Abstract
Isometamidium Chloride (ISM) is one of the principal drugs used to counteract Trypanosoma congolense infection in livestock, both as a prophylactic as well as a curative treatment. However, numerous cases of ISM resistance have been reported in different African regions, representing a significant constraint in the battle against Animal African Trypanosomiasis.
In order to identify genetic signatures associated with ISM resistance in T. congolense, the sensitive strain MSOROM7 was selected for induction of ISM resistance in a murine host. Administered ISM concentrations in immune-suppressed mice were gradually increased from 0.001 mg/kg to 1 mg/kg, the maximal dose used in livestock. As a result, three independent MSOROM7 lines acquired full resistance to this concentration after five months of induction, and retained this full resistant phenotype following a six months period without drug pressure. In contrast, parasites did not acquire ISM resistance in immune-competent animals, even after more than two years under ISM pressure, suggesting that the development of full ISM resistance is strongly enhanced when the host immune response is compromised. Genomic analyses comparing the ISM resistant lines with the parental sensitive line identified shifts in read depth at heterozygous loci in genes coding for different transporters and transmembrane products, and several of these shifts were also found within natural ISM resistant isolates. These findings suggested that the transport and accumulation of ISM inside the resistant parasites may be modified, which was confirmed by flow cytometry and ex vivo ISM uptake assays that showed a decrease in the accumulation of ISM in the resistant parasites.
Keywords: Trypanosoma congolense, Isometamidium Chloride, Drug resistance, Whole Genome Sequencing, Allele frequency shifts, Transmembrane transporter proteins
Graphical abstract
Highlights
-
•
Impaired host immunity promotes ISM resistance acquisition in T. congolense.
-
•
Fixation of non-synonymous SNPs in transporter genes may induce ISM resistance.
-
•
The accumulation of ISM is reduced in resistant T. congolense.
1. Introduction
African trypanosomes are protozoan parasites transmitted by tsetse flies and responsible for African Trypanosomiasis in both humans (HAT) and animals (AAT). T. congolense, T. vivax and T. brucei brucei are the major pathogens responsible for AAT, a disease also known as Nagana and considered as one of the principal causes of malnutrition and poverty in 37 countries in sub-Saharan Africa (Mattioli et al., 2004). Nagana puts 55 million cattle at risk and leads to the death of three million animals every year, inflicting a direct annual loss of US$ 1.0–1.2 billion in cattle production (Hursey and Slingenbergh, 1995, Mattioli et al., 2004, Cecchi and Mattioli, 2009, Cecchi et al., 2014).
Isometamidium Chloride (ISM) and Diminazene Aceturate (DA) are the main drugs currently available to cure AAT (Giordani et al., 2016). ISM belongs to the phenanthridine drug group of which many members are documented to be mutagenic (Uilenberg, 1998), and DA is an aromatic diamidine. About 35 million doses of trypanocidal drugs are administered every year in an attempt to control the disease (Geerts and Holmes, 1998, Mattioli et al., 2004). However, those compounds have been in use for over 50 years (Lourie and Yorke, 1939, Watkins and Woolfe, 1952, Brown et al., 1961, Sahin et al., 2014), and numerous cases of ISM and DA resistance and treatment failures have been reported in 17 African countries, representing a serious problem in the battle against AAT (Sinyangwe et al., 2004, Delespaux et al., 2008a, Delespaux et al., 2008b, Moti et al., 2012, Mungube et al., 2012, Sow et al., 2012, Dagnachew et al., 2015, Moti et al., 2015).
ISM and DA accumulate inside the cytoplasm and mitochondrion of trypanosomes and have a common target: the mitochondrial DNA (Shapiro and Englund, 1990). ISM also enters the nucleus where it binds to the nuclear DNA (Kaminsky et al., 1997). The mitochondrial DNA, known as kinetoplast DNA (kDNA), represents about 10–20% of the genome of the trypanosomes (Daniels et al., 2010) and consists of a giant network of interlocked DNA rings (Liu et al., 2005). ISM selectively inhibits the kDNA type II topoisomerase, a protein involved in the replication of the mitochondrial DNA, which leads to the shrinking and disappearance of the kDNA network, and ultimately to cell death (Shapiro and Englund, 1990, Wang and Englund, 2001, Roy Chowdhury et al., 2010). DA has high affinity for A-T base pairs and binds to circular double strand DNA to induce complete and irreversible loss of the kDNA network (Brack et al., 1972, Mahler and Perlman, 1973, Gonzalez et al., 1997).
While the mechanisms of ISM and DA resistance have been described in few trypanosome species (Brun and Lun, 1994, Neupert, 1997, Schnaufer et al., 2005, Dean et al., 2013, Gould and Schnaufer, 2014, Baker et al., 2015), little is known for T. congolense. Here, the identification of a GAA insertion in a gene coding for an ATP-binding cassette (ABC) transporter (i.e. energy-dependent transport of various substrates across biological membranes) in a range of ISM resistant parasites led to the hypothesis of a potential involvement of ABC transporters in ISM resistance (Delespaux et al., 2005). However, this GAA insertion was not shared by all ISM resistant T. congolense strains, suggesting that other mechanisms of drug resistance acquisition exist. The mitochondrial membrane potential (ΔΨm) was also suggested to be an important determinant of the rate and accumulation of ISM inside the mitochondrion, with a diminished ΔΨm resulting in a decreased ISM accumulation inside the mitochondrion and drug resistance (Wilkes et al., 1997). Active extrusion by plasma membrane transporters has also been proposed to contribute to the intracellular ISM accumulation level. Here, significant differences in drug transport were suggested to be correlated with the resistant phenotype (Sutherland et al., 1992).
To facilitate the identification of genomic determinants and mechanisms associated with drug resistance, Whole Genome Sequencing (WGS) technologies have been used in numerous studies, including some on protozoan parasites such as Leishmania spp (Downing et al., 2011, Mondelaers et al., 2016) and Plasmodium falciparum (Miotto et al., 2015) allowing the successful identification of the genomic signatures of drug resistance in these pathogenic organisms. Therefore, we have used this WGS methodology to investigate the genomic changes that occur during the experimental induction of ISM resistance in the initially sensitive MSOROM7 T. congolense strain. In three independent experiments, immune-suppressed mice were treated with increasing doses of ISM until the parasites acquired resistance to 1 mg/kg ISM, the maximal dose used in livestock. Flow cytometry and ex vivo uptake/extrusion assays revealed a reduced accumulation of ISM in the three independently induced resistant parasite lines. WGS analysis comparing these ISM resistant lines with the sensitive parental line identified allele frequency shifts in a variety of genes of which some were coding for different transporters and transmembrane proteins. In complement, we performed a similar WGS comparison of 50 T. congolense field isolates with a documented ISM susceptibility profile.
2. Methods
2.1. Ethics statement
The animal work was approved by the Institute of Tropical Medicine (ITM) animal ethics committee, clearance numbers DG020 and VP2014-2.
2.2. In vivo drug susceptibility testing
To evaluate the susceptibility of a T. congolense strain to ISM, six mice were injected with the strain of interest at a concentration of 105 parasites (bloodstream forms)/mouse. Twenty-four hours later, each mouse was treated with a single ISM dose of 1 mg/kg. The trypanosome presence in these mice was monitored for two months by microscopical examination and PCR analysis using the primers and protocol described in (Cox et al., 2005). At the end of the experiment a strain was considered sensitive to 1 mg/kg ISM when all mice (6/6) were cured, and was considered resistant when five or six relapses (reappearance of parasitemia) were observed. A strain showed partial resistance (intermediate profile) when one to four mice relapsed following drug administration (Eisler et al., 2001).
2.3. Effect of ISM pressure on the genome of T. congolense
To obtain insights into the genomic consequences of an ISM drug pressure on a parasite cell population, we maintained the ISM resistant isolate TRT15 in OF-1 mice for 10 passages under two different conditions: without drug pressure versus with a continuous ISM pressure. TRT15 is a T. congolense isolate that was originally collected from cattle in 1996 in Zambia (Delespaux et al., 2008a) (Table S1). To start the experiment, mice were infected by intraperitoneal injection with 105 parasites/mouse. When the level of parasitemia in the blood reached 7.9 × 106 parasites/ml, the mouse was either treated with 1 mg/kg ISM or left untreated. When parasitemia reached 1.3 × 108 parasites/ml, blood was collected and purified using an anion exchanger Diethylaminoethyl (DEAE)-cellulose resin suspended in Phosphate Saline Glucose (PSG) buffer, pH 8 (Lanham and Godfrey, 1970) to obtain parasites for a new infection (passage). Parasites were collected at the start of the experiment and after passages 5 and 10 for DNA extraction and sequencing (see below).
2.4. ISM resistance induction experiment
The sensitive T. congolense strain KTT/MSOROM7C1 (further referred as MSOROM7) was chosen to induce ISM resistance in immune-suppressed OF-1 mice (Fig. 1). MSOROM7 is a clone derived from the Zambian isolate KTT/MSOROM7 that was obtained from cattle in 2003 (Masumu et al., 2009). This clone was generated by infecting an OF-1 mouse with a single parasite using the micro-drop method (Masumu et al., 2009). Then, the parasites were passaged only once before the start of the ISM resistance induction experiment. Therefore, we can assume that the parasites were isogenic at the start of this experiment.
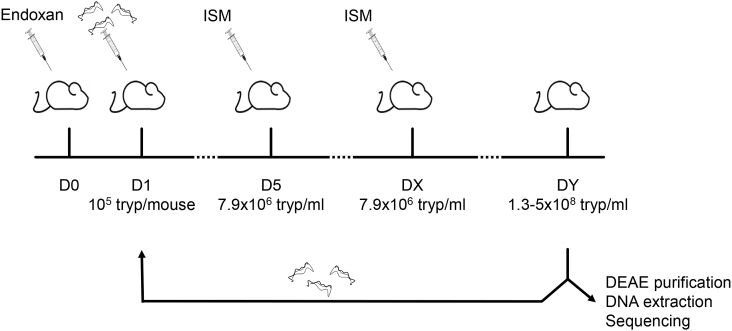
Fig. 1.
Induction of ISM resistance, experimental procedure. One day prior to infection with the sensitive strain MSOROM7, mice were injected with Endoxan® (D0). When parasitemia reached the concentration of 7.9 × 106/ml (D5), mice were treated with 0.001 mg/kg ISM. Mice became microscopically non-parasitemic but parasitemia reappeared within a few days (Dx), and mice were treated with an increased dose of ISM (0.005 mg/kg ISM). When parasitemia reappeared and reached 1.3–5x108/ml (Dy), mice were euthanized and parasites were collected to inject a new group of naïve mice that were first treated with 0.005 mg/kg ISM, and then with 0.01 mg/kg ISM. This cycle was repeated using increasing doses of ISM (0.02; 0.04 and 1 mg/kg) until reaching resistance to 1 mg/kg ISM.
One day prior to infection (D0), three naïve mice were injected intraperitoneally with 200 mg/kg cyclophosphamide (Endoxan®, Baxter) in order to obtain immune-suppression. The next day (D1), these mice were infected intraperitoneally with a starting dose of 105 bloodstream form trypanosomes. When the level of parasitemia in the blood reached 7.9 × 106 trypanosomes/ml, mice were treated with 0.001 mg/kg ISM (i.e. first ISM treatment) and became microscopically non-parasitemic within a few days. When parasitemia in blood reappeared and reached 7.9 × 106 trypanosomes/ml (DX), mice were injected with an increased dose of ISM (0.005 mg/kg). As soon as the parasitemia reappeared and reached a level between 1.3 × 108 and 5 × 108 trypanosomes/ml (i.e. peak of parasitemia; DY), blood from each individual mouse was collected on a DEAE resin for parasite isolation and DNA extraction. A part of the collected blood was also used to infect a new group of three Endoxan® treated mice with a starting dose of 105 trypanosomes. These newly infected mice received two ISM injections: a first injection with 0.005 mg/kg ISM, and after parasites reappearance, a second injection with 0.01 mg/kg ISM (Ndoutamia et al., 1993). This infection-ISM treatment cycle was repeated several times with increasing doses (0.02; 0.04 and 1 mg/kg ISM) until full resistance to 1 mg/kg ISM was acquired. Using this methodology, three independent ISM resistant lines were obtained: MSOROM7-G; MSOROM7-H and MSOROM7-I. After each treatment cycle (annotated as passages one, two, three, four and five; or referred to as MSOROM7-G2, -G3, -G4, -G; MSOROM7-H2; -H3, -H4, -H; and MSOROM7-I2, -I3, -I4, -I; Table S1) parasites were collected for DNA extraction and sequencing (see below).
A similar resistance induction experiment in immune-competent OF-1 mice (i.e. no Endoxan® treatment) failed to give rise to fully resistant parasites.
2.5. Bioinformatic and statistical analyses
DNA extraction, sequencing and bioinformatic analyses were performed as described earlier (Tihon et al., 2017). In short, reads were mapped against the reference genome T. congolense IL3000 (Jackson et al., 2012) with SMALT v7.4 (http://sourceforge.net/projects/smalt/), and duplicate reads were filtered with SAMtools V1.0 (Li et al., 2009), resulting in an average read depth of 30.9× per TRT15 sample and 33.4× per MSOROM7 sample. Single-Nucleotide-Polymorphisms (SNPs) and insertions/deletions smaller than 15 bp in length (indels) were called using the Genome Analysis Toolkit v3.4 (GATK) (McKenna et al., 2010, DePristo et al., 2011) and annotated using SnpEff (Cingolani et al., 2012) based on the gene annotation file of T. congolense IL3000 (http://tritrypdb.org/common/downloads/release-6.0/TcongolenseIL3000/gff/data/). Gene deletions were measured by both the normalized read depth and the fraction of the gene covered by reads (length). We considered a gene to be totally deleted (i.e. gene deletion) when 75% or more of its depth coverage was missing. In the depth analysis, we used MSOROM7-G4 (i.e. mean depth coverage of 20×) instead of MSOROM7-G (i.e. mean depth coverage of 13×) to estimate long deletions in samples with sufficient depth coverage, and this choice was made because MSOROM7-G4 was as ISM resistant as MSOROM7-G (see results). Gene duplications were measured solely by the normalized read depth. To provide statistical significance for the gene duplication analysis, we calculated the differences between the sensitive and resistant lines and calculated the standard z scores. We converted the standard z scores to p-values using python scipy survival function. See (Tihon et al., 2017) for details on all computational parameters and filtering criteria for each of the above mentioned computational analyses.
We checked whether novel SNPs and small indels appeared in TRT15 and MSOROM7 over the course of the experiments. As this was not the case (see results), we checked whether there was a change in allele frequencies at heterozygous loci between the different passages of TRT15 and MSOROM7. To this end, we calculated the proportion of reads that support a given allele at a given SNP site, providing an estimate of the read depth allele frequency. For TRT15 we compared the read depth allele frequencies between passage 0 (start of the experiment) and passages 5 (1 mg/kg ISM) and 10 (1 mg/kg ISM). For MSOROM7 we compared the read depth allele frequencies between passage 0 and passages 2 (0.01 mg/kg ISM), 3 (0.02 mg/kg ISM), 4 (0.04 mg/kg ISM) and 5 (1 mg/kg ISM). If a sample at a given passage did not have sufficient read depth coverage (i.e. less than 10× coverage) at a given SNP site, then this site was removed from further analysis. In order to measure statistical significance we calculated standard deviations σ and a corresponding z-score of the differences in read depth allele frequencies between the passages as mentioned above. Finally, we only retained those sites for which the read depth allele frequency shifted of at least 0.25 and for which the z-score was at least 3. Shifts in read depth allele frequencies higher than 0.8 were considered homozygous, which was similar to the definition of (Nielsen et al., 2011) where a homozygous genotype was described as a proportion of an allele of less than 0.2 or larger than 0.8.
A similar approach was used to identify changes in allele frequencies between drug resistant and drug sensitive isolates from natural populations. To this end, we used published sequence data from 50 T. congolense isolates from 10 different countries in sub-Saharan Africa (Tihon et al., 2017). Population genomics analyses revealed two clusters: the TC1 cluster contained 21 parasites from The Gambia, Mali, Burkina Faso, Togo, Cameroon, Ethiopia and Uganda and the TC2 cluster contained three parasites from Congo, Tanzania and Zambia (Tihon et al., 2017). The remaining 25 parasites from Zambia and one parasite from Burkina Faso were of uncertain ancestry, showing signatures of introgressive hybridization (Tihon et al., 2017). The ISM sensitivity of all these isolates were determined in previous studies (Geigy and Kauffmann, 1973, Pinder and Authie, 1984, Clausen et al., 1992, Afewerk et al., 2000, Knoppe, 2002, Delespaux et al., 2005, Mamoudou et al., 2006, Mamoudou et al., 2008, Masumu et al., 2009, Moti et al., 2012): 30 strains were resistant to 1 mg/kg ISM, 14 were sensitive to 1 mg/kg ISM and six showed an intermediate profile with partial resistance to 1 mg/kg ISM (Table S1). Within each group (i.e. TC1, TC2 and Zambia) we compared the averaged read depth allele frequencies between resistant and sensitive isolates. Differences in the averaged allele frequencies ≥0.8 between the resistant and sensitive strains were classified as homozygous. Differences in the allele frequencies greater than 0.25 (but lower than 0.8) were classified as heterozygous. We selected genes with the averaged allele frequency shift higher than 0.25 between resistance and sensitive strains with a p value ≤ 0.05 (Mann-Whitney-Wilcoxon test). For a more targeted analysis, we focused on a set of 185 genes including some playing critical roles in drug resistance in various pathogens (Table S6) to further investigate allele frequency shifts between sensitive and resistant field isolates. This analysis identified 217 alleles in a total of 54 genes whose averaged allele frequency shifted ≥0.25 between the sensitive and resistant parasites of each population (Table S7).
The dataset generated and analyzed during the current study is available in EBI under the accession number PRJEB21066, http://www.ebi.ac.uk/ena/data/view/PRJEB21066.
2.6. ISM accumulation and extrusion
The accumulation of ISM in the sensitive and resistant T. congolense MSOROM7 lines as well as in the ISM resistant field isolate TRT1 was first monitored by flow cytometry, using the fluorescent properties of the drug (ISM: λex: 374 nm; λem: 590 nm). For an unknown reason, the field isolate TRT15 was very sensitive to the DEAE purification and many cells were lost during the isolation procedure, precluding the collection of a sufficient amount of parasites to perform the experiment. To overcome this issue, we performed the flow cytometric analysis on the ISM resistant field isolate TRT1 that is genetically nearly identical to the TRT15 strain (Tihon et al., 2017).
DEAE-column purified parasites from infected mouse blood were maintained in a cold PSG buffer pH 8. For each ISM condition, 106 trypanosomes were suspended in ice-cold PSG containing 0–10 μg/ml ISM supplemented with the Fixable Viability Stain (FVS, BD Horizon - λex: 649 nm; λem: 660 nm) and incubated for 30 min at 4 °C in the dark. Cells were washed twice in PSG and were analyzed with a BD FACSVerse flow cytometer and the BD FACSuite software v1.0.3.
The accumulation and extrusion of ISM in the MSOROM7 sensitive and resistant lines (except MSOROM7-H for which no sufficient amount of parasites could be harvested) and in the field isolate TRT1 were also measured following the protocol described in (Eze et al., 2016). The ISM fluorescence was measured using a VICTOR Multilabel Plate Reader (PerkinElmer) (ISM: λex: 374 nm; λem: 590 nm) and quantified using a standard curve of known ISM concentrations (i.e. the fluorescence of known concentrations of ISM (i.e. ISM in assay buffer) were determined using the VICTOR Multilabel Plate Reader to make a standard curve of known concentrations).
2.7. Real time qPCR
We identified allele frequency shifts in the gene TcIL3000_8_1200 coding for the drug/metabolite transporter (DMT) in the resistant MSOROM7-H line compared with the sensitive parental line (see results). In addition, the DMT gene had two copies in the sensitive parasites and one of these copies disappeared in the resistant lines (see results). To identify a potential link between allele frequency shifts, copy number variations and gene expression level, the expression level of the DMT gene TcIL3000_8_1200 was evaluated in MSOROM7 and in the resistant MSOROM7 lines challenged or not with 1 mg/kg ISM. PCR primers were designed with Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) and evaluated with OligoAnalyzer (https://eu.idtdna.com/calc/analyzer). Each primer was blasted against trypanosome genomes (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to evaluate their specificity. RNA extractions were conducted with TRIzol Reagent (Invitrogen), and the cDNA synthesis was performed using the SensiMix™ SYBR® No-ROX Kit (Bioline) according to manufacturer's instructions. The qPCR reactions were conducted on a LightCycler 480 (Roche) with SYBRGreen I master mix (Qiagen) and 500 nM of each primer. The cycling conditions were as followed: 95 °C for 10 s; 40 cycles at 95 °C for 10 s; 52 °C for 10 s; and 72 °C for 30s.
We selected TERT and GAPDH as reference genes after assessing the stability of the gene expression level of eight putative housekeeping genes using BioGazelle qBase + v1.5 software (Hellemans et al., 2007). The primers used were:
DMT (gene of interest) F: 5′-ACATCGGGCATTTGGACCTT-3′; R: 5′-CAGTGCACCAACGAAAGCAA-3′
TERT (housekeeping gene) F: 5′-TTTCGCCCTCGTTTTCCTCA-3′; R: 5′-AGAAATCACGACCACACGCT-3′
GAPDH (housekeeping gene) F: 5′-CGTTGAGACGGAGTGCTTGA-3′; R: 5′-GATGGACTTCTCGGCACTGA-3′
2.8. Statistical analyses of laboratory experiments
For the ISM accumulation/extrusion and gene expression level experiments, two-tailed unpaired Mann-Whitney-t test were carried out in GraphPad Prism 5. Data were represented as means ± standard error of the mean. P values ≤ 0.05 were considered to be statistically significant.
3. Results
3.1. Genomic changes in a resistant parasite under drug pressure
To evaluate the impact of a continuous ISM pressure on the T. congolense genome we maintained the drug resistant field isolate TRT15 for 10 passages in mice with 1 mg/kg ISM (TRT15p10-ISM). Although infected mice became microscopically non-parasitemic after each round of ISM treatment, parasitemia in mice reappeared within less than a week.
Bioinformatic analyses revealed a total of 157,797 SNPs and 3098 indels in TRT15 in comparison to the T. congolense IL3000 reference genome when SNP calling was done with the combined sequence data of all TRT15 samples obtained during the experiment. Following drug pressure, we could not detect any novel SNPs nor indels appearing in TRT15. However, we found that the read depth frequency of alleles at heterozygous loci changed over time (Table 1). A shift in read depth allele frequency of at least 0.5 was observed at 11 loci after 5 passages and at 62 loci after 10 passages under drug pressure, while no such shifts were observed when TRT15 was not subjected to drug pressure (Table 1). Of these 62 loci, 17 shifts resulted in read depth allele frequencies larger than 0.8 following drug pressure, but these were mostly found within genes coding for hypothetical proteins (results not shown).
Table 1.
Number of allele frequency shifts identified in each experiment. In the first experiment, we maintained the drug resistant field isolate TRT15 for 5 and 10 passages in mice either with 1 mg/kg ISM (i.e. TRT15p5-ISM and TRT15p10-ISM, respectively) or without the drug pressure (i.e. TRT15p5 and TRT15p10, respectively). In the second experiment, three independent ISM resistant lines were generated starting from the same initial MSOROM7 clone, namely: MSOROM7-G, MSOROM7-H and MSOROM7-I. In the last experiment, we compared the resistant and sensitive field isolates of each natural populations identified in (Tihon et al., 2017).
| TRT15 compared to | Number of shifts |
|||
|---|---|---|---|---|
| TRT15p5 | TRT15p10 | TRT15p5-ISM | TRT15p10-ISM | |
| Shift ≥0.50 | 0 | 0 | 11 | 62 |
| Shift ≥0.333 | 2 | 2 | 93 | 499 |
| Shift ≥0.25 |
25 |
27 |
248 |
941 |
|
MSOROM7 compared to |
MSOROM7-G |
MSOROM7-H |
MSOROM7-I |
|
| Shift ≥0.50 | 163 | 4 | 26 | |
| Shift ≥0.333 | 1496 | 190 | 627 | |
| Shift ≥0.25 |
1784 |
972 |
1955 |
|
|
Resistant compared to Sensitive |
TC1 |
TC2 |
Zambia |
|
| Shift ≥0.50 | 2 | 12 | 3 | |
| Shift ≥0.333 | 10 | 32 | 42 | |
| Shift ≥0.25 | 30 | 70 | 132 | |
While the continuous ISM pressure did not induce novel SNPs nor indels it caused copy number variations. Compared to the TRT15 genome before drug pressure we identified a total of 36 and 116 gene duplications and 7 and 38 gene deletions in TRT15 after 5 and 10 passages under drug pressure, respectively (Table S2) (Fig. S1 A and B). In contrast, only 19 and 13 gene duplications and 3 and 13 gene deletions were identified after 5 and 10 passages without drug pressure, respectively (Table S2) (Fig. S1 A and B).
3.2. Induction of drug resistance in a sensitive parasite
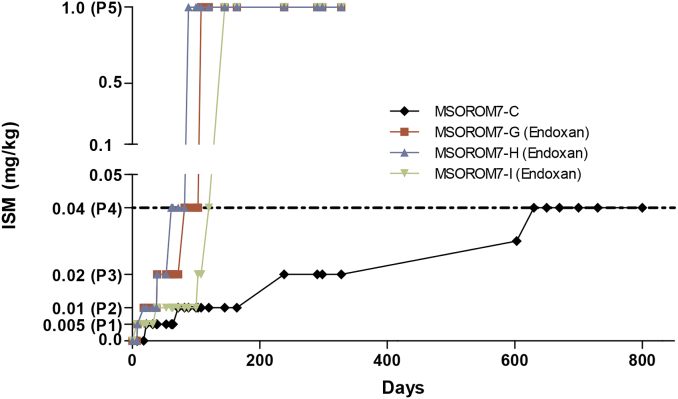
Three independent ISM resistant lines were generated in the murine host (i.e. immune-suppressed) starting from the sensitive clone MSOROM7 by a gradual ISM induction pressure. These three lines were referred to as MSOROM7-G, MSOROM7-H and MSOROM7-I. Full resistance to 1 mg/kg ISM was acquired after three to five months under increasing drug pressure (i.e. five passages in the murine host) (Fig. 2). MSOROM7-H acquired full resistance more rapidly, already after 88 days under ISM challenge, while MSOROM7-G and MSOROM7-I acquired full resistance after 108 and 145 days, respectively. Notably, once ISM resistance reached a threshold value of 0.04 mg/kg (i.e. passage four), these parasites were also fully resistant to the 1 mg/kg ISM dose (i.e. passage five) (Fig. 2). All three cell lines maintained their acquired ISM resistant phenotype when grown in immune-competent mice for six months without any drug pressure.
Fig. 2.
Induction of ISM resistance in the immune-suppressed murine host (Endoxan®treated). The T. congolense sensitive strain MSOROM7 acquired ISM resistance between three to five months in three independent experiments. In total, five ISM-dose passages in the murine hosts were sufficient to acquire full ISM resistance: passage one (P1): resistant to 0.005 mg/kg ISM; passage two (P2): resistant to 0.01 mg/kg ISM; passage three (P3): resistant to 0.02 mg/kg ISM; passage four (P4): resistant to 0.04 mg/kg ISM; passage five (P5): resistant to 1 mg/kg ISM. The three independently ISM resistant lines that acquired full resistance after passage five (1 mg/kg ISM) are referred to as MSOROM7-G, MSOROM7-H and MSOROM7-I. In immune-competent mice, the acquisition of fully resistant parasites failed: only one parasite strain (MSOROM7-C) acquired a partial resistance to 0.04 mg/kg ISM after a period of >600 days.
In contrast, the same induction set-up in immune-competent mice resulted in only one line (MSOROM7-C) acquiring partial ISM resistance to 0.04 mg/kg after 20 months of drug pressure with the reappearance of parasitemia only observed in 1/3 of the ISM-treated animals. However, this line did not acquire full resistance to 1 mg/kg ISM after more than two years of drug pressure (Fig. 2). All other attempts to acquire resistant parasites in the immune-competent mice failed because of parasite clearance following drug administration.
3.3. Genomic differences between sensitive and resistant parasites
Bioinformatic analyses revealed a total of 93,850 SNPs in comparison to the T. congolense IL3000 reference genome when SNP calling was done with a combined sequence dataset of all MSOROM7 samples that were obtained during the experiment. We also identified 1786 small indels in the MSOROM7 lines compared to the reference genome IL3000, of which 86 in coding regions, including 51 frameshift mutations. Following drug resistance induction, we examined genomic changes but did not identify any novel SNPs nor indels appearing in any of the MSOROM7 ISM resistant lines. However, we did find many genomic changes in the read depth frequency of allele at heterozygous loci (Table 1), while similar shifts were not observed for indel sites (results not shown).
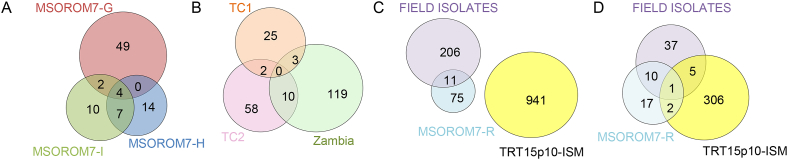
A total of 1784; 972 and 1955 loci showed an allele frequency shift larger than 0.25 (z score ≥ 3) in MSOROM7-G, MSOROM7-H and MSOROM7-I respectively (Table 1). Of these, 55, 25 and 23 resulted in a read depth frequency larger than 0.8 in MSOROM7-G, MSOROM7-H and MSOROM7-I lines, respectively (Table S3), of which four were common among the three resistant MSOROM7 lines but none were common with the resistant TRT15p10-ISM line (Fig. 3A and C; Table S3).
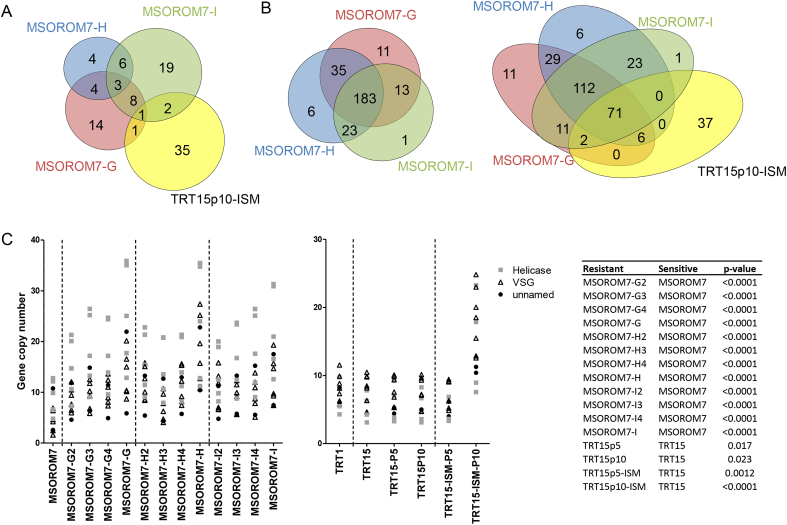
Fig. 3.
Number of frequency shifts in the alternate alleles identified in: A) the MSOROM7 resistant lines compared with the original MSOROM7 sensitive line. Four homozygous changes in nucleotide sequences were common to all resistant strains and included one synonymous change in the gene TcIL3000.11.2040 encoding a hypothetical protein, and three non-coding changes (see Table S3: Red); B) Between the resistant and sensitive parasites of each T. congolense populations (i.e. TC1, TC2 and Zambia) identified in (Tihon et al., 2017); C) Between the resistant and sensitive field isolates (i.e. combined TC1, TC2 and Zambian strains), between the MSOROM7 resistant lines compared with the sensitive MSOROM7, and between TRT15p10-ISM compared with TRT15. In total, 11 shifts were common between the resistant field isolates and the MSOROM7 resistant lines, but no common shifts were identified between the field isolates, the MSOROM7 resistant lines and TRT15p10-ISM D) The allele frequency shifts identified in these experiments were found in a total of 53 genes in the field-isolated strains, in 30 genes in the MSOROM7 resistant lines, and in 314 genes in the TRT15p10-ISM isolate.
In MSOROM7-G, four out of 55 allele frequency shifts were detected at non-synonymous positions in the ABC transporter genes TcIL3000_7_4990 and TcIL3000_10_9180, while one shift was detected at a non-synonymous position in the gene TcIL3000.11.12160, an orthologue of the T. b. brucei gene Tb11.01.3390 encoding the mitochondrial Type II topoisomerase (Table S3: Bold).
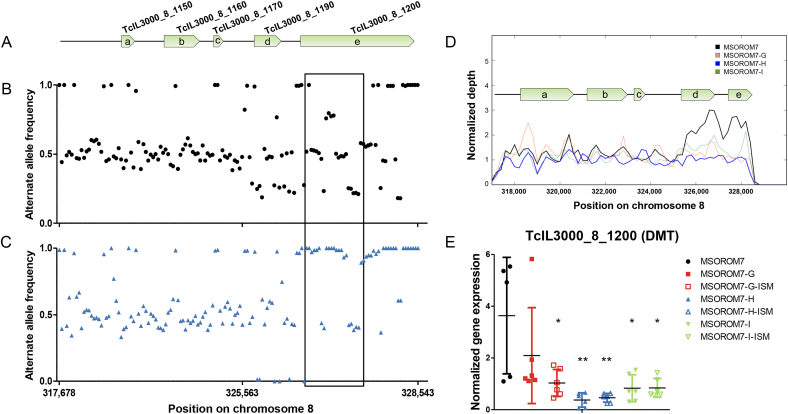
In MSOROM7-H, we discovered that 10 out of 25 shifts were located in the gene TcIL3000_8_1200 that encodes for the permease of the drug/metabolite transporter (DMT). Eight of these shifts were detected at non-synonymous positions and reached a read depth frequency larger than 0.8 at passage three and remained there until the end of the experiment (Table 2, Table S3: Bold). We closely examined all SNPs including SNP clusters (i.e. which were excluded from the original analysis, see (Tihon et al., 2017)) in the chromosomal region spanning the DMT gene. In the sensitive MSOROM7, we observed that the alternate allele frequencies of the genes TcIL3000_8_1190 (block ‘d’ in Fig. 4A and D) and TcIL3000_8_1200 (block ‘e’ in Fig. 4A and D) were of 0.25, 0.5, 0.75 or 1 (Fig. 4B). Read depth analysis of this genomic region revealed the presence of two copies of both genes in the sensitive strain (Fig. 4D), possibly explaining this peculiar tetrasomic-like alternate allele frequency pattern. In addition, we identified that almost all alleles within the DMT gene shifted to higher frequencies in MSOROM7-H compared with MSOROM7, while this was not the case for alleles found upstream of the gene (Fig. 4C). We further observed that in MSOROM7-H the normalized read depth started to decrease from passage two until full resistance, revealing the presence of only one copy of both genes in this resistant line (Fig. 4D). Similar observations were made for MSOROM7-G and MSOROM7-I (Fig. 4D) and for the resistant TRT15 (data not shown) that also had only one copy of the DMT gene. We examined the expression level of the DMT gene and observed a significant downregulation in all induced resistant lines but MSOROM7-G (p > 0.05) compared with MSOROM7 (Fig. 4E). Interestingly, MSOROM7-H, which had more definitive allele frequency shifts at non-synonymous positions, showed the lowest expression of the DMT gene (Fig. 4E).
Table 2.
Alternate allele frequencies (including these of SNP clusters) in the gene TcIL3000_8_1200 coding for the DMT product, as detected by WGS in the sensitive strain MSOROM7 and in the intermediate and resistant MSOROM7-H parasites. The alternate allele frequencies ranged from 0.5 (i.e. heterozygous SNP) to 1 (i.e. homozygous SNP). Alternate allele frequencies of 0.8 or higher are shown in bold and illustrate the homozygous SNPs identified in the intermediate and resistant MSOROM7-H parasites compared with the MSOROM7 sensitive line. Ref: Reference allele. NYS: Non-synonymous SNP; SYN: Synonymous SNP. P2: Resistant to 0.01 mg/kg ISM; P3: Resistant to 0.02 mg/kg ISM; P4: Resistant to 0.04 mg/kg ISM; P5: Resistant to 1 mg/kg ISM.
| Impact | position (bp) | Ref. | SNP | Alternate allele frequencies |
|||||
|---|---|---|---|---|---|---|---|---|---|
| MSOROM7 | MSOROM7-H |
||||||||
| P2 | P3 | P4 | P5 | ||||||
| NSY | 77 | G | A | 0.5 | 0.3 | 1.0 | 1.0 | 1.0 | |
| SNP cluster | NSY | 122 | G | A | 0.5 | 0.4 | 0.8 | 0.9 | 1.0 |
| SNP cluster | NSY | 124 | T | G | 0.5 | 0.4 | 0.9 | 0.9 | 1.0 |
| SNP cluster | NSY | 128 | G | T | 0.5 | 0.4 | 0.9 | 1.0 | 1.0 |
| NSY | 224 | G | A | 0.5 | 0.4 | 0.8 | 1.0 | 1.0 | |
| NSY | 401 | T | C | 0.5 | 0.6 | 0.8 | 0.9 | 1.0 | |
| SNP cluster | NSY | 419 | A | C | 0.5 | 0.5 | 0.8 | 0.9 | 1.0 |
| SNP cluster | NSY | 422 | A | G | 0.5 | 0.5 | 0.8 | 0.8 | 1.0 |
| SNP cluster | SYN | 423 | G | A | 0.5 | 0.5 | 0.7 | 0.8 | 0.9 |
| SYN | 483 | A | G | 0.5 | 0.5 | 0.9 | 0.9 | 0.9 | |
| SYN | 696 | C | T | 0.6 | 0.5 | 0.9 | 0.8 | 0.9 | |
| NSY | 697 | G | C | 0.6 | 0.5 | 0.9 | 0.8 | 0.9 | |
| NSY | 709 | G | T | 0.6 | 0.5 | 0.9 | 0.9 | 0.9 | |
| NSY | 731 | C | G | 0.6 | 0.5 | 0.9 | 0.9 | 0.9 | |
| NSY | 733 | A | G | 0.6 | 0.5 | 1.0 | 0.9 | 0.9 | |
| NSY | 791 | C | T | 0.6 | 0.5 | 0.9 | 0.9 | 1.0 | |
| SNP cluster | SYN | 855 | T | C | 0.5 | 0.3 | 1.0 | 1.0 | 1.0 |
| SNP cluster | NSY | 856 | A | G | 0.4 | 0.3 | 1.0 | 1.0 | 1.0 |
| SNP cluster | SYN | 862 | C | T | 0.5 | 0.3 | 1.0 | 1.0 | 1.0 |
Fig. 4.
Alternate allele frequencies (including these of SNP clusters) in the TcIL3000_8_1200 gene and its surrounding region in MSOROM7 and MSOROM7-H. Note that the TcIL3000_8_1200 gene is longer in the MSOROM7 lines compared with the reference TcIL3000 genome because of a frameshift indel at the gene position 823. A) Genomic region analyzed. a. TcIL3000_8_1150 b. TcIL3000_8_1160 c. TcIL3000_8_1170 d. TcIL3000_8_1190 e. TcIL3000_8_1200. This figure is based on the number of SNPs found in this genomic region to highlight the distribution of SNPs. The scale does not reflect the physical size of these genes. A gap in the reference genome after the gene TcIL3000_8_1200 prevented further analysis of the downstream region of the gene. B) Alternate allele frequencies in the sensitive MSOROM7; C) Alternate allele frequencies in the resistant MSOROM7-H. Each dot/triangle represents one allele position. Only the frequencies of the alternate alleles located in the TcIL3000_8_1200 gene shifted and became fixed in the resistant population (black box) D) Normalized read depth. In the MSOROM7 sensitive strain, the normalized read depth indicates the presence of two copies of the genes TcIL3000_8_1190 and TcIL3000_8_1200. These gene duplications were confirmed by sequencing the MSOROM7 sensitive strain with TruSeq DNA libraries using the methodology described in (Dumetz et al., 2017) (data not shown). The normalized read depth decreased in the resistant lines, suggesting the loss of one of these copies in response to drug pressure. E) Expression level of the DMT gene TcIL3000_8_1200 (i.e. evaluation of the mRNA expression level) in MSOROM7 sensitive and resistant lines challenged or not with 1 mg/kg ISM by Real-Time qPCR. Average ± standard error of the mean; *: P < 0.05; **: P < 0.01 (n = 6).
In MSOROM7-I, notable allele frequency shifts included three non-synonymous SNPs in the genes TcIL3000_8_410 and TcIL3000_10_13700 encoding transmembrane proteins (Table S3: Bold).
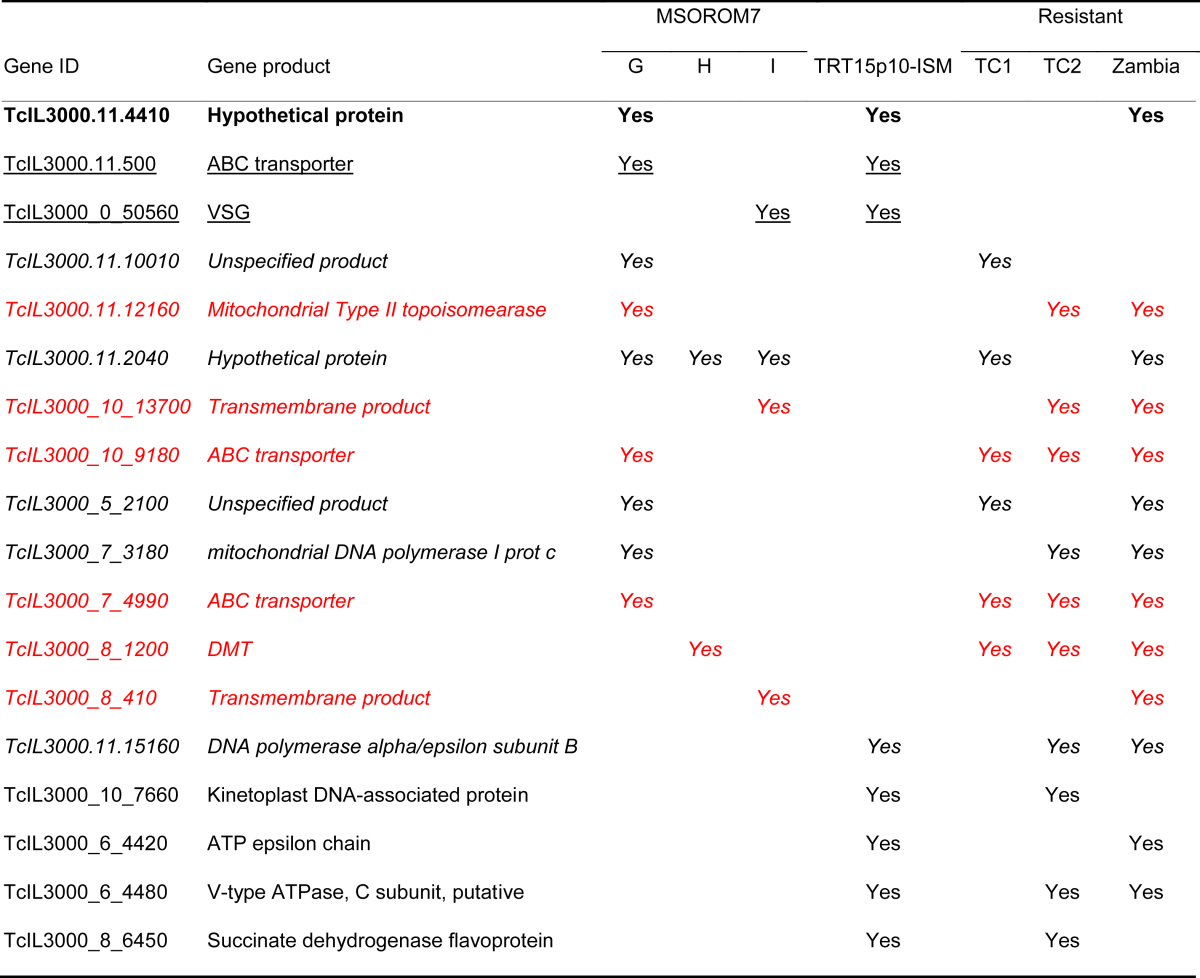
Six of the genes identified in these ISM resistant lines (i.e. TcIL3000_7_4990, TcIL3000_10_9180, TcIL3000.11.12160, TcIL3000_8_1200, TcIL3000_8_410 and TcIL3000_10_13700) also showed shifts larger than 0.25 in T. congolense natural populations (i.e. TC1, TC2 and Zambia; see (Tihon et al., 2017)) when the read depth allele frequencies were compared between ISM sensitive and ISM resistant parasites (Table 3: Red). Shifts in the allele frequency were also found in additional common genes in drug resistant MSOROM7, the natural resistant isolates and/or TRT15 maintained for 10 passages under drug pressure (Table 3).
Table 3.
List of genes in which allele frequency shifts were detected in two or more independent experiments (i.e. selection experiment; TRT15 experiment; comparison between resistant and sensitive field isolates). Allele frequency shifts in the gene TcIL3000.11.4410 were identified in all three experiments (bold). Two genes showed allele frequency shifts in the MSOROM7 resistant lines and TRT15p10-ISM isolate (underlined). Allele frequency shifts were detected in 10 common genes between the MSOROM7 resistant lines and the field isolates (italic; red). An additional four genes showed allele frequency shifts in both TRT15p10-ISM and the field isolates.
While we did not identify novel SNPs nor indels in the MSOROM7 resistant lines, we observed new structural variations of the genome. Gene copy number analysis revealed 31 gene deletions in MSOROM7-G4, 17 gene deletions in MSOROM7-H and 39 gene deletions in MSOROM7-I compared with the MSOROM7 sensitive line (Table S4). Overall, 22 gene deletions were shared between two or more resistant lines, which were found in the gene TcIL3000_0_00780 encoding a transposase and in genes coding for unspecified products (Fig. 5A; Table S4: Bold). Interestingly, four of the deletions identified in the TRT15p10-ISM were common to gene deletions detected in one or more MSOROM7 resistant lines (Fig. 5A; Table S4: Red), but these affected genes are coding for unspecified products. We also identified 58 gene deletions in the resistant TC2 field isolates compared to their sensitive counterparts, three of which were also observed in the resistant MSOROM7 lines (i.e. non coding RNA genes; Table S4: Green, Fig. S1 C).
Fig. 5.
Structural variations: gene deletions and gene duplications in the resistant MSOROM7 lines and TRT15p10-ISM compared with MSOROM7. A) Number of long deletions detected in the MSOROM7 resistant lines compared with the MSOROM7 sensitive line, and number of long deletions detected in TRT15p10-ISM compared with TRT15. B) Number of genes with at least one additional copy in the MSOROM7 resistant lines compared with the MSOROM7 sensitive line, and in TRT15p10-ISM compared with TRT15. C) Changes in gene copy number of 13 genes coding for VSGs (i.e. 5 genes), Helicases (i.e. 5 genes) and unnamed products (i.e. 3 genes) in the MSOROM7 intermediate and resistant lines compared with the MSOROM7 sensitive line, and in TRT15p10-ISM compared with TRT15, and statistical significance (complete list Table S5).
Furthermore, 272 genes had at least one additional copy in the MSOROM7 resistant lines compared with the sensitive line (Fig. 5B; Table S5). The gene copy number of 79 of these 272 genes also increased in TRT15p10-ISM compared with TRT15 (Fig. 5C for a representative example; Table S5: Bold). Some of the highlights of these 272 genes were 115 genes coding for VSGs or VSG associated proteins; five genes coding for cell surface expressed proteins; six genes coding for ATP-dependent dead h RNA helicases (RNA helicases); seven genes coding for retroposon hot spot proteins and 11 genes coding for the transferrin receptor ESAG6. In addition, the duplication of seven genes was common between TC1 or Zambian resistant field isolates and one or more MSOROM7 resistant lines (Fig. S1 D; Table S5: Underlined).
3.4. Reduced ISM accumulation in resistant parasites
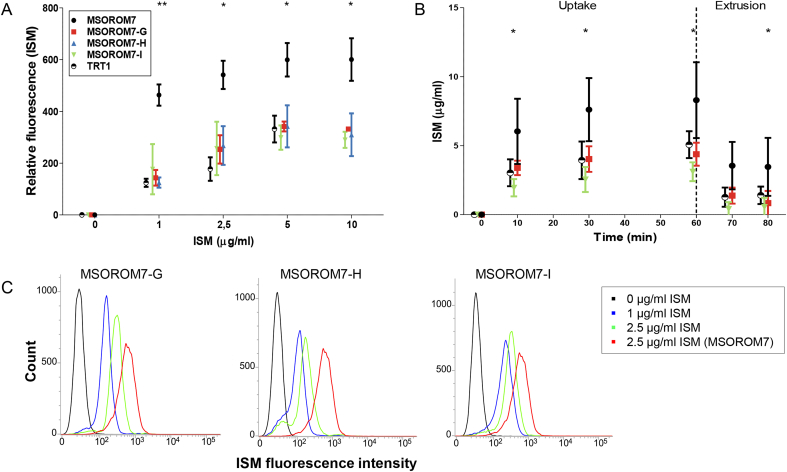
A significant decrease in ISM accumulation was observed by flow cytometry in all resistant trypanosomes compared with the sensitive MSOROM7 parental line (Fig. 6A and C). The accumulation of ISM in the sensitive and resistant parasites gradually increased as ISM concentrations augmented, until reaching a plateau at a concentration of ISM of 5 μg/ml in the incubation medium. At this concentration, the sensitive cells displayed a mean ISM fluorescent signal of 599 (standard deviation = 64.6) compared to 327 (standard deviation = 52.6) for the resistant parasites. ISM exposure started to kill parasites at a concentration of 5 μg/ml and higher, as indicated by the increased signal of the Fixable Viability Stain (FVS) (data not shown). No differences in the accumulation of ISM could be detected between the three induced-resistant lines. A similar decrease in ISM accumulation was also observed for the ISM resistant field isolate TRT1 (Fig. 6A).
Fig. 6.
Accumulation and uptake of ISM in the sensitive and resistant MSOROM7 lines and in the resistant field isolate TRT1. A) and C) Evaluation of ISM accumulation by flow cytometry (n = 3). A significant decrease in the accumulation of ISM is observed in all resistant strains compared with the sensitive MSOROM7 parental line. B) Measurement of the uptake and extrusion of ISM by an ex vivo fluorescent assay (n = 4, except MSOROM7 where n = 7). The uptake of ISM was significantly reduced in MSOROM7-G, MSOROM7-I and TRT1 (p < 0.05) compared with the sensitive MSOROM7 parental line. After 60 min (dashed line) parasites were placed in a drug free medium for 10 and 20 min to evaluate ISM extrusion. Average ± standard error of the mean; *: P < 0.05, **: P < 0.01.
Then, we assessed the dynamics of the accumulation and extrusion of ISM in the sensitive and resistant parasites and observed a fast accumulation of ISM within the first 10 min of incubation for all cell lines (Fig. 6B). After this initial period, the ISM accumulation remained at the same level over time. The observed ISM accumulation in the resistant MSOROM7-G, MSOROM7-I and TRT1 was significantly lower than for the sensitive MSOROM7 line (Fig. 6B). For both sensitive and resistant parasites, the concentration of ISM accumulating inside trypanosomes exceeded the initial starting concentration of 1 μg/ml, suggesting the involvement of an active transport of ISM. Finally, the extrusion of ISM by the parasites displayed a similar profile for all the strains (Fig. 6B).
4. Discussion
The drug ISM plays a key role in the treatment of T. congolense infections. However, little information is available about its cellular uptake and accumulation, its mechanisms of action and development of drug resistance in this parasite, impeding a proper management of ISM resistance in the field. Here, we used WGS to study the genetic changes that may underlie the acquisition of ISM resistance within an in vivo model.
We demonstrated the importance of the immune system of the host in preventing the development of ISM resistance. In an immune-competent host, sensitive parasites failed to acquire resistance to 1 mg/kg ISM after more than two years under drug pressure. In immune-suppressed animals, sensitive parasites evolved to fully drug resistant parasites after three to five months of drug pressure. Similar observations were previously described in T. b. evansi parasites (Osman et al., 1992, Mutugi et al., 1994), and it has been hypothesized that trypanocidal drugs, rather than killing parasites, induce restrictions in the replication of the trypanosomes that will then be eliminated by the immune system of the host (Geerts et al., 2001). This mechanism could explain partly why trypanosomes can overcome an ISM pressure in immune-deficient animals. The link between a weakened host immune system and drug resistance acquisition in protozoan parasites is further highlighted by the observation that impaired immunity resulted in treatment failures and drug resistance to visceral leishmaniasis and malaria in HIV co-infected patients (Troya et al., 2008, Mondal et al., 2010, Menendez et al., 2011). In the field, impaired animal immunity could result from injuries, infections with other pathogens such as intestinal worms, malnutrition and stressful situations. Proper livestock health management is therefore highly important to lower the probability of ISM resistance development.
Within the drug resistant parasites, gene deletions and duplications could be detected in contrast to novel SNPs or indels when comparing them with the drug sensitive parasites. Moreover, no significant changes were observed within the genes coding for the F1-subunit γ and other proteins interfering with the mitochondrial F1FO-ATP synthase, suggesting that T. congolense developed different ISM resistance mechanisms than T. b. brucei (i.e. in vitro-induced resistance), T. b. evansi and T. b. equiperdum (Brun and Lun, 1994, Dean et al., 2013, Gould and Schnaufer, 2014, Baker et al., 2015). In addition, the resistant MSOROM7 T. congolense parasites were still fully capable of completing successfully their life cycle in the tsetse fly vector (data not shown), indicating the presence of a functional kDNA. Interestingly, we identified significant shifts in allele frequency among heterozygous loci associated with non-synonymous changes in genes coding for ABC transporters, DMT and transmembrane products. Specifically on the DMT gene, we observed a strong increase in the frequency of 19 alternate alleles at heterozygous sites, resulting in the fixation of 14 non-synonymous changes in nucleotide sequence in MSOROM7-H. This fixation probably resulted from the deletion of one of the two copies of the DMT gene that were present in the original sensitive genome. In order to explain the adjacent alternate allele frequencies of 0.25, 0.5 and 0.75 in the sensitive strain (see Fig. 4B), one must assume that both copies show different genotypes, as explained in the following theoretical example: (i) BB/BA/BB (three sites, B and A standing for alternate and the reference alleles, respectively) and (ii) AA/AA/BA, hereby explaining an alternate allele frequency of 0.5/0.25/0.75 for the respective three sites. The loss of the second gene in the resistant MSOROM7-H would lead to a unique BB/BA/BB, with alternate allele frequencies of 1/0.5/1, resulting in fixation of a series of alternate alleles.
The loss of one DMT gene copy was also observed in MSOROM7-G and MSOROM7-I. However, since we did not observe any fixation of alternate alleles at non-synonymous positions, we concluded that the first copy of the gene was lost in these two resistant lines.
Interestingly, the expression level of the DMT product was significantly downregulated in the resistant parasites, especially in MSOROM7-H where allele frequency shifts at non-synonymous positions were observed. This suggests that the combination of the loss of one copy of the DMT genes and the fixation of non-synonymous mutations had a higher impact on the gene expression level. This may have contributed to the observed decrease in ISM accumulation within the drug resistant parasites.
Members of the DMT superfamily have been involved in antibiotics resistance in Staphylococcus aureus (Grinius and Goldberg, 1994) and Escherichia coli (Yerushalmi et al., 1995), and in chloroquine resistance in P. falciparum (Martin and Kirk, 2004). Similarly, an important role of ABC transporters in drug resistance has previously been proposed in T. congolense (Delespaux et al., 2005) and has been documented in many other organisms including T. brucei, T. cruzi, P. falciparum, Schistosoma mansoni, and Leishmania parasites (Ouellette et al., 2001, Perez-Victoria et al., 2002, Shahi et al., 2002, Alibu et al., 2006, Sanderson et al., 2009, Franco et al., 2015, Garg and Goyal, 2015, Pinto-Almeida et al., 2015, Veiga et al., 2016). Changes in other transport proteins also conferred resistance in Trypanosoma spp. (Barrett et al., 1995, Matovu et al., 2003, de Koning et al., 2004, Witola et al., 2004, Munday et al., 2015). The potential link between the observed allele frequency shifts and ISM transport modifications still has to be confirmed, but the genes listed in this paper represent promising targets for further functional experimental analyses.
Our genomic analyses also revealed that new gene deletions and gene amplifications may possibly play a role in ISM resistance. However, it is difficult to establish a link between ISM resistance and the observed gene deletions/amplifications due to relatively poor annotation of the T. congolense genome. The fact that 79 gene amplifications were common between the experimentally induced MSOROM7 resistant parasites and the TRT15 parasites under drug pressure for 10 passages may suggest that these changes in gene copy numbers reflect the mutagenic properties of ISM. This is possibly the consequence of the high affinity of ISM for double-stranded DNA molecules (Wilkes et al., 1995) that leads to its accumulation in both the mitochondrion and nucleus of the parasites where it induces double strand breaks and subsequently inhibits the kDNA (Shapiro and Englund, 1990) and DNA (Kaminsky et al., 1997) replications. To overcome DNA damages and maintain its integrity, cells possess a DNA repair machinery that consists of a complex of distinct repair pathways (reviewed in (Genois et al., 2014)). This includes a double strand breaks (DSB) repair that involves both homologous and non-homologous recombination (Genois et al., 2014). The potential involvement of the DSB repair in response to an ISM pressure could explain the observed shifts in read depth allele frequency (i.e. homologous recombination where the sister chromatid acts as repair template) and CNVs (i.e. homologous recombination between homologous repeats to amplify or delete DNA sequences) (Genois et al., 2014). This hypothesis is supported by the observation of the deletion of the DMT gene (Fig. 4) and other gene deletions and duplications (Fig. 5), possibly as the result of DSB repair events covering these genes. In addition, allele frequency shifts and CNVs were also observed in the TRT15 isolate under drug pressure but not in the absence of drug pressure, further suggesting that ISM induces strong mutagenic effects.
The potential involvement of the DNA repair machinery in drug resistance mechanisms has been suggested in P. falciparum where the DNA repair machinery was upregulated to respond to exogenous stress (i.e. DNA damaging agents) and promoted cell survival (Gupta et al., 2016), and in Leishmania parasites where the amplification of P-glycoprotein genes through homologous recombination conferred drug resistance (Ouellette et al., 1991, Grondin et al., 1993). Similar mechanisms could potentially exist in T. congolense in response to the DNA damaging drug ISM to promote cell survival and drug resistance.
Other genome modifying mechanisms such as point mutations and sexual recombination could also play a role in some of the genomic modifications observed in the resistant parasites. However, there is currently no evidence about a potential recombination cycle in bloodstream form T. congolense parasites (i.e. meiosis is taking place in the tsetse fly salivary glands in T. brucei (Gibson et al., 2008, Peacock et al., 2011)).
5. Conclusion
It is clear that the acquisition of ISM resistance in T. congolense is a complex phenomenon and that multiple mechanisms could lead to the acquisition of the drug resistant phenotype known as the ‘many roads to drug resistance’ concept (Hefnawy et al., 2017). While we demonstrated that the immune system of the host plays a key role in hindering the acquisition of ISM resistance, we were able to induce drug resistance in a sensitive isolate in an immune-suppressed host. Subsequent genomic analyses revealed that non-synonymous mutations within genes encoding ABC transporters, DMT and some ‘unidentified’ transmembrane proteins may explain the development of drug resistance. The next step would be to perform functional analyses and confirm the involvement of these genes in ISM resistance.
Acknowledgements
We thank Guy Caljon for helping with the flow cytometry experiments.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2017.10.002.
Contributor Information
Eliane Tihon, Email: etihon@itg.be.
Hideo Imamura, Email: himamura@itg.be.
Frederik Van den Broeck, Email: fvandenbroeck@itg.be.
Lieve Vermeiren, Email: lvermeiren@itg.be.
Jean-Claude Dujardin, Email: jcdujardin@itg.be.
Jan Van Den Abbeele, Email: jvdabbeele@itg.be.
Funding sources
This work was financed by the EU- Global Program on Agricultural Research for Development, Grant reference number: TRYRAC/DCI-FOOD/2011/279-754, and the Department of Economy, Science and Innovation in Flanders, ITM-SOFIB GemiNI (755044).
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Afewerk Y., Clausen P.H., Abebe G., Tilahun G., Mehlitz D. Multiple-drug resistant Trypanosoma congolense populations in village cattle of Metekel district, north-west Ethiopia. Acta Trop. 2000;76:231–238. doi: 10.1016/s0001-706x(00)00108-x. [DOI] [PubMed] [Google Scholar]
- Alibu V.P., Richter C., Voncken F., Marti G., Shahi S., Renggli C.K., Seebeck T., Brun R., Clayton C. The role of Trypanosoma brucei MRPA in melarsoprol susceptibility. Mol. Biochem. Parasitol. 2006;146:38–44. doi: 10.1016/j.molbiopara.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Baker N., Hamilton G., Wilkes J.M., Hutchinson S., Barrett M.P., Horn D. Vacuolar ATPase depletion affects mitochondrial ATPase function, kinetoplast dependency, and drug sensitivity in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9112–9117. doi: 10.1073/pnas.1505411112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M.P., Zhang Z.Q., Denise H., Giroud C., Baltz T. A diamidine-resistant Trypanosoma equiperdum clone contains a P2 purine transporter with reduced substrate affinity. Mol. Biochem. Parasitol. 1995;73:223–229. doi: 10.1016/0166-6851(95)00120-p. [DOI] [PubMed] [Google Scholar]
- Brack C., Delain E., Riou G., Festy B. Molecular organization of the kinetoplast DNA of Trypanosoma cruzi treated with berenil, a DNA interacting drug. J. Ultrastruct. Res. 1972;39:568–579. doi: 10.1016/s0022-5320(72)90122-0. [DOI] [PubMed] [Google Scholar]
- Brown K.N., Hill J., Holland A.E. Anti-trypanosomal activity of certain phenyldiazoamino- and phenylazoamino-phenanthridinium compounds. Br. J. Pharmacol. Chemother. 1961;17:396–405. doi: 10.1111/j.1476-5381.1961.tb01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R., Lun Z.R. Drug sensitivity of Chinese Trypanosoma evansi and Trypanosoma equiperdum isolates. Veterinary Parasitol. 1994;52:37–46. doi: 10.1016/0304-4017(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Cecchi G., Mattioli R.C. 2009. Geospatial Datasets and Analyses for an Environmental Approach to African Trypanosomiasis. Programme against African Trypanosomiasis. [Google Scholar]
- Cecchi G., Paone M., Feldmann U., Vreysen M.J., Diall O., Mattioli R.C. Assembling a geospatial database of tsetse-transmitted animal trypanosomosis for Africa. Parasites Vectors. 2014;7:39. doi: 10.1186/1756-3305-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen P.H., Sidibe I., Kabore I., Bauer B. Development of multiple drug resistance of Trypanosoma congolense in Zebu cattle under high natural tsetse fly challenge in the pastoral zone of Samorogouan, Burkina Faso. Acta Trop. 1992;51:229–236. doi: 10.1016/0001-706x(92)90041-u. [DOI] [PubMed] [Google Scholar]
- Cox A., Tilley A., McOdimba F., Fyfe J., Eisler M., Hide G., Welburn S. A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp. Parasitol. 2005;111:24–29. doi: 10.1016/j.exppara.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Dagnachew S., Terefe G., Abebe G., Barry D., McCulloch R., Goddeeris B. In vivo experimental drug resistance study in Trypanosoma vivax isolates from tsetse infested and non-tsetse infested areas of Northwest Ethiopia. Acta Trop. 2015;146:95–100. doi: 10.1016/j.actatropica.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.P., Gull K., Wickstead B. Cell biology of the trypanosome genome. Microbiol. Mol. Biol. Rev. MMBR. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning H.P., Anderson L.F., Stewart M., Burchmore R.J., Wallace L.J., Barrett M.P. The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: additional insights on diamidine resistance in african trypanosomes. Antimicrob. Agents Chemother. 2004;48:1515–1519. doi: 10.1128/AAC.48.5.1515-1519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S., Gould M.K., Dewar C.E., Schnaufer A.C. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespaux V., Dinka H., Masumu J., Van den Bossche P., Geerts S. Five-fold increase in Trypanosoma congolense isolates resistant to diminazene aceturate over a seven-year period in Eastern Zambia. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2008;11:205–209. doi: 10.1016/j.drup.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Delespaux V., Geysen D., Majiwa P.A., Geerts S. Identification of a genetic marker for Isometamidium Chloride resistance in Trypanosoma congolense. Int. J. Parasitol. 2005;35:235–243. doi: 10.1016/j.ijpara.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Delespaux V., Geysen D., Van den Bossche P., Geerts S. Molecular tools for the rapid detection of drug resistance in animal trypanosomes. Trends Parasitol. 2008;24:236–242. doi: 10.1016/j.pt.2008.02.006. [DOI] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T., Imamura H., Decuypere S., Clark T.G., Coombs G.H., Cotton J.A., Hilley J.D., de Doncker S., Maes I., Mottram J.C. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21:2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumetz F., Imamura H., Sanders M., Seblova V., Myskova J., Pescher P., Vanaerschot M., Meehan C.J., Cuypers B., De Muylder G. Modulation of aneuploidy in Leishmania donovani during adaptation to different in vitro and in vivo environments and its impact on gene expression. mBio. 2017;8 doi: 10.1128/mBio.00599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisler M.C., Brandt J., Bauer B., Clausen P.H., Delespaux V., Holmes P.H., Ilemobade A., Machila N., Mbwambo H., McDermott J. Standardised tests in mice and cattle for the detection of drug resistance in tsetse-transmitted trypanosomes of African domestic cattle. Veterinary Parasitol. 2001;97:171–182. doi: 10.1016/s0304-4017(01)00415-0. [DOI] [PubMed] [Google Scholar]
- Eze A.A., Gould M.K., Munday J.C., Tagoe D.N., Stelmanis V., Schnaufer A., De Koning H.P. Reduced mitochondrial membrane potential is a late adaptation of Trypanosoma brucei brucei to isometamidium preceded by mutations in the gamma subunit of the F1Fo-ATPase. PLoS Negl. Trop. Dis. 2016;10:e0004791. doi: 10.1371/journal.pntd.0004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco J., Ferreira R.C., Ienne S., Zingales B. ABCG-like transporter of Trypanosoma cruzi involved in benznidazole resistance: gene polymorphisms disclose inter-strain intragenic recombination in hybrid isolates. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015;31:198–208. doi: 10.1016/j.meegid.2015.01.030. [DOI] [PubMed] [Google Scholar]
- Garg M., Goyal N. MAPK1 of Leishmania donovani modulates antimony susceptibility by downregulating P-glycoprotein efflux pumps. Antimicrob. Agents Chemother. 2015;59:3853–3863. doi: 10.1128/AAC.04816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts S., Holmes P.H. Drug management and parasite resistance in bovine trypanosomiasis in Africa. Food Agri. Organ. U. N. 1998;31 [Google Scholar]
- Geerts S., Holmes P.H., Eisler M.C., Diall O. African bovine trypanosomiasis: the problem of drug resistance. Trends Parasitol. 2001;17:25–28. doi: 10.1016/s1471-4922(00)01827-4. [DOI] [PubMed] [Google Scholar]
- Geigy R., Kauffmann M. Sleeping sickness survey in the Serengeti area (Tanzania) 1971. I. Examination of large mammals for trypanosomes. Acta trop. 1973;30:12–23. [PubMed] [Google Scholar]
- Genois M.M., Paquet E.R., Laffitte M.C., Maity R., Rodrigue A., Ouellette M., Masson J.Y. DNA repair pathways in trypanosomatids: from DNA repair to drug resistance. Microbiol. Mol. Biol. Rev. MMBR. 2014;78:40–73. doi: 10.1128/MMBR.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W., Peacock L., Ferris V., Williams K., Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasites Vectors. 2008;1:4. doi: 10.1186/1756-3305-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani F., Morrison L.J., Rowan T.G., HP D.E.K., Barrett M.P. The animal trypanosomiases and their chemotherapy: a review. Parasitology. 2016;143:1862–1889. doi: 10.1017/S0031182016001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V.M., Perez J.M., Alonso C. The berenil ligand directs the DNA binding of the cytotoxic drug Pt-berenil. J. Inorg. Biochem. 1997;68:283–287. doi: 10.1016/s0162-0134(97)00111-6. [DOI] [PubMed] [Google Scholar]
- Gould M.K., Schnaufer A. Independence from Kinetoplast DNA maintenance and expression is associated with multidrug resistance in Trypanosoma brucei in vitro. Antimicrob. Agents Chemother. 2014;58:2925–2928. doi: 10.1128/AAC.00122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinius L.L., Goldberg E.B. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 1994;269:29998–30004. [PubMed] [Google Scholar]
- Grondin K., Papadopoulou B., Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D.K., Patra A.T., Zhu L., Gupta A.P., Bozdech Z. DNA damage regulation and its role in drug-related phenotypes in the malaria parasites. Sci. Rep. 2016;6:23603. doi: 10.1038/srep23603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefnawy A., Berg M., Dujardin J.C., De Muylder G. Exploiting knowledge on Leishmania drug resistance to support the quest for new drugs. Trends Parasitol. 2017;33:162–174. doi: 10.1016/j.pt.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hursey B.S., Slingenbergh J. Food and Africulture Organization of the United Nations; 1995. The Tsetse Fly and its Effects on Agriculture in Sub-Saharan Africa. [Google Scholar]
- Jackson A.P., Berry A., Aslett M., Allison H.C., Burton P., Vavrova-Anderson J., Brown R., Browne H., Corton N., Hauser H. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky R., Schmid C., Lun Z.R. Susceptibility of dyskinetoplastic Trypanosoma evansi and T. equiperdum to Isometamidium Chloride. Parasitol. Res. 1997;83:816–818. doi: 10.1007/s004360050346. [DOI] [PubMed] [Google Scholar]
- Knoppe T.N. Freie Universität Berlin; 2002. Drug Sensitivity of Trypanosoma Congolense (BRODEN, 1904) Stocks, Isolated from Cattle in Burkina Faso, West Africa; p. 122. [Google Scholar]
- Lanham S.M., Godfrey D.G. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 1970;28:521–534. doi: 10.1016/0014-4894(70)90120-7. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu Y., Motyka S.A., Agbo E.E., Englund P.T. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lourie E.M., Yorke W. Studies in chemotherapy. XXI.-The trypanocidal action of certain aromatic diamidines. Ann. Trop. Med. Parasit. 1939;33:289–304. [Google Scholar]
- Mahler H.R., Perlman P.S. Induction of respiration deficient mutants in Saccharomyces cerevisiae by berenil. I. Berenil, a novel, non-intercalating mutagen. Mol. General Genet. MGG. 1973;121:285–294. doi: 10.1007/BF00433228. [DOI] [PubMed] [Google Scholar]
- Mamoudou A., Delespaux V., Chepnda V., Hachimou Z., Andrikaye J.P., Zoli A., Geerts S. Assessment of the occurrence of trypanocidal drug resistance in trypanosomes of naturally infected cattle in the Adamaoua region of Cameroon using the standard mouse test and molecular tools. Acta Trop. 2008;106:115–118. doi: 10.1016/j.actatropica.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Mamoudou A., Zoli A., Tanenbe C., Andrikaye J.P., Bourdanne, Marcotty T., Delespaux V., Clausen P.H., Geerts S. Evaluation sur le terrain et sur souris de la resistance des trypanosomes des bovins du plateau de l'Amaoua au Cameroun a l’aceturate de diminazene et au chlorure d’isometamidium. Rev. Elev. Med. Vet. Pays Trop. 2006;59:6. [Google Scholar]
- Martin R.E., Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- Masumu J., Geysen D., Van den Bossche P. Endemic type of animal trypanosomiasis is not associated with lower genotype variability of Trypanosoma congolense isolates circulating in livestock. Res. Vet. Sci. 2009;87:265–269. doi: 10.1016/j.rvsc.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matovu E., Stewart M.L., Geiser F., Brun R., Maser P., Wallace L.J., Burchmore R.J., Enyaru J.C., Barrett M.P., Kaminsky R. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryot. Cell. 2003;2:1003–1008. doi: 10.1128/EC.2.5.1003-1008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli R.C., Feldmann U., G. H, W. W, J. J, J. S Tsetse and trypanosomiasis intervention policies supporting sustainable animal-agricultural development. Food, Agric. Environ. 2004;2:5. [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez C., Serra-Casas E., Scahill M.D., Sanz S., Nhabomba A., Bardaji A., Sigauque B., Cistero P., Mandomando I., Dobano C. HIV and placental infection modulate the appearance of drug-resistant Plasmodium falciparum in pregnant women who receive intermittent preventive treatment. Clin. Infect. Dis. 2011;52:41–48. doi: 10.1093/cid/ciq049. [DOI] [PubMed] [Google Scholar]
- Miotto O., Amato R., Ashley E.A., MacInnis B., Almagro-Garcia J., Amaratunga C., Lim P., Mead D., Oyola S.O., Dhorda M. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S., Bhattacharya P., Ali N. Current diagnosis and treatment of visceral leishmaniasis. Expert Rev. Anti Infect. Ther. 2010;8:919–944. doi: 10.1586/eri.10.78. [DOI] [PubMed] [Google Scholar]
- Mondelaers A., Sanchez-Canete M.P., Hendrickx S., Eberhardt E., Garcia-Hernandez R., Lachaud L., Cotton J., Sanders M., Cuypers B., Imamura H. Genomic and molecular characterization of miltefosine resistance in Leishmania infantum strains with either natural or acquired resistance through experimental selection of intracellular amastigotes. PLoS One. 2016;11:e0154101. doi: 10.1371/journal.pone.0154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moti Y., De Deken R., Thys E., Van Den Abbeele J., Duchateau L., Delespaux V. PCR and microsatellite analysis of diminazene aceturate resistance of bovine trypanosomes correlated to knowledge, attitude and practice of livestock keepers in South-Western Ethiopia. Acta Trop. 2015;146:45–52. doi: 10.1016/j.actatropica.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Moti Y., Fikru R., Van Den Abbeele J., Buscher P., Van den Bossche P., Duchateau L., Delespaux V. Ghibe river basin in Ethiopia: present situation of trypanocidal drug resistance in Trypanosoma congolense using tests in mice and PCR-RFLP. Vet. Parasitol. 2012;189:197–203. doi: 10.1016/j.vetpar.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Munday J.C., Settimo L., de Koning H.P. Transport proteins determine drug sensitivity and resistance in a protozoan parasite, Trypanosoma brucei. Front. Pharmacol. 2015;6:32. doi: 10.3389/fphar.2015.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungube E.O., Vitouley H.S., Allegye-Cudjoe E., Diall O., Boucoum Z., Diarra B., Sanogo Y., Randolph T., Bauer B., Zessin K.H. Detection of multiple drug-resistant Trypanosoma congolense populations in village cattle of south-east Mali. Parasites Vectors. 2012;5:155. doi: 10.1186/1756-3305-5-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutugi M.W., Boid R., Luckins A.G. Experimental induction of suramin-resistance in cloned and uncloned stocks of Trypanosoma evansi using immunosuppressed and immunocompetent mice. Trop. Med. Parasitol. 1994;45:232–236. [PubMed] [Google Scholar]
- Ndoutamia G., Moloo S.K., Murphy N.B., Peregrine A.S. Derivation and characterization of a quinapyramine-resistant clone of Trypanosoma congolense. Antimicrob. Agents Chemother. 1993;37:1163–1166. doi: 10.1128/aac.37.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Paul J.S., Albrechtsen A., Song Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011;12:443–451. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman A.S., Jennings F.W., Holmes P.H. The rapid development of drug-resistance by Trypanosoma evansi in immunosuppressed mice. Acta Trop. 1992;50:249–257. doi: 10.1016/0001-706x(92)90081-8. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Hettema E., Wust D., Fase-Fowler F., Borst P. Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M., Legare D., Papadopoulou B. Multidrug resistance and ABC transporters in parasitic protozoa. J. Mol. Microbiol. Biotechnol. 2001;3:201–206. [PubMed] [Google Scholar]
- Peacock L., Ferris V., Sharma R., Sunter J., Bailey M., Carrington M., Gibson W. Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3671–3676. doi: 10.1073/pnas.1019423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria J.M., Di Pietro A., Barron D., Ravelo A.G., Castanys S., Gamarro F. Multidrug resistance phenotype mediated by the P-glycoprotein-like transporter in Leishmania: a search for reversal agents. Curr. Drug Targets. 2002;3:311–333. doi: 10.2174/1389450023347588. [DOI] [PubMed] [Google Scholar]
- Pinder M., Authie E. The appearance of isometamidium resistant Trypanosoma congolense in West Africa. Acta Trop. 1984;41:247–252. [PubMed] [Google Scholar]
- Pinto-Almeida A., Mendes T., Armada A., Belo S., Carrilho E., Viveiros M., Afonso A. The role of efflux pumps in Schistosoma mansoni praziquantel resistant phenotype. PLoS One. 2015;10:e0140147. doi: 10.1371/journal.pone.0140147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Chowdhury A., Bakshi R., Wang J., Yildirir G., Liu B., Pappas-Brown V., Tolun G., Griffith J.D., Shapiro T.A., Jensen R.E. The killing of African trypanosomes by ethidium bromide. PLoS Pathog. 2010;6:e1001226. doi: 10.1371/journal.ppat.1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin A., Asencio C., Izotte J., Pillay D., Coustou V., Karembe H., Baltz T. The susceptibility of Trypanosoma congolense and Trypanosoma brucei to Isometamidium Chloride and its synthetic impurities. Veterinary Parasitol. 2014;203:270–275. doi: 10.1016/j.vetpar.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Sanderson L., Dogruel M., Rodgers J., De Koning H.P., Thomas S.A. Pentamidine movement across the murine blood-brain and blood-cerebrospinal fluid barriers: effect of trypanosome infection, combination therapy, P-glycoprotein, and multidrug resistance-associated protein. J. Pharmacol. Exp. Ther. 2009;329:967–977. doi: 10.1124/jpet.108.149872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A., Clark-Walker G.D., Steinberg A.G., Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahi S.K., Krauth-Siegel R.L., Clayton C.E. Overexpression of the putative thiol conjugate transporter TbMRPA causes melarsoprol resistance in Trypanosoma brucei. Mol. Microbiol. 2002;43:1129–1138. doi: 10.1046/j.1365-2958.2002.02831.x. [DOI] [PubMed] [Google Scholar]
- Shapiro T.A., Englund P.T. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl. Acad. Sci. U. S. A. 1990;87:950–954. doi: 10.1073/pnas.87.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinyangwe L., Delespaux V., Brandt J., Geerts S., Mubanga J., Machila N., Holmes P.H., Eisler M.C. Trypanocidal drug resistance in eastern province of Zambia. Vet. Parasitol. 2004;119:125–135. doi: 10.1016/j.vetpar.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Sow A., Sidibe I., Bengaly Z., Marcotty T., Sere M., Diallo A., Vitouley H.S., Nebie R.L., Ouedraogo M., Akoda G.K. Field detection of resistance to Isometamidium Chloride and diminazene aceturate in Trypanosoma vivax from the region of the Boucle du Mouhoun in Burkina Faso. Vet. Parasitol. 2012;187:105–111. doi: 10.1016/j.vetpar.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Sutherland I.A., Mounsey A., Holmes P.H. Transport of isometamidium (Samorin) by drug-resistant and drug-sensitive Trypanosoma congolense. Parasitology. 1992;104(Pt 3):461–467. doi: 10.1017/s0031182000063721. [DOI] [PubMed] [Google Scholar]
- Tihon E., Imamura H., Dujardin J.C., Van Den Abbeele J., Van den Broeck F. Discovery and genomic analyses of hybridization between divergent lineages of Trypanosoma congolense, causative agent of Animal African Trypanosomiasis. Mol. Ecol. 2017 doi: 10.1111/mec.14271. [DOI] [PubMed] [Google Scholar]
- Troya J., Casquero A., Refoyo E., Fernandez-Guerrero M.L., Gorgolas M. Long term failure of miltefosine in the treatment of refractory visceral leishmaniasis in AIDS patients. Scand. J. Infect. Dis. 2008;40:78–80. doi: 10.1080/00365540701466215. [DOI] [PubMed] [Google Scholar]
- Uilenberg G. In: AaC Protection, editor. vol. 2016. Food and Agriculture Organization (FAO) of the United Nations; 1998. (A Field Guide for the Diagnosis, Treatment and Prevention of African Animal Trypanosomosis). [Google Scholar]
- Veiga M.I., Dhingra S.K., Henrich P.P., Straimer J., Gnadig N., Uhlemann A.C., Martin R.E., Lehane A.M., Fidock D.A. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016;7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Englund P.T. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins T.I., Woolfe G. Effect of changing the quaternizing group on the trypanocidal activity of dimidium bromide. Nature. 1952;169:506–507. doi: 10.1038/169506a0. [DOI] [PubMed] [Google Scholar]
- Wilkes J.M., Mulugeta W., Wells C., Peregrine A.S. Modulation of mitochondrial electrical potential: a candidate mechanism for drug resistance in African trypanosomes. Biochem. J. 1997;326(Pt 3):755–761. doi: 10.1042/bj3260755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes J.M., Peregrine A.S., Zilberstein D. The accumulation and compartmentalization of Isometamidium Chloride in Trypanosoma congolense, monitored by its intrinsic fluorescence. Biochem. J. 1995;312(Pt 1):319–327. doi: 10.1042/bj3120319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola W.H., Inoue N., Ohashi K., Onuma M. RNA-interference silencing of the adenosine transporter-1 gene in Trypanosoma evansi confers resistance to diminazene aceturate. Exp. Parasitol. 2004;107:47–57. doi: 10.1016/j.exppara.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Yerushalmi H., Lebendiker M., Schuldiner S. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 1995;270:6856–6863. doi: 10.1074/jbc.270.12.6856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.