOs used, non-specific interactions are listed in supplementary table 9 and proteins removed after RNAse treatment are provided in supplementary table 10.
Normalized spectral abundance factor (NSAF) was calculated for each protein and was used for comparison between different samples. For protein interaction assays a ratio ≥ 1.5 or ≤ 0.67 in triplicate samples was considered as biological change. Cytoscape 3.4.0 was used for network clustering and visualization46
Comparison to previously reported interactions
Previously reported protein-protein interactions were extracted from BioGRID and Mentha16 protein-protein interaction databases and then manually curated to generate a composite of the two databases. Interactions for Dlg4 were manually curated through the removal of interactions identified in Arbuckle et al., 201047. Dlg4 interactions described in Fernandez et al., 200910 were restricted to those found only in the tandem affinity purifications. We also manually added Cyfip1 protein-protein interactions described in De Rubeis et al., 201348 as these were not present in the BioGRID or Mentha databases. Comparisons between the data reported in the current study and of previously reported interactions were carried out using those interactions identified in homo sapiens, mus musculus, rattus norvegicus, or a combination of all three species. In addition, the method of detection for each previously reported protein-protein interaction was extracted if the interaction was reported in BioGRID or manually curated if the interaction was reported in Mentha. Interactions that were determined using a single domain of a particular protein were not included.
The Jaccard similarity between domains
With the domain-complex association matrix C = (cij), with cij taking values 0 or 1 with cij = 1 indicating the j-th complex contains the i-th domain, the Jaccard similarity between domain i and domain i′ is defined as sij, = |ci ∩ ci′|/ci ∪ ci′|, where ci is the set of complexes containing domain i. If two domains belong to the same complex, the similarity of the two domains equals 1. The dissimilarity can thus be defined as dii = 1 − sjj′. Based on the dissimilarity, hierarchical clustering with average linkage is then used to cluster the domains. We also used the same approach to cluster the complexes by comparing the domain architectures of the complexes.
Testing for enrichment of protein domains within complexes
For a given complex, we only consider domains occurring at least one time in the complex. For a given domain, we test the hypothesis if the domain is enriched in the complex. The statistical significance, the p-value, is calculated as the conditional probability that the domain occurs at least the observed number of times given that it occurs at least once. More precisely, let N be the total number of proteins and M be the number of proteins containing the domain. Let n be the number of proteins in the complex. Under the null hypothesis that the domain is not enriched in the complex, the number of proteins containing the domain follows a hypergeometric distribution H(M, N, n). Let m be the number of proteins in the complex that contain the domain. Then the p-value is calculated by:
We used Bonferroni procedure to adjust for multiple testing. That is, only domains with p-value less than α/T is considered significant, where T is the number of different domains analyzed across all reported complexes (n = 320).
Testing for co-expression using the Brainspan database
For each gene complex we considered three parameters: a) number of genes in the complex, b) the minimum and maximum expression of genes in the complex, c) the correspondent age (lifespan) where the complex was discovered.
C_k = the number of genes in the complex
C_min, C_max = the minimum and maximum expression of genes in the complex
C_L = the relevant age (lifespan) where the complex was discovered
For a given C_L the average pairwise squared sample correlation (ASC) was calculated across all genes in the complex. Additionally, pseudo-complexes were created by randomly sampling k genes from C_L without replacement with expression values between C_min and C_max. In each case 10,000 trials were performed and the ASC across each pseudo-complex was calculated to provide the empirical p-value.
We used 4–7 individuals per developmental stage, analyzing up to 16 regions per individual. Every sample was a region from an individual and we analyzed correlation across individuals, across the whole dataset (person and region), for each gene in the cluster. We calculated min/max expression in each cluster and randomly sampled more groups with min/max bounds equal to our cluster and calculated average pairwise correlation. Variation between regions was not significant and therefore allowed the use of different brain regions per individual to increase sample size.
e14: (second trimester): 76 samples [5 individuals, 18 regions] [A1C=5,AMY=5,CBC=5,DFC=5,HIP=5,IPC=2,ITC=5,M1C=3,M1CS1C=2,MD=5, MFC=5,MGE=2,OFC=5,S1C=5,STC=3,STR=3,V1C=3,VFC=5]
p7: (Late-Fetal/Neonatal): 39 samples [4 individuals, 16 regions] [A1C=3,AMY=3,CBC=3,DFC=3,HIP=2,IPC=2,ITC=2,M1C=2,MD=2,MFC=2,OFC =2,S1C=2,STC=4,STR=2,V1C=3,VFC=2]
p14: (Early-Childhood): 59 samples [4 individuals, 16 regions] [A1C=4,AMY=4,CBC=4,DFC=3,HIP=4,IPC=4,ITC=4,M1C=4,MD=3,MFC=3,OFC =3,S1=4,STC=4,STR=3,V1C=4,VFC=4]
Adult: (YoungAdult/Adult): 111 samples [7 individuals, 16 regions] [A1C=7,AMY=7,CBC=7,DFC=7,HIP=7,IPC=7,ITC=7,M1C=7,MD=7,MFC=6,OFC =7,S1C=7,STC=7,STR=7,V1C=7,VFC=7]
DFC - dorsolateral prefrontal cortex, VFC - ventrolateral prefrontal cortex, OFC - orbital frontal cortex, IPC - posteroventral (inferior) parietal cortex, ITC - inferolateral temporal cortex (area TEv, area 20), A1C - primary auditory cortex (core), AMY - amygdaloid complex, CBC - cerebellar cortex, HIP - hippocampus (hippocampal formation), M1C - primary motor cortex (area M1, area 4), MFC - anterior (rostral) cingulate (medial prefrontal) cortex, MD - mediodorsal nucleus of thalamus, S1C - primary somatosensory cortex (area S1, areas 3,1,2), STR – striatum, V1C - primary visual cortex (striate cortex, area V1/17).
Human genetics datasets and enrichment analysis
De novo coding mutations on exome sequenced parent-proband trio cohorts from SCZ, ASD, DD and ID were used for analysis, using probands from unaffected siblings and from probands diagnosed with congenital heart disease, but without any associated neurodevelopmental or syndromic phenotype (CHD-NS), as controls. We also analyzed de novo protein truncating variants (PTVs) found in patients with SCZ32.
We collected de novo mutations from SCZ (7 studies), ASD (6 studies), DD (2 studies), ID (3 studies), and CHD (2 studies) (see Supplementary Table 7- Summary Information). Unaffected siblings and control trios were also available from a subset of these studies (8 studies). For CHD, we split the full CHD cohort into those presenting with additional disease phenotypes (Syndromic CHD or CHD-S) and those with isolated CHD phenotypes (Non-syndromic CHD or CHD-NS). All de novo mutation lists were re-annotated using Variant Effect Predictor (VEP) version 8149. We filtered de novo mutations present in the non-psychiatric cohort of the Exome Aggregation Consortium (ExAC), which improves our likelihood of selecting pathogenic variants, as evidenced by the mutation burden in disease cohorts relative to controls being restricted to variants absent from ExAC50. Gene-set enrichment is tested using a 2-sided binomial exact test, with the theoretical expectation determined by the aggregated mutation expectation in the gene set51. This test explicitly controls for the enrichment in mutational burden, as enrichment is relative to the total number of mutations observed rather than the number of trios analyzed. We focused our enrichment tests on protein-truncating and missense mutations, and the combination of both (nonsynonymous mutations). To correct for multiple testing, we estimated the family-wise error rate across all protein networks using simulated de novo variants. Each simulation randomly drew mutations for each mutation class using per-gene probabilities from the mutation model, and the family-wise error used is the 5th percentile of minimum p-values across all simulations. The multiple testing correction thresholds for p-value significance are as follows: PTV = 2e-3, missense = 9e-4, and nonsynonymous = 1.4e-3. (references are listed in the main manuscript)
SyPPRes
The phenotype-based prioritization of candidate genes for human diseases within Protein interactions and human whole-genome and whole-exome sequencing studies is integrated within the Phenolyzer platform. Description of the algorithim and scoring system is described in Yang et al., 201535 SyPPRes: http://neurocomplex.usc.edu/
Statistics
Statistical methods used to test for protein domain enrichment, co-expression using the Brainspan database, and enrichment within human genetics datasets are detailed above. For western blot quantitation, statistical analysis was performed using unpaired t-tests and results are expressed as mean ± s.e.m. No statistical methods were used to pre-determine sample size. Our sample sizes are similar to those reports in previous publications3,10,37,52,53. Data distribution was assumed to be normal but this was not formally tested. Data collection and analysis were not performed blind to the conditions of the experiments. No randomization was used. No data points were excluded from the analyses performed.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (ref = PMID:27683222) with the dataset identifier PXD006277.
Datasets for individual protein complexes can also be downloaded at: SyPPRes: http://neurocomplex.usc.edu/
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Child Health and Human Development (MH104603-01A1; to M.P.C.) and NIH grant MH108728 (to K.W. and H.Y). Simons Foundation Autism Research Initiative (SFARI) grants 248429 and 345034 as well as DOD CDMRP AR110189 (to T.A.), B.N. thanks support from the Stanley Center for Psychiatric Research.
Bibliography
- 1.Bayes A, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nature neuroscience. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins MO, et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. Journal of neurochemistry. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 3.Coba MP, et al. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Science signaling. 2009;2:ra19. doi: 10.1126/scisignal.2000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins MO, et al. Proteomic analysis of in vivo phosphorylated synaptic proteins. The Journal of biological chemistry. 2005;280:5972–5982. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- 5.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nature neuroscience. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 6.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 7.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harbor perspectives in biology. 2011:3. doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. The Journal of cell biology. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naisbitt S, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 13.Vinade L, et al. Affinity purification of PSD-95-containing postsynaptic complexes. Journal of neurochemistry. 2003;87:1255–1261. doi: 10.1046/j.1471-4159.2003.02091.x. [DOI] [PubMed] [Google Scholar]
- 14.Grant SG. Synaptopathies: diseases of the synaptome. Current opinion in neurobiology. 2012 doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calderone A, Castagnoli L, Cesareni G. mentha: a resource for browsing integrated protein-interaction networks. Nature methods. 2013;10:690–691. doi: 10.1038/nmeth.2561. [DOI] [PubMed] [Google Scholar]
- 17.Bornigen D, et al. Concordance of gene expression in human protein complexes reveals tissue specificity and pathology. Nucleic Acids Res. 2013;41:e171. doi: 10.1093/nar/gkt661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor IW, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nature biotechnology. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 19.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin J, et al. Eukaryotic protein domains as functional units of cellular evolution. Science signaling. 2009;2:ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- 21.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids research. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014 doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgieva L, et al. De novo CNVs in bipolar affective disorder and schizophrenia. Human molecular genetics. 2014;23:6677–6683. doi: 10.1093/hmg/ddu379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirov G, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Human molecular genetics. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular psychiatry. 2011 doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. American journal of human genetics. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nature neuroscience. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavlowsky A, Chelly J, Billuart P. Emerging major synaptic signaling pathways involved in intellectual disability. Molecular psychiatry. 2012;17:682–693. doi: 10.1038/mp.2011.139. [DOI] [PubMed] [Google Scholar]
- 30.Xie Z, et al. Receptor Tyrosine Kinase MET Interactome and Neurodevelopmental Disorder Partners at the Developing Synapse. Biological psychiatry. 2016 doi: 10.1016/j.biopsych.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endele S, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nature genetics. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 32.Genovese G, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nature neuroscience. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zybailov B, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. Journal of proteome research. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nature methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014 doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, et al. Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Science signaling. 2016;9:rs8. doi: 10.1126/scisignal.aaf6716. [DOI] [PubMed] [Google Scholar]
- 38.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peca J, Feng G. Cellular and synaptic network defects in autism. Current opinion in neurobiology. 2012;22:866–872. doi: 10.1016/j.conb.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattheisen M, et al. Genome-wide association study in obsessive-compulsive disorder: results from the OCGAS. Molecular psychiatry. 2015;20:337–344. doi: 10.1038/mp.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elia J, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nature genetics. 2012;44:78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 44.Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome research. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter H, Hofree M, Ideker T. Genotype to phenotype via network analysis. Current opinion in genetics & development. 2013;23:611–621. doi: 10.1016/j.gde.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nature genetics. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Analytical chemistry. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 49.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosmicki JA, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nature genetics. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nature genetics. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coba MP, et al. TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:13987–13999. doi: 10.1523/JNEUROSCI.2433-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuthbert PC, et al. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:2673–2682. doi: 10.1523/JNEUROSCI.4457-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (ref = PMID:27683222) with the dataset identifier PXD006277.
Datasets for individual protein complexes can also be downloaded at: SyPPRes: http://neurocomplex.usc.edu/