Abstract
Widespread resistance towards antimony and reports of relapses following miltefosine treatment has severely affected the management of visceral leishmaniasis (VL) in the Indian subcontinent. Paromomycin (PMM), an aminoglycoside antibiotic, has been licensed for VL treatment in India in 2007. Although its use is still restricted in the field, unraveling the molecular mechanism of resistance towards PMM is the key to preserve the drug. In this study, PMM resistant lines were selected up to 100 μM of PMM in three distinct field isolates of Leishmania donovani at promastigote stage. The resistance induced at promastigote level was also evident in amastigotes which showed 6 fold decreases in PMM susceptibility. Comparative transcriptome profiling of PMM resistant (PMM-R) and the corresponding PMM sensitive (PMM-S) parasites revealed modulated expression of 500 genes (1.5 fold cut off) in PMM-R parasites. Selected genes were validated for their modulated expression by quantitative real-time PCR. Functional classification and pathway analysis of modulated genes indicated probable adaptations in drug resistant lines which included a) reduced oxidative phosphorylation; b) increased glycosomal succinate fermentation and substrate level phosphorylation; c) dependency on lipids and amino acids for energy generation; d) reduced DNA synthesis and increased DNA damage repair and e) decreased protein synthesis and degradation. Interestingly, PMM-R parasites showed a marked increase in PMM susceptibility in presence of verapamil and amlodipine, antagonists of Ca2+ channel that are also modulators of ABC transporters. Moreover, infection of macrophages by PMM-R parasites led to modulated nitric oxide (NO) levels while reactive oxygen species (ROS) level remained unaltered. The present study highlights the putative mechanisms of PMM resistance in Leishmania.
Keywords: Leishmania donovani, Drug resistance, Paromomycin, Transcriptome, ABC transporters, Nitric oxide, Visceral leishmaniasis
Graphical abstract
1. Introduction
Visceral leishmaniasis (VL) or kala-azar is the most severe form of leishmaniasis and may be fatal, if left untreated. In the absence of any vaccine, control of VL relies mainly on chemotherapy. Existing drugs for VL have serious drawbacks in terms of safety, efficacy, cost, and development of resistance. Miltefosine (MIL) was considered as one of the main pillars of VL elimination program in India; however, reports of relapses following MIL treatment and decline in its efficacy raised concerns regarding its utility in VL control (Bhandari et al., 2012, Sundar et al., 2012, Rijal et al., 2013). In a recent study the declining efficacy of MIL for the treatment of post kala-azar dermal leishmaniasis (PKDL) patients, considered to be a major reservoir for transmission of the disease, has been reported (Ramesh et al., 2015). Another drug, amphotericin B, is associated with toxicity (Srivastava et al., 2011) and its liposomal formulation (AmBisome) has been recommended as a first line treatment of VL in the Indian subcontinent (World Health Organisation, 2010).
Paromomycin (PMM) is another treatment option for VL control in the Indian subcontinent and has been found to be effective as monotherapy as well as in combination with other drugs (Sinha et al., 2011, Sundar et al., 2011, Rahman et al., 2017). Development of resistance towards aminoglycosides such as PMM has been frequently encountered in a variety of bacteria and PMM resistance in Leishmania can readily be induced in vitro (El-On et al., 1991, Maarouf et al., 1998, Hendrickx et al., 2012). A report of increased ED90 towards PMM in L.donovani isolates from VL cases residing in a high endemic region is alarming (Prajapati et al., 2012), suggesting that PMM is at considerable risk for the development of dug resistance. Therefore, this issue needs to be proactively addressed in laboratory studies.
Previous studies on PMM resistance in Leishmania parasites suggest that drug resistance is associated with increased membrane fluidity, reduced accumulation of drug (Jhingran et al., 2009, Bhandari et al., 2014), altered mitochondrial energy metabolism (Maarouf et al., 1997) and increased tolerance to the host defence mechanisms (Bhandari et al., 2014). In a proteomic study, a number of proteins showed differential expression in experimental PMM resistant (PMM-R) parasites as compared to the wild type (Chawla et al., 2011).
In the present study, gene expression profiling of experimentally selected PMM-R and the corresponding PMM sensitive (PMM-S) parasites was carried out for identification of genes responsible for possible mechanisms of PMM resistance, using oligonucleotide array representing genomic sequences of L. infantum and L. major (Leprohon et al., 2009). Differentially modulated genes and pathways that may contribute to PMM resistance were identified. Role of ABC transporters was further explored in PMM resistance using verapamil and amlodipine that are Ca2+ channel antagonists and ABC transporter modulators. Production of nitric oxide (NO) and reactive oxygen species (ROS) by PMM-S/PMM-R parasites upon infection to the host macrophages was also studied.
2. Materials and methods
2.1. Culture of parasites
Three distinct clinical isolates of L. donovani, were obtained from bone marrow aspirates of VL patient, as described in our previous studies (Bhandari et al., 2014, Deep et al., 2017). One of these isolates (MHOM/IN/2000/K133) was obtained from antimony responsive VL patient reporting to Safdarjung Hospital, New Delhi and the other two isolates (MHOM/IN/09/BHU573/0 and MHOM/IN/09/BHU568/0) were obtained from antimony unresponsive patients that reported to Kala-Azar Medical Research Centre, Muzaffarpur, Bihar.
The study was approved by the Ethics Committee of the Institute of Medical Sciences, Banaras Hindu University (Dean, 2007–08/42, dated 15-05-2008), Varanasi and Institutional Ethics committee of Safdarjung Hospital & VMMC (VMMC/SJH/PROJECT/22-10-2012/7), New Delhi, India. Written informed consent was obtained from patients.
Promastigotes were cultured at 25 °C in M199 medium with 25 mM HEPES (pH7.4) supplemented with 10% FBS, 100 IU penicillin G and 100 μg/ml of streptomycin.
2.2. Selection of PMM resistant parasites
PMM resistance was artificially selected in cloned lines (derived by limited dilution cloning) of all the three isolates. Parasites were gradually exposed to increasing PMM concentration (25–100 μM) at promastigote stage. At each step, parasites were cultured for at least 5–8 passages to attain steady and optimal cell growth. The concentration of drug was increased only when the adapted isolate showed comparable growth kinetics to wild type culture as described previously (Bhandari et al., 2014).
2.3. Drug susceptibility assay at promastigote stage
Drug susceptibility assays at promastigote stage were performed as previously described (Deep et al., 2017). Briefly, log-phase promastigotes were harvested and counted in a counting chamber. Promastigotes were seeded into 96-well plates at 5 × 105 parasites/well and incubated with 200 μl medium alone or serial dilutions of PMM (1.9 μM–2000 μM). 50 μl resazurin (Sigma Aldrich, USA) [0.0125% (w/v) in PBS] was added to each well after72-h incubation at 25 °C and the plates were incubated further for 18 h. Cell viability was measured fluorimetrically (λex 550 nm; λem 590 nm, Infinite M200, Tecan, Switzerland). The results were expressed as the percentage reduction in the parasite viability compared to that in untreated control wells. 50% inhibitory concentration (IC50) and 90% inhibitory concentration (IC90) was calculated using sigmoidal regression analysis. All experiments were performed at least thrice in quadruplicates.
2.4. Drug susceptibility assay at intracellular amastigote stage
In vitro PMM susceptibility was assessed at intracellular amastigote level by following macrophage-amastigote model as previously described (Deep et al., 2017). Briefly, the mice peritoneal exudates derived macrophages (PECs) were infected with late log phase promastigotes at a ratio of 10: 1 (parasite: macrophage), plated into 8 well chamber slides and incubated for 16 h at 37 °C in 5% CO2. Excess, non-adhered promastigotes were removed by washing and infected cells were re-incubated for 48 h with different dilutions of PMM (0, 10, 20, 40, 80, 120 and 150 μM). Macrophages were then examined for intracellular amastigotes after staining with Diff-Quik solutions. The number of L. donovani amastigotes was counted in 100 macrophages, at 1000× magnification. The survival rate of parasites relative to untreated macrophages was calculated. IC50 and IC90 were determined by sigmoidal regression analysis. The assays were performed in duplicate and repeated at least thrice.
2.5. Oligonucleotide array
Single-color microarray-based gene expression profiling was performed using a high-density Leishmania multispecies 60-mer oligonucleotide microarray slide [8 × 15K format] representing the entire genome of L. infantum and L. major. The microarray chip, printed by Agilent Technologies, USA, included a total of 9233 Leishmania-specific genes including 540 control probes as described earlier (Rochette et al., 2008, Leprohon et al., 2009, Kulshrestha et al., 2014).
2.6. RNA extraction and evaluation
Total RNA was extracted from 108 late log phase promastigotes using Trizol reagent as described by the manufacturer. RNA clean up was performed using RNeasy Plus mini kit (Qiagen, USA) according to manufacturer's instructions. The purified RNA was quantified using Nanodrop by estimating the absorbance at 260 and 280 nm. The quality and integrity of RNA was assessed on RNA 6000 Nano Assay Chips on Bioanalyzer 2100 (Agilent Technologies, USA) (Kulshrestha et al., 2014).
2.7. RNA labelling, amplification, hybridization and data analysis
500 ng of total RNA was reverse transcribed using oligo dT primer tagged to T7 promoter sequence. cDNA thus obtained was converted to cRNA using T7 RNA polymerase enzyme and Cy3 dye. 600 ng of Cy3 labeled cRNA was hybridized on the array (AMADID: 27511) using the Gene Expression Hybridization kit (Agilent Technologies, USA) in Sure hybridization Chambers (Agilent) at 65° C for 16 h. Hybridized slides were washed using Agilent Gene Expression wash buffers (Agilent Technologies, USA) and scanned on a G2505C scanner (Agilent Technologies, USA). Images thus obtained were quantified using Agilent's Feature Extraction Software Version-10.7. Feature extracted raw data was analyzed using GeneSpring GX12.6.1 microarray data and pathway analysis tool. Quartile (75th percentile) normalization was performed. Storey and bootstrapping analysis was performed for multiple testing corrections. All microarray data is available on the GEO NCBI database in the MIAME format; http://www.ncbi.nlm.nih.gov/geo with the GEO accession number GSE74208. DNA microarray data were analyzed by custom R programs to illustrate the expression profile of PMM-R L. donovani by extrapolating on a chromosome map of L. infantum. Gene ontology annotation for functional classification of modulated genes was performed using GeneDB, BLAST2GO, and AmiGO databases. The pathway analysis was carried out using GeneSpring GX12.6.7 and KEGG pathway analysis tool. Interaction of genes and identification of interacting partners of differentially modulated genes in PMM-R parasites was studied using String 9.01 database (Kulshrestha et al., 2014). Biological and statistical criteria were used to identify differentially expressed genes between PMM-S and PMM-R lines. Biological threshold of differential expression was 1.5 (over-expression) or 0.66 (under-expression). Statistically significant differentially expressed genes were determined by t-test (unpaired) for two groups; (p value cutoff) < 0.05.
2.8. Quantitative real time PCR
A total of 15 genes (8 up- and 7 down-regulated) were selected to validate their modulated expression in PMM-R lines by quantitative real time PCR (Q-RT-PCR). Comparative gene expression analysis was carried out using all the three PMM-S and PMM-R parasites. Total RNA was isolated from promastigotes using TRIzol reagent (Invitrogen, USA) and reverse transcribed to cDNA. Equal amounts of cDNA were run in triplicate and amplified in 25 μl reactions containing 1 × Fast SYBR Green master mix (Applied Biosystems, USA), 100 ng/ml forward and reverse primers. Reactions were carried out using ABI 7500 Real time PCR system (Applied Biosystems, USA). Three independent RNA preparations were used for each Q-RT-PCR experiment. Gene expression levels were normalized using constitutively expressed genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cystathionine-β-synthase (CBS). Quantification of the relative changes in target gene expression was calculated using 2−ΔΔCt method. (Decuypere et al., 2008, Kumar et al., 2012, Bhandari et al., 2014). The list of primers, designed using Primer express software version 3.0 (Applied Biosystems, USA) and used for real time PCR with name and functional relevance of the gene is given in Table S1.
2.9. PMM susceptibility in presence of modulators of ABC transporters
Susceptibility towards PMM was determined in all the three PMM-R and PMM-S parasites in presence of verapamil and amlodipine. At promastigote stage, both PMM-S and PMM-R isolates were incubated with different drug concentrations in presence of 8 μM of verapamil or 0.5 μg/ml of amlodipine and IC50 was determined as mentioned above in section 2.3. Similarly, at intracellular amastigote stage, PMM susceptibility was determined by incubating the infected macrophages (both by WT and PMM-R parasites) with different drug dilutions of PMM as described in section 2.4 and verapamil (8 μM) or amlodipine (1 μg/ml). At this concentration, the modulators alone were not lethal to PMM-S/PMM-R isolates (data not shown). IC50 was calculated by the sigmoidal regression analysis.
2.10. Assessment of NO production in PMM-S/PMM-R parasites infected macrophages
NO production in infected macrophages was determined as previously described (Kulshrestha et al., 2011). The mice PECs, stimulated with 1 μg/ml of lipopolysaccharide (LPS) from Escherichia coli, were infected with PMM-S or PMM-R parasites as described in section 2.4 and incubated for 48 h. The NO levels were estimated in cell culture supernatant by reducing the nitrate (accumulated over 48 h) to nitrite with nitrate reductase and measuring the nitrite concentration colorimetrically by Griess reaction. The amount of nitrite accumulated was calculated from a standard curve constructed with different concentrations of sodium nitrite (in a linear range between 10 and 80 μM).
2.11. Assessment of intracellular ROS production in PMM-S/PMM-R parasites infected macrophages
Intracellular ROS production was assessed as described previously (Deep et al., 2017) with modifications. The mice PECs infected with PMM-S/PMM-R parasites as described in section 2.4, were incubated for 48 h without or with PMM (13 μM). Intracellular ROS levels (expressed as mean fluorescence intensity unit) were measured fluorimetrically at 495 nm excitation and 527 nm emission wavelength (Infinite M200, Tecan, Switzerland) using H2DCFDA dye.
3. Results
3.1. Selection of PMM resistance in indian isolates
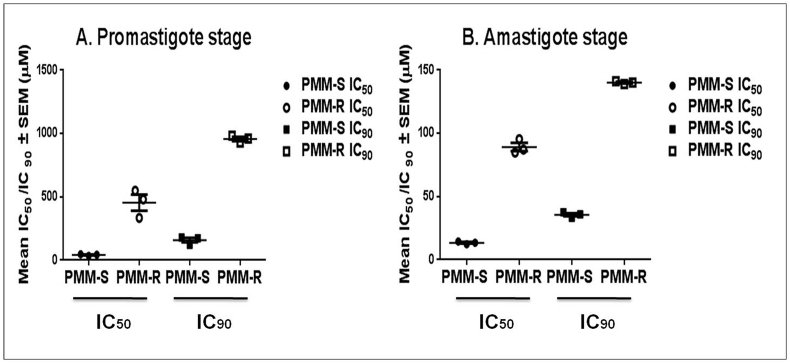
Cloned parasite lines of the three distinct isolates of L. donovani were successfully adapted step by step for PMM tolerance up to 100 μM (61.5 μg/ml) of drug concentration, achieving stable PMM resistance in 32 weeks. Above this concentration, the parasites were unable to show similar growth kinetics as their corresponding wild type parasites. PMM susceptibility was determined at both promastigote and intracellular amastigote stages for the three PMM-S and PMM-R parasites. Drug resistant parasites showed a significant increase in IC50 and IC90 values as compared to their corresponding wild type parental parasites at both the life stages. In promastigotes, the mean IC50 values were >10 fold increased (454.38 ± 109.59 μM vs 41.71 ± 6.48 μM) and mean IC90 were >6 fold increased (957.2 ± 27.83 μM vs 158.31 ± 31.13 μM) (Fig. 1A) in comparison with the parent strain. In amastigotes, there was 6 fold increase of mean IC50 (58.51 ± 0.16 μM vs 9.77 ± 0.61 μM) and a >4 fold increase of mean IC90 (124.7 ± 2.01 μM vs 26.50 ± 1.05 μM) (Fig. 1B). All drug resistant parasites exhibited similar morphology as compared to the wild type parental lines and the drug-resistance phenotype was stable in the absence of PMM (followed up to 12 passages).
Fig. 1.
Drug susceptibility of PMM-R and the corresponding WT parasites: Sensitivity profile of three PMM resistant and their respective wild type isolates towards PMM at A. promastigote and B. amastigote stage. IC50 and IC90 values are the mean of three independent experiments performed in triplicate.
3.2. Comparative transcriptome profiling of genes in PMM-S and PMM-R
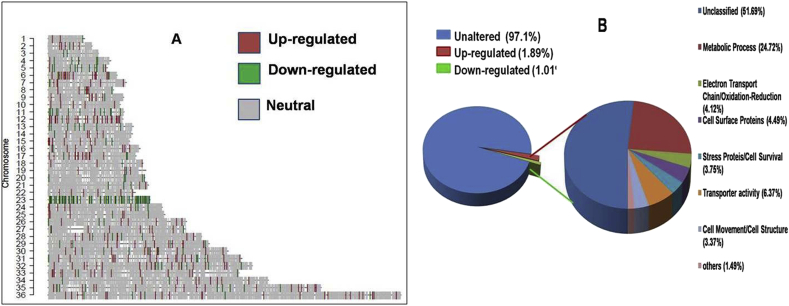
Comparative transcriptome profiling of PMM-S (K133 WT) and PMM-R isolate (K133 PMM-R) was performed using single color DNA microarray technology. The plot of log2 transformed expression ratio of K133 PMM-R (red line) compared to K133 WT (green line) as a function of the chromosomal location of microarray probes is shown in Fig S1. Based on 1.5 fold cut off and p value < 0.05 in PMM-R parasites, the plot revealed that 500 genes (approx. 5.45%) were differentially modulated, of which, 327 genes were up-regulated and 173 genes were down-regulated. A few genes showed ≥4 fold up- or down-regulation in drug resistant mutants (Fig S1). The pattern of global mRNA expression is shown in Table S2. To analyse gene expression level on genomic scale, we generated chromosome map (Fig. 2A) using Custom R program. Analysis of chromosome map identified that higher number of up-regulated genes were located on chromosomes 6, 12, 32, 35 and 36. The maximum number of up-regulated genes was on chromosome12, 50% of which were hypothetical proteins. Other genes located on chromosome 12 were glucose-6-phosphate isomerase (involved in glycolysis), NADH-flavin oxidoreductase, (involved in fatty acid metabolism) and alanine aminotransferase (responsible for conversion of alanine to pyruvate). Up-regulated genes included glutamine synthetase (glutamine synthesis) and folate/biopterin transporter (folate metabolism) on chromosome 6, chaperone protein DNAj, ubiquitin conjugating enzyme and chloride channel protein on chromosome 32, SNF-7 like protein and amino acid permease on chromosome 35, and exoribonuclease 2, ubiquitin protein ligase, nucleoporin and GPI-α mannosyltransferaseon chromosome 36.There were no up regulated genes on chromosome 3, 13, 20 and 23. The maximum number of down-regulated genes were located on chromosome 23 in PMM-R isolates that included the Cu2+ ion transporter (Linj.23.0870), hydrogen ion transmembrane transporter (Linj.23.1910), genes for DNA replication (Linj.23.1640), protein synthesis (Linj.23.0340), galactose/sucrose metabolism (Linj.23.1070), fatty acid biosynthesis (Linj.23.1560) fatty acid β oxidation (Linj.23.0860) and glutathione metabolism (Linj.23.0050) (Fig. 2A).
Fig. 2.
A. Comparative gene expression of PMM-S and PMM-R parasite on chromosomal map. Chromosome map for expression data was generated using Custom R program. Red and green lines indicate up regulated and down-regulated genes in PMM-R parasite, respectively, while gray lines show the genes that are equally expressed in both samples and white regions represent those not hybridized to the probe. Chromosomes showing high up regulation in PMM resistance included Chromosome 12 and Chromosom 36 whereas high down regulation is evident on Chromosome 23. B. Distribution of genes differentially modulated during PMM resistance in Leishmania donovani according to gene ontology (GO) functional categories. Overall distribution of genes shows total number of unique genes up-regulated or down-regulated is 2.9%. GO categories of genes differentially expressed in PMM-R parasite suggest that genes belonging to metabolic process, electron transport chain/oxidation-reduction, cell surface proteins, stress proteins, cell movement and transporter activity were affected. Unclassified proteins include hypothetical proteins (proteins with unknown function and not tested experimentally) and proteins with no GO categories (unclassified) that have been experimentally characterized. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Pathway analysis
Both up- and down-regulated genes were analyzed using BLAST2GO, AmiGO and KEGG databases to identify the pathways involved in PMM resistance. The percentage of differentially expressed genes according to GO function categories is shown in Fig. 2B. The fold change in expression of >2 fold up- and down-regulated genes with their functional categories is listed in Table S3. According to functional description and interacting partner of specific genes at protein level (String 9.01 database), a total of 129 genes were classified into eight different categories as discussed below.
3.3.1. Glycolysis, substrate level phosphorylation & oxidative phosphorylation
PMM-R parasites have been reported to show increased expression of several enzymes involved in glycolytic pathway (Chawla et al., 2011). In the present study, up-regulation of 2 enzymes of glycolytic pathway, glucose-6-phosphate isomerase and phosphoglycerate mutase enzymes was observed.
Among the enzymes that take part in TCA cycle, isocitrate dehydrogenase involved in synthesis of α-ketoglutarate was up-regulated (1.5 fold), while succinate dehydrogenase involved in conversion of succinate to fumarate was down-regulated (2.17 fold). Glycosomal phosphoenol pyruvate carboxykinase (2.13 fold), which converts phosphoenol pyruvate to oxaloacetate and cytosolic malate dehydrogenase (2.80 fold), that reversibly catalyzes the inter-conversion of malate and oxaloacetete, were up-regulated. These enzymes are involved in glycosomal succinate fermentation.
A down-regulation of 2.23 fold in NADH-ubiquinone oxidoreductase (Complex system I) was observed.
3.3.2. Lipid metabolism
Lipase involved in lipid metabolic process was up-regulated (2.12 fold). Another gene coding for choline/ethanolamine phosphotransferase, involved in synthesis of phosphatidylcholine, was up-regulated (2.54 fold). A down-regulation (2.22 fold) in lathosterol oxidase like protein involved in biosynthesis of sterol was observed. 3-ketoacyl-CoA thiolase that catalyzes β-oxidation of long chain fatty acid showed down-regulation (2.10 fold) in expression.
3.3.3. Down-regulation of genes involved in DNA synthesis & translation machinery
Genes responsible for DNA replication, DNA polymerase θ (LinJ.23.1640), H1 histone like protein (LinJ.33.3390) and endonuclease/exonuclease activity (LinJ.28.1000) were down-regulated. Genes involved in translational machinery like tryptophenyl t-RNA synthetase involved in aminoacyl-tRNA biosynthesis during translation, 5.8 S ribosomal RNA and 28 S ribosomal RNA were also down-regulated. On the other hand a gene encoding a hypothetical protein (LinJ.26.1540), interacting with other proteins responsible for DNA damage repair, was up-regulated. Up-regulation of exoribonuclease and RNA binding protein (LinJ.09.0100) was also observed in PMM resistance.
3.3.4. Modulation in protein/amino acid metabolic processes
Genes encoding phosphoglycan beta 1,3 galactosyltransferase (involved in glycosylation of proteins) and ubiquitin-conjugating enzyme protein (involved in post-translational protein modification) were up-regulated. We also observed up-regulation of SNF7 like proteins (LinJ.35.0580) which are involved in protein sorting and interact with proteins responsible for vesicular protein trafficking. A number of peptidases (carboxypeptidase and metallopeptidase) involved in protein degradation were down-regulated.
3.3.5. Modulated expression of transporters
ATP Binding Cassette transporters (ABC transporters) ABC1 and ABCA7 and ABCC5 showed higher mRNA expression in PMM-R parasites. Interestingly, an ABC transporter, ABCB3 was down regulated. Nucleoside transporter1, vacuolar proton translocating ATPase and Cu2+ ion transporter showed down-regulated expression in PMM resistant isolates. On the other hand, Folate/Biopterin transporter, glucose transporter, P-type ATPase showed up-regulated expression.
3.3.6. Cellular components/cell movement/cell signalling
Expression of genes coding for cellular movement like paraflagellar rod component, and dynein like protein were up-regulated, while genes for coronin and centrin were down-regulated in PMM-R isolates. Ca2+ dependent signalling, which contributes to microtubule formation during cell division, was up-regulated. Protein phosphatase 2c like protein, serine/threonine phosphatase and a protein kinase was up-regulated in PMM-R.
3.3.7. Proteins involved in intracellular survival
Isoprenyl cysteine alpha-carbonyl methylesterase (ecotin putative), inhibitor of serine peptidase, that helps parasites to avoid killing by host macrophages (Faria et al., 2011) was up-regulated. Up-regulation was also observed for tryparedoxin-like protein and leishmanolysin, GP63, having protein tyrosine phosphatase activator activity. Peroxidoxin (peroxiredoin), trypanothione synthetase and pteridine reductase 1 (PTR1) were down-regulated.
3.3.8. Cell surface proteins
Genes encoding cell surface proteins, such as GPI anchored proteins, amastin like proteins and ppg3 related protein, exhibited up-regulation in PMM-R isolates.
3.4. Validation of modulated gene expression with Q-RT-PCR
A total of 15 genes indicative to play a role in PMM resistance were selected to validate their expression by Q-RT-PCR in three PMM-S and PMM-R parasites. Fold change in gene expression of all the three PMM-R/PMM-S obtained by Q-RT-PCR was compared with the fold difference in gene expression obtained by microarray experiments (Fig S2). Results obtained by Q-RT-PCR were in agreement with the transcriptomic data derived by microarray experiments for all the selected genes in all the three PMM-R vs PMM-S parasites.
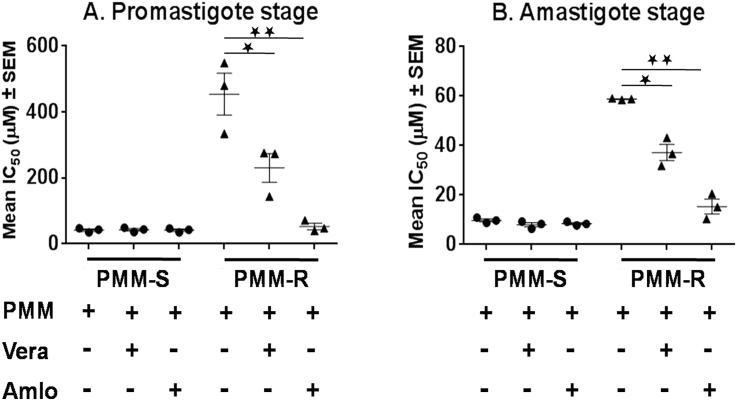
3.5. Reversion of PMM resistance phenotype in presence of modulators of ABC transporters
We determined the susceptibility towards PMM in presence of modulators of ABC transporters, verapamil and amlodipine. All the three PMM-R isolates showed a significant decrease of approx. 2 fold in the IC50 in presence of verapamil and more than 7 fold decreases in IC50 in presence of amlodipine at promastigote stage (Fig. 3A). Partial reversion of resistant property was also evident at intracellular amastigote stage as the IC50 of PMM for the resistant strains decreased approx. 1.4–1.8 fold in presence of verapamil and approx. 2.8 to >5 fold in presence of amlodipine (Fig. 3B). However, there was no significant alteration in IC50 of PMM-S strains in presence of verapamil or amlodipine at either promastigote or amastigote stage (Fig. 3A and B).
Fig. 3.
Susceptibility of PMM-R isolates in presence of inhibitors to ABC transporter.In vitro susceptibility towards PMM in presence of verapamil (Vera) and amlodipine (Amlo) in three different PMM-Rand WT isolates at A. Promastigote stage and B. Amastigote stage. Each individual value represents mean fold change in IC50 ± SD of the results from three separate assays in duplicates. * represents p = 0.05, ** represents p = 0.01, *** represents p = 0.001.
3.6. Modulation in NO but not ROS production in PMM-S vs PMM-R infected macrophages
NO and ROS levels were measured in PMM-S/PMM-R infected mice PECs. NO levels were significantly increased in infected macrophages either with PMM-S or PMM-R parasites as compared to un-infected (UI) macrophages (p < 0.5). PMM-R parasites showed a significant decrease of 2.13 ± 0.17 fold (p < 0.5) in NO level in LPS stimulated macrophages as compared to the PMM-S parasites (Fig S3A).
No significant difference was observed in ROS levels in LPS stimulated macrophages when infected with PMM-S vs PMM-R parasites in presence/absence of PMM (Fig S3B).
4. Discussion
PMM is an effective treatment option for antileishmanial therapy and emergence of resistance towards this drug can only be appropriately judged upon its regular use in the field. However, understanding the alterations in the parasite behaviour associated with PMM resistance is essential to assess the propensity for emergence and spread of resistant strains (Hendrickx et al., 2015). To understand the mechanism of resistance towards PMM, drug resistant parasite lines were selected in vitro and mRNA expression profiling was compared between PMM-S and PMM-R parasites. This allowed the identification of a number of genes that may play an important role in generation of resistant phenotype. We adapted three different field isolates for PMM resistance and validated modulated expression of selected genes in all the three parasites in comparison with their respective wild type parental strains.
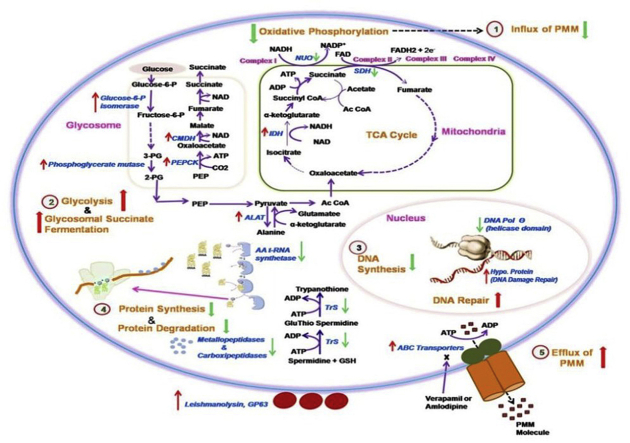
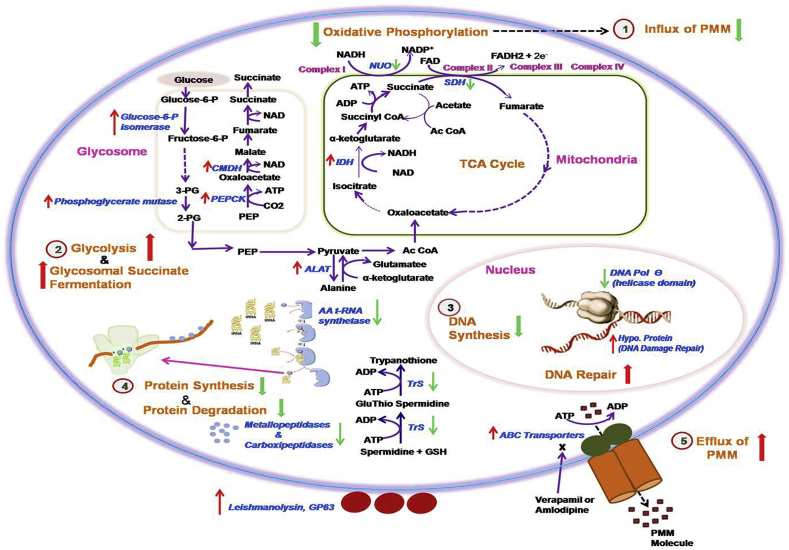
Internalization of aminoglycoside antibiotic across the cytoplasmic membrane in bacteria requires metabolic energy from electron transport chain in an oxygen dependent manner (Bryan and Kwan, 1983). A significant inhibition of PMM accumulation has been observed in presence of an inhibitor of electron transport chain (Jhingran et al., 2009). Further, in a recent study, decreased expression of proteins involved in oxidative phosphorylation has been reported in PMM-R parasites (Chawla et al., 2011). In our study, genes coding for enzyme succinate dehydrogenase involved in TCA cycle and complex II, and NADH ubiquinone oxidoreductase involved in complex I of oxidative phosphorylation were down-regulated, indicating down regulation of aerobic respiratory system. It may be an adaptation of PMM-R isolates to facilitate reduced internalization of PMM within the cell. In presence of down-regulated oxidative phosphorylation, there may be up-regulated substrate level phosphorylation in PMM-R parasites for energy generation, as evident by the up-regulation of enzymes of glycosomal succinate fermentation. Succinate fermentation was suggested as the major pathway for regenerating glycosomal pools of ATP/NAD+ in T. brucei procyclic stages (Creek et al., 2015). Predicted changes in metabolic fluxes in PMM-R L.donovani are shown in Fig. 4.
Fig. 4.
Predicted adaptations in L.donovani in PMM resistance. Genes altered in PMM-R parasites are represented here. Genes marked with up and down arrow represent respectively the up-regulated genes and the down-regulated genes in PMM-R parasites. 1,2,3,4 and 5 are probable adaptations in PMM-R parasites.1. Down-regulation of oxidative phosphorylation leads to reduced influx of PMM drug, 2. Enzyme for glycolysis and glycosomal succinate fermentation were up-regulated indicating increased substrate level phosphorylation, 3. Down-regulated DNA polymerase θ suggesting reduced DNA synthesis. Hypo. protein responsible for DNA damage repair were up-regulated 4.Tryptophenyl-t-RNA synthatase and several ribosomal proteins were down-regulated suggesting reduced protein synthesis, Down-regulation of metallo-and carboxipeptidases indicated reduced protein degradation, 5. Up-regulation of ABC transporters and reversion of resistant phenotype in presence of modulators, verapamil or amlodipine suggested probable role of ABC transporters in selection of PMM resistance. Abbreviations used are as follows: 2-PG, 2-phosphoglycerate; 3-PG, 3-phosphoglycerate; AA t-RNA Synthetase, Aminoacyl t-RNA Synthetase; Ac CoA, Acetyl Coenzyme A; ALAT, Alanine aminotransferase; CMDH, Cytosolic malate dehydrogenase; DNA pol. θ, DNA Polymerase θ; Fructose-6-P, Fructose-6-phospahet; Glucose-6-P, Glucose-6-phosphate; GSH, Glutathione; Hypo. Protein, Hypothetical protein; IDH, Isocitrate dehydrogenase; NUO, NADH ubiquinone oxidoreductase; PEP, Phosphoenol pyruvate; PEPCK, Phosphoenol pyruvate carboxykinase; SDH, Succinate dehydrogenase; TrS, Trypanothione synthetase.
We observed up-regulation of enzymes involved in glycolytic pathway indicating up-regulated glycolysis in PMM-R isolates. Further, up-regulation of lipase and alanine aminotransferase was observed in PMM-R parasites suggesting that PMM-R parasites may rely on amino acid and lipid metabolism for energy generation.
In PMM-R mutant parasites down-regulation of DNA polymerase θ (helicase domain only) suggested reduction in DNA replication. Further, drug resistant parasites showed down-regulation of genes responsible for exo/endonuclease activity and up-regulation of genes for DNA damage repair and that may recompense the decreased DNA synthesis and damage due to drug pressure.
A decrease in translation machinery in PMM-R parasites was observed as evident by down-regulation of tryptophenyl t-RNA synthetase, 28S ribosomal RNA and 5.8S ribosomal RNA. Interestingly, our results were contrary to that observed in comparative proteomics study of WT and PMM-R parasites by Chawla et al. (2011) where they reported increased expression of translational machinery. Therefore, it would be interesting to explore comparative mechanism of gene regulation at transcription and translation level in L. donovani parasites in presence of PMM drug pressure. Further, we observed down-regulation of a number of metallo- and carboxy-peptidases which are responsible for protein degradation. This may be a compensatory approach of drug resistant parasites to overcome reduced translational machinery.
Overexpression of ABC transporters has been involved in the development of drug resistance in L. donovani and L. infantum (Leprohon et al., 2006, Kumar et al., 2012, Purkait et al., 2012). Potential role of two ABC transporters, MRPA (ABCC subfamily) and MDR1 (ABCB subfamily) has been reported in drug resistance. MRPA has been found to be involved in Sb(v) resistance in axenic and intracellular amastgotes of L. infantum (El Fadili et al., 2005). Another ABC transporter, ABCG4 (ABCG subfamily) has been found to be involved in miltefosine resistance (Castanys-Muñoz et al., 2007). Analysis of microarray data identified up-regulation of ABC transporters ABC1 & ABCA7 (ABCA subfamily) & ABCC5 (ABCC subfamily) in PMM-R parasites. Recently we have reported the overexpression of MDR1 and MRPA in paromomycin resistance in Indian isolates (Bhandari et al., 2014). In this study, the use of modulators of ABC transporters, verapamil or amlodipine, resulted in a significant increase in PMM susceptibility of PMM-R parasites, suggesting the possible role of ABC transporters in selection of PMM-R phenotypes. However, in PMM-R parasites, specific targets of these modulators need to be explored.
Several genes responsible for intracellular survival of Leishmania parasite showed modulated expression. Up-regulated ecotin gene suggested that PMM-R mutants may have better survival than PMM-S parasites against killing by host macrophages. GP63, a metallo-peptidase which is a protein tyrosine phosphatase activator, was up-regulated in PMM-R parasites. Tyrosine phosphatase is a negative regulator of NO production by host macrophages upon Leishmania infection (Forget et al., 2006). PMM induce NO production in host macrophages, although parasite killing mechanism of the drug is independent of NO (Kulshrestha et al., 2011). Decreased NO levels by PMM-R isolates infected macrophages as compared to PMM-S isolates infected macrophages suggested that PMM-R parasites successfully modulated NO production within host macrophages. Trypanothione synthetase and peroxiredoxin were down-regulated in PMM-R mutants, suggesting reduced defence against oxidative stress. Further, similar ROS levels in PMM-S vs PMM-R infected macrophages with or without PMM validated mechanism of action or resistance to PMM was not associated with ROS as reported in previous studies (Bhandari et al., 2014, Moreira et al., 2011).
Our results indicated reduced intracellular concentration of PMM in PMM-R phenotypes by decreased drug internalization and increased drug efflux as reported in bacterial aminoglycoside resistance (Moore et al., 1999). PMM resistant Leishmania parasites share some mechanisms of resistance with MIL-R and SAG resistant (SAG-R) parasites like reduced drug internalization and increased drug efflux due to up-regulation of ABC transporters (Legare et al., 2001, Kulshrestha et al., 2014). We also observed some unique adaptations in PMM-R parasites. MIL-R and SAG-R parasites exhibit decreased drug induced ROS accumulation (Deep et al., 2017, Moreira et al., 2011) and thus are tolerant to ROS induced stress, whereas, PMM-R parasites did not show any tolerance towards oxidative stress. Unlike MIL-R, PMM-R parasites are not only more tolerant to nitrosative stress (Bhandari et al., 2014) but also modulate macrophage NO production for better survival in host. Further, transcriptome profiling indicated metabolic evolution of PMM-R parasites by depending on glycolysis, glycosomal succinate fermentation and substrate level phosphorylation for energy generation.
Overall, our results suggest that the mechanism of resistance towards PMM is a multifactorial phenomenon (Fig. 4). A number of genes/pathways responsible for PMM resistance, identified in this study by genomic microarray need to be functionally characterized at both promastigote and amastigote stages and/or PMM resistant clinical isolates in order to confirm their role in PMM resistance.
Acknowledgement
We are thankful to Prof Marc Ouellette, Research Centre in Infectious Diseases, Faculty of Medicine, Laval University,Quebec, Canada, for kindly sharing the microarray design with us. The work has been funded by Indian Council of Medical Research grant No. 5/8-7(102)/2012-ECD-II. We report no conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2017.10.004.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Bhandari V., Kulshrestha A., Deep D.K., Stark O., Prajapati V.K., Ramesh V., Sundar S., Schonian G., Dujardin J.C., Salotra P. Drug susceptibility in Leishmania isolates following miltefosine treatment in cases of visceral leishmaniasis and post kala-azar dermal leishmaniasis. PLoS Negl. Trop. Dis. 2012;6:e1657. doi: 10.1371/journal.pntd.0001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari V., Sundar S., Dujardin J.C., Salotra P. Elucidation of cellular mechanisms involved in experimental paromomycin resistance in Leishmania donovani. Antimicrob. Agents Chemother. 2014;58(5):2580–2585. doi: 10.1128/AAC.01574-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan L.E., Kwan S. Roles of ribosomal binding, membrane potential and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 1983;23:835–845. doi: 10.1128/aac.23.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanys-Muñoz E., Alder-Baerens N., Pomorski T., Gamarro F., Castanys S. A novel ATP-binding cassette transporter from Leishmania is involved in transport of phosphatidylcholine analogues and resistance to alkyl-phospholipids. Mol. Microbiol. 2007;64(5):1141–1153. doi: 10.1111/j.1365-2958.2007.05653.x. [DOI] [PubMed] [Google Scholar]
- Chawla B., Jhingran A., Panigrahi A., Stuart K.D., Madhubala R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin -susceptible -resistant Leishmania donovani. PLoS One. 2011;6(10):e26660. doi: 10.1371/journal.pone.0026660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D.J., Mazet M., Achcar F., Anderson J., Kim D., Kamour R., Morand P., Millerioux Y., Biran M., Kerkhoven E.J., Chokkathukalam A., Weidt S.K., Burgess K.E.V., Breitling R., Watson D.G., Bringaud F., Barrett M.P. Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 2015;11(3):e1004689. doi: 10.1371/journal.ppat.1004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decuypere S., Vanaerschot M., Rijal S., Yardley V., Maes L., de Doncker S., Chappuis F., Dujardin J.C. Gene expression profiling of Leishmania (Leishmania) donovani: overcoming technical variation and exploiting biological variation. Parasitology. 2008;135:183–194. doi: 10.1017/S0031182007003782. [DOI] [PubMed] [Google Scholar]
- Deep D.K., Singh R., Bhandari V., Verma A., Sharma V., Wajid S., Sundar S., Ramesh V., Dujardin J.C., Salotra P. Increased miltefosine tolerance in clinical isolates of Leishmania donovani is associated with reduced drug accumulation, increasedinfectivity and resistance to oxidative stress. PLoS Negl. Trop. Dis. 2017;11(6):e0005641. doi: 10.1371/journal.pntd.0005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fadili K., Messier N., Leprohon P., Roy G., Guimond C., Trudel N., Saravia N.G., Papadopoulou B., Le´gare´ D., Ouellette M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob. Agents Chemother. 2005;49(5):1988–1993. doi: 10.1128/AAC.49.5.1988-1993.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-On J., Sulitzeanu A., Schnur L.F. Leishmania major: resistance of promastigotes to paromomycin, and susceptibility of amastigotes to paromomycin-methylbenzethonium chloride ointment. Ann. Trop. Med. Parasit. 1991;85:323–328. doi: 10.1080/00034983.1991.11812568. [DOI] [PubMed] [Google Scholar]
- Faria M.S., Reis F.C.G., Azevedo-Pereira R.L., Morrison L.S., Mottram J.C., Lima A.P.C.A. Leishmania inhibitor of serine peptidase prevents TLR4 activation by neutrophil elastase promoting parasite survival in murine macrophages. J. Immunol. 2011;186(1):411–422. doi: 10.4049/jimmunol.1002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget G., Gregory D.J., Whitcombe L.A., Olivier M. Role of host protein tyrosine phosphatase SHP-1 in Leishmania donovani-Induced inhibition of nitric oxide production. Infect. Immun. 2006;74(11):6272–6279. doi: 10.1128/IAI.00853-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx S., daLuz R.A.I., Bhandari V., Kuypers K., Shaw C.D., Lonchamp J., Salotra P., Carter K., Sundar S., Rijal S., Dujardin J.C., Cos P., Maes L. Experimental induction of paromomycin resistance in antimony-resistant strains of L. donovani: outcome dependent on in vitro selection protocol. PLoS Negl. Trop. Dis. 2012;6(5):1–7. doi: 10.1371/journal.pntd.0001664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx S., Leemans A., Annelies Mondelaers A., Rijal S., Khanal B., Dujardin J.C., Delputte P., Cos P., Maes L. Comparative fitness of a parent Leishmania donovani clinical isolate and its experimentally derived paromomycin-resistant strain. PLoS One. 2015;10(10):e0140139. doi: 10.1371/journal.pone.0140139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhingran A., Chawla B., Saxena S., Barrett M.P., Madhubala R. Paromomycin: uptake and resistance in Leishmania donovani. Mol. Biochem. Parasitol. 2009;164:111–117. doi: 10.1016/j.molbiopara.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshrestha A., Singh R., Kumar D., Negi N.S., Salotra P. Antimony-resistant clinical isolates of Leishmania donovani are susceptible to paromomycin and sitamaquine. Antimicrob. Agents Chemother. 2011;55(6):2916–2921. doi: 10.1128/AAC.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshrestha A., Sharma V., Singh R., Salotra P. Comparative transcript expression analysis of miltefosine-sensitive and miltefosine-resistant Leishmania donovani. Parasitol. Res. 2014;113:1171–1184. doi: 10.1007/s00436-014-3755-6. [DOI] [PubMed] [Google Scholar]
- Kumar D., Singh R., Bhandari V., Kulshrestha A., Negi N.S., Salotra P. Biomarkers of antimony resistance: need for expression analysis of multiple genes to distinguish resistance phenotype in clinical isolates of Leishmania donovani. Parasitol. Res. 2012;111:223–230. doi: 10.1007/s00436-012-2823-z. [DOI] [PubMed] [Google Scholar]
- Legare D., Cayer S., Singh A.K., Richard D., Papadopoulou B., Ouellette M. ABC proteins of Leishmania. J. Bioenerg. Biomembr. 2001;33:469–474. doi: 10.1023/a:1012870904097. [DOI] [PubMed] [Google Scholar]
- Leprohon P., Legare D., Girard I., Papadopoulou B., Ouellette M. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryot. Cell. 2006;5:1713–1725. doi: 10.1128/EC.00152-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P., Légaré D., Raymond F., Madore E., Hardiman G., Corbeil J., Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarouf M., de Kouchkovsky Y., Brown S., Petit P.X., Robert-Gero M. In vivo interference of paromomycin with mitochondrial activity of Leishmania. Exp. Cell Res. 1997;232:339–348. doi: 10.1006/excr.1997.3500. [DOI] [PubMed] [Google Scholar]
- Maarouf M., Adeline M.T., Solignac M., Vautrin D., Robert-Gero M. Development and characterization of paromomycin-resistant Leishmania donovani promastigotes. Parasite. 1998;5:167–173. doi: 10.1051/parasite/1998052167. [DOI] [PubMed] [Google Scholar]
- Moore R.A., DeShazer D., Reckseidler S., Weissman A., Woods D.E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira W., Leprohon P., Ouellette M. Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis. 2011;2:e201. doi: 10.1038/cddis.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati V.K., Mehrotra S., Gautam S., Rai M., Sundar S. In vitro Antileishmanial drug susceptibility of clinical isolates from patients with Indian visceral leishmaniasis–Status of Newly Introduced Drugs. Am. J. Trop. Med.Hyg. 2012;87(4):655–657. doi: 10.4269/ajtmh.2012.12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkait B., Kumar A., Nandi N., Sardar A.H., Das S., Kumar S., Pandey K., Ravidas V., Kumar M., De T., Singh D., Das P. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob. Agents Chemother. 2012;56:1031–1041. doi: 10.1128/AAC.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R., Vishal Goyal V., Haque R., Jamil K., Faiz A., Samad R., Ellis S., Balasegaram M., Margriet den Boer M., Rijal S., Strub-Wourgaft N., Alves F., Alvar J., Sharma B. Safety and efficacy of short course combination regimens with AmBisome, miltefosine and paromomycin for the treatment of visceral leishmaniasis (VL) in Bangladesh. PLoS Negl. Trop. Dis. 2017;11(5):e0005635. doi: 10.1371/journal.pntd.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V., Singh R., Avishek K., Verma A., Deep D.K., Verma S., Salotra P. Decline in clinical efficacy of oral miltefosine in treatment of Post Kala-azar Dermal Leishmaniasis (PKDL) in India. PLoS Negl. Trop. Dis. 2015;9:e0004289. doi: 10.1371/journal.pntd.0004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal S., Ostyn B., Uranw S., Rai K., Bhattarai N.R., Dorlo T.P., Beijnen J.H., Vanaerschot M., Decuypere S., Dhakal S.S., Das M.L., Karki P., Singh R., Boelaert M., Dujardin J.C. Increasing failure of miltefosine in the treatment of kala-azar in Nepal and the potential role of parasite drug resistance, re-infection or noncompliance. Clin. Infect. Dis. 2013;56:1530–1538. doi: 10.1093/cid/cit102. [DOI] [PubMed] [Google Scholar]
- Rochette A., Raymond F., Ubeda J.M., Smith M., Messier N., Boisvert S., Rigault P., Corbeil J., Ouellette M., Papadopoulou B. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. doi: 10.1186/1471-2164-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P.K., Jha T.K., Thakur C.P., Nath D., Mukherjee S., Aditya A.K., Sundar S. Phase 4 pharmacovigilance trial of paromomycin injection for the treatment of visceral leishmaniasis in India. J. Trop. Med. 2011;2011:645203. doi: 10.1155/2011/645203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P., Prajapati V.K., Rai M., Sundar S. Unusual case of resistance to amphotericin B in visceral leishmaniasis in a region in India where leishmaniasis is not endemic. J. Clin. Microbiol. 2011;49:3088–3091. doi: 10.1128/JCM.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S., Sinha P.K., Rai M., Verma D.K., Nawin K., Alam S., Chakravarty J., Vaillant M., Verma N., Pandey K., Kumari P., Lal C.S., Arora R., Sharma B., Ellis S., Strub-Wourgaft N., Balasegaram M., Olliaro P., Das P., Modabber F. Comparison of short-course multidrug treatment with standard therapy for visceral leishmaniasis in India: an open-label, noninferiority, randomised controlled trial. Lancet. 2011;377:477–486. doi: 10.1016/S0140-6736(10)62050-8. [DOI] [PubMed] [Google Scholar]
- Sundar S., Singh A., Rai M., Prajapati V.K., Singh A.K., Ostyn B., Boelaert M., Dujardin J.C., Chakravarty J. Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin. Infect. Dis. 2012;55:543–550. doi: 10.1093/cid/cis474. [DOI] [PubMed] [Google Scholar]
- World Health Organisation . 2010. “Control of the Leishmaniasis,” Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniasis.http://whqlibdoc.who.int/trs/WHO_TRS_949_eng.pdf Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.