Abstract
Phialophora as defined by its type species P. verrucosa is a genus of Chaetothyriales, and a member of the group known as ‘black yeasts and relatives’. Phialophora verrucosa has been reported from mutilating human infections such as chromoblastomycosis, disseminated phaeohyphomycosis and mycetoma, while morphologically similar fungi are rather commonly isolated from the environment. Phenotypes are insufficient for correct species identification, and molecular data have revealed significant genetic variation within the complex of species currently identified as P. verrucosa or P. americana. Multilocus analysis of 118 strains revealed the existence of five reproductively isolated species apparently having different infectious potentials. Strains of the sexual morph Capronia semiimmersa cluster within P. americana. The newly defined taxa differ markedly in their predilection for the human host.
Keywords: Chaetothyriales, chromoblastomycosis, phaeohyphomycosis, Phialophora, phylogeny, taxonomy
INTRODUCTION
Phialophora verrucosa is the type species of the genus Phialophora, which belongs to the family Herpotrichiellaceae (Chaetothyriales) comprising the black yeasts and relatives. This phylogenetic affiliation excludes numerous species that have been classified in older literature in Phialophora on the basis of the combination of morphological characters of a melanised thallus and one-celled, sticky conidia that are produced through large phialidic collarettes in a poorly differentiated conidial apparatus. Gams (2000) provided an overview of phialophora-like fungi and found that according to current standards belong in nine orders of Ascomycota; for nearly all of these, separate generic names are available at present.
Numerous asexual species in the Chaetothyriales classified in Cladophialophora, Exophiala or Fonsecaea show presence of phialophora-like synasexual morphs on nutritionally poor media, demonstrating the taxonomic coherence of species belonging to this order (De Hoog et al. 1999). Such phialidic synasexual morphs are also known in Cladophialophora carrionii, the agent of human chromoblastomycosis in arid climates and one of the nearest neighbours of P. verrucosa in molecular phylogeny. Although strictly monomorphic for phialides, P. verrucosa phylogenetically belongs to a group as a whole as the ‘carrionii-clade’ with Cladophialophora carrionii as the core species.
Several other but unrelated monomorphic phialophora-like lineages are known in the Chaetothyriales. Phialophora europaea, P. reptans, known from superficial skin infections in humans (Saunte et al. 2012), P. attae and P. capiguarae, from ant nests (Attili-Angelis et al. 2014), P. sessilis from inert surfaces (Caretta et al. 2006, Zhuang et al. 2010), P. livistonae, from living plant leaves (Crous et al. 2012) and P. oxyspora all are members of the ‘europaea-clade’ (De Hoog et al. 2011, Feng et al. 2012). This clade was given family status as Cyphellophoraceae by Réblová et al. (2013) and as a consequence some of the member species were reclassified in Cyphellophora.
As a result of the above rearrangements, the genus Phialophora, for which the Index Fungorum lists 92 species names (as per 01-01-2016), from a phylogenetic viewpoint is restricted to P. verrucosa and its sister species Phialophora americana, as they both cluster in the ‘carrionii-clade’. Species of this clade, i.e. Cladophialophora carrionii, Cl. samoensis and P. verrucosa have been reported from mutilating cases of chromoblastomycosis, disseminated phaeohyphomycosis and mycetoma, which all can be chronic and refractory to therapy (McGinnis 1983, Turiansky et al. 1995, Hofmann et al. 2005, Seyedmousavi et al. 2014). Phialophora americana, a sister species of P. verrucosa is mostly regarded as being environmental. Also Cl. carrionii has an environmental sibling, viz. Cladophialophora yegresii (De Hoog et al. 2007). The bipartition clinical / environmental is however ambiguous. Phialophora verrucosa was first reported as a human pathogen a century ago (Lane 1915, Medlar 1915a, b), but fungi under this name have also been isolated from natural soils and plant debris (Gezuele et al. 1972). For most of these reports no material is known to be preserved and misidentifications with numerous phialophora-like fungi may have been concerned (Gams 2000, Lopez Martinez & Mendez Tovar 2007).
Recent studies have proven that molecular techniques have a higher precision in segregating phenotypically similar species that may differ in pathogenicity (Marimón et al. 2006, 2007). In black yeasts and allied fungi, molecular siblings may differ significantly in virulence; compare for example the neurotrope Cladophialophora bantiana and the gasoline-associated fungus Cl. psammophila (Badali et al. 2011). Internal transcribed spacer (ITS) sequencing is effective for species identification among black yeasts, as has been proven with the aid of multilocus studies (Zeng & De Hoog 2008, Heinrichs et al. 2012). No multi-locus verification is available for the P. verrucosa / P. americana complex (Untereiner et al. 2008). Molecular typing of mitochondrial DNA using restriction fragment length polymorphisms (RFLP) suggested that P. verrucosa comprised three groups, while analyses of group 1 introns in the 28S ribosomal RNA gene divided the species into five genotypes (Yamagishi et al. 1997, Takizawa et al. 2011). Given this genetic variation a study of phylogenetic relationships is overdue.
Patients infected by P. verrucosa showed significant differences in treatment outcomes. This may be due to hidden genetic immune disorders of the host, but the possibility that different Phialophora species were concerned cannot be excluded (Tong et al. 2013, Wang et al. 2014). The aim of the present study was to explore the taxonomy of the P. verrucosa complex and to determine whether genetic diversity was associated with differences in pathogenicity. Sequence analyses of the ribosomal internal transcribed spacers (ITS), and partial β-tubulin (BT2), translation elongation factor 1 alpha (TEF1) and the small and large subunits of the nuclear ribosomal RNA (SSU / LSU) regions were used alone or in combination. Additionally, phenotypic characters of morphology and physiology were included along with ecological data.
MATERIALS AND METHODS
Strains studied
One hundred and twenty-six isolates that were initially identified as P. verrucosa based on morphology from across the world and including 32 from clinical samples, 89 from the environment, and five from unknown sources were analysed (Table 1). Strains were obtained from the Research Center for Medical Mycology at Peking University from 1997 to 2014, and from the reference collection of the Centraalbureau voor Schimmelcultures Fungal Biodiversity Centre (CBS), Utrecht, The Netherlands. Phialophora americana, Capronia semiimmersa, Ca. svrcekiana, Cl. carrionii and Cl. yegresii were also included in the study.
Table 1.
List of strains analysed with isolation data.
| Species | Culture no.1 | Other reference | Source | Geography | GenBank numbers2 |
References | |
|---|---|---|---|---|---|---|---|
| ITS | BT2 | ITS | |||||
| P. americana | CBS 400.67 | Soil | Brazil | EU514695 | EU514708 | Untereiner et al. (2008) | |
| CBS 281.35 | ATCC 4806; IMI 021191; MUCL 41728; NIH 8719; UAMH 9609 | Chromoblastomycosis, verrucous | USA | EU514694 | EU514707 | Untereiner et al. (2008) | |
| NYS 323-90 | – | – | U31840 | – | Yan et al. (1995) | ||
| CBS 221.97 | CDC B-2723; IHM 1700; MUCL 40613 | – | Uruguay | U31839 | KU306350 | Yan et al. (1995) | |
| CBS 220.97 | ATCC 51962; CDC 5; MUCL 40612 | Tree | Virginia, USA | U31837 | KU306348 | Yan et al. (1995) | |
| P. americana, originally identified as Capronia semiimmersa | UAMH 10875 (T) | CDC 10; Conant 333 | Woodpulp | USA | EU514696 | EU514712 | Untereiner et al. (2008) |
| UAMH 10876 | C.J.K. Wang 1050; WUC 402 | Wood | USA | EU514697 | EU514713 | Untereiner et al. (2008) | |
| MUCL 40572 | AFTOL 658 | – | France | AF050259 | EU514703 | Untereiner & Naveau (1999), Untereiner et al. (2008) | |
| MUCL 39979 | Rotten wood | USA | AF050260 | EU514702 | Untereiner & Naveau (1999), Untereiner et al. (2008) | ||
| P. americana, originally identified as Capronia svrcekiana | UAMH 10874 | Wood | Czech Republic | EU514693 | EU514706 | Untereiner et al. (2008) | |
| UAMH 10873 | Wood | Czech Republic | EU514692 | EU514705 | Untereiner et al. (2008) | ||
| UAMH 10872 | Wood | Czech Republic | EU514691 | EU514704 | Untereiner et al. (2008) | ||
| P. americana, originally identified as P. verrucosa | BMU 01246 | CBS 140292 | Chromoblastomycosis | North China | KF881941 | KF971741 | This study |
| BMU 01244 | CBS 140291; DCU-600, ATCC 38561, IFM 4928 | Subcutaneous cyst | Japan | AB190375 | KF971743 | Iwatsu & Miyaji (1978); this study | |
| IFM 5089 | Human | Japan | AB550776 | – | Takizawa et al. (2011) | ||
| CBS 225.97 | CDC B-2152 | Keratomycosis | Texas, USA | U31847 | KU306353 | Yan et al. (1995) | |
| FMC 2214 | Human | Colombia | AF397136 | – | Heinrichs et al. (2012) | ||
| BMU 00125 | CBS 140309 | Tree bark | Jiamusi, northeast China | KF881947 | KF971748 | This study | |
| BMU 05998 | CBS 140311 | Soil of patient’s garden | Hebei, north China | KF881949 | KF971750 | This study | |
| BMU 05996 | Tree bark of patient’s garden | Hebei, north China | KF881950 | KF971751 | This study | ||
| BMU 04541 | CBS 140312 | Leaf of Changbaishan | Changchun, northeast China | KF881951 | KF971752 | This study | |
| BMU 00131 | Dead wood | Beijing, north China | KF881952 | KF971753 | This study | ||
| BMU 06000 | Soil of patient’s garden | Hebei, north China | KF881953 | KF971766 | This study | ||
| BMU 00132 | Wheat | Jiamusi, northeast China | KF881954 | KF971754 | This study | ||
| BMU 05997 | Soil of patient’s garden | Hebei, north China | KF881956 | KF971756 | This study | ||
| BMU 04493 | Soil | Changchun, northeast China | KF881958 | KF971758 | This study | ||
| BMU 04507 | Tree bark | Changchun, northeast China | KF881961 | KF971762 | This study | ||
| BMU 04522 | Leaf | Changchun, northeast China | KF881962 | KF971763 | This study | ||
| BMU 00121 | Leaf | Jiamusi, northeast China | KF881963 | KF971764 | This study | ||
| BMU 00101 | CBS 140307 | Soil | Xingjiang, northwest China | KJ700965 | KM658141 | This study | |
| BMU 00107 | Soil | Xian, northwest China | KJ700946 | KM658122 | This study | ||
| BMU 00109 | Soil | Xian, northwest China | KJ700945 | KM658121 | This study | ||
| BMU 00110 | Soil | Xian, northwest China | KJ700949 | KM658125 | This study | ||
| BMU 00111 | Soil | Xian, northwest China | KJ700951 | KM658127 | This study | ||
| BMU 00114 | Soil | Haerbin, northeast, China | KJ700956 | KM658132 | This study | ||
| BMU 00117 | Soil | Xian, northwest China | KJ700962 | KM658138 | This study | ||
| BMU 00118 | Soil | Xian, northwest China | KJ700964 | KM658140 | This study | ||
| BMU 00147 | Soil | Xian, northwest China | KJ700957 | KM658133 | This study | ||
| BMU 00170 | Soil | Beijing, north China | KJ700944 | KM658120 | This study | ||
| BMU 00206 | Soil | Beijing, north China | KJ700958 | KM658134 | This study | ||
| BMU 00432 | Soil | Jiamusi, northeast China | KJ700967 | KM658143 | This study | ||
| BMU 04506 | CBS 140329 | Soil | Changchun, northeast China | KJ700955 | KM658131 | This study | |
| BMU 04524 | Soil | Changchun, northeast China | KJ700963 | KM658139 | This study | ||
| BMU 04528 | Soil | Changchun, northeast China | KJ700950 | KM658126 | This study | ||
| BMU 04532 | Soil | Changchun, northeast China | KJ700968 | KM658144 | This study | ||
| BMU 04538 | Soil | Changchun, northeast China | KJ700954 | KM658130 | This study | ||
| BMU 04554 | Tree bark | Changchun, northeast China | KJ700959 | KM658135 | This study | ||
| BMU 07607 | Soil | Shanghai, east China | KJ700969 | KM658145 | This study | ||
| BMU 07608 | Soil | Shanghai, east China | KJ700970 | KM658146 | This study | ||
| BMU 07617 | Coal | Shanghai, east China | KJ700976 | KM658082 | This study | ||
| BMU 07625 | CBS 140305 | Leaf | Huangzhou, east China | KJ700981 | KM658086 | This study | |
| BMU 07626 | CBS 140327 | Leaf | Changsha, central China | KJ700982 | KM658089 | This study | |
| BMU 07645 | Leaf | Chongqing, southwest China | KJ700985 | KM658092 | This study | ||
| BMU 07650 | Leaf | Chongqing, southwest China | KJ700987 | KM658094 | This study | ||
| BMU 07653 | Leaf | Chongqing, southwest China | KJ700988 | KM658095 | This study | ||
| BMU 07640 | Leaf | Chongqing, southwest China | KJ700993 | KM658102 | This study | ||
| BMU 07641 | CBS 140313 | Leaf | Chongqing, southwest China | KJ700994 | KM658103 | This study | |
| BMU 07647 | Leaf | Chongqing, southwest China | KJ700996 | KM658105 | This study | ||
| BMU 07652 | Leaf | Chongqing, southwest China | KJ700997 | KM658106 | This study | ||
| BMU 07660 | Leaf | Chongqing, southwest China | KJ701000 | KM658109 | This study | ||
| BMU 07693 | Leaf | Chongqing, southwest China | KJ701005 | KM658081 | This study | ||
| BMU 07696 | CBS 140315 | Wood | Lijiang, southwest China | KJ701009 | KM658116 | This study | |
| BMU 07695 | CBS 140301 | Decaying wood | Lasa, southwest China | KJ701010 | KM658117 | This study | |
| BMU 07610 | Bamboo | Shanghai, east China | KJ701011 | KM658118 | This study | ||
| CBS 840.69 | FMR 3247; MUCL 15537; LM 342; VTT D-96477; A. Salonen No 501 | Decaying timber | Finland | AF050283 | EU514711 | Untereiner & Naveau (1999), Untereiner et al. (2008) | |
| IFM 41871 | Soil | Colombia | AB550778 | – | Takizawa et al. (2011) | ||
| gi281331169 | Japanese flounder | Japan | AB538235 | – | |||
| CBS 102234 | Decaying trunk (Gochnathia polymorpha) | Brazil | KU306358 | KU306351 | |||
| P. chinensis, originally identified as P. verrucosa | BMU 02669 | CBS 140300 | Chromoblastomycosis | Guangdong, south China | KF881930 | KF971731 | This study |
| BMU 01890 (T) | CBS 140326 | Chromoblastomycosis | Guangdong, south China | KF881964 | KF971765 | This study | |
| IFM 51934 | Human | China | AB550779 | – | Takizawa et al. (2011) | ||
| BMU 00441 | CBS 140310 | Wood | Haikou, south China | KF881948 | KF971749 | This study | |
| BMU 00127 | Tree bark | Haikou, south China | KF881955 | KF971755 | This study | ||
| BMU 00447 | CBS 140308 | Bark | Zhanjiang, south China | KF881957 | KF971757 | This study | |
| BMU 00104 | Soil | Xian, northwest China | KJ700960 | KM658136 | This study | ||
| BMU 00112 | Soil | Haerbin, northeast China | KJ700966 | KM658142 | This study | ||
| BMU 00150 | CBS 140306 | Soil | Haerbin, northeast China | KJ700947 | KM658123 | This study | |
| BMU 01057 | CBS 140328 | Soil | Xian, northwest China | KJ700953 | KM658129 | This study | |
| BMU 07609 | Wood | Shanghai, east China | KJ700971 | KM658147 | This study | ||
| BMU 07612 | Wood | Shanghai, east China | KJ700972 | KM658148 | This study | ||
| BMU 07613 | CBS 140314 | Wood | Shanghai, east China | KJ700973 | KM658149 | This study | |
| BMU 07615 | Bamboo | Shanghai, east China | KJ700974 | KM658150 | This study | ||
| BMU 07616 | Soil | Shanghai, east China | KJ700975 | KM658088 | This study | ||
| BMU 07621 | Soil | Guangzhou, south China | KJ700978 | KM658083 | This study | ||
| BMU 07622 | Soil | Guangzhou, south China | KJ700979 | KM658084 | This study | ||
| BMU 07623 | Banyan leaves | Guangzhou, south China | KJ700980 | KM658085 | This study | ||
| BMU 07642 | Leaf | Chongqing, southwest China | KJ700983 | KM658090 | This study | ||
| BMU 07643 | Leaf | Chongqing, southwest China | KJ700984 | KM658091 | This study | ||
| BMU 07649 | Leaf | Chongqing, southwest China | KJ700986 | KM658093 | This study | ||
| BMU 07654 | Leaf | Chongqing, southwest China | KJ700989 | KM658096 | This study | ||
| BMU 07627 | Soil | Nanning, south China | KJ700990 | KM658097 | This study | ||
| BMU 07629 | Soil | Nanning, south China | KJ700991 | KM658098 | This study | ||
| BMU 07636 | Dead wood | Nanning, south China | KJ701012 | KM658099 | This study | ||
| BMU 07637 | Dead wood | Nanning, south China | KJ700992 | KM658100 | This study | ||
| BMU 07661 | CBS 140304 | Wheat straw | Guangzhou, south China | KJ701013 | KM658101 | This study | |
| BMU 07646 | Leaf | Chongqing, southwest China | KJ700995 | KM658104 | This study | ||
| BMU 07656 | CBS 140303 | Leaf | Chongqing, southwest China | KJ700998 | KM658107 | This study | |
| BMU 07657 | Leaf | Chongqing, southwest China | KJ700999 | KM658108 | This study | ||
| BMU 07630 | Soil | Nanning, south China | KJ701001 | KM658110 | This study | ||
| BMU 07639 | Molded leaf | Nanning, south China | KJ701002 | KM658151 | This study | ||
| BMU 07664 | CBS 140302 | Molded leaf | Nanning, south China | KJ701003 | KM658111 | This study | |
| BMU 07692 | Leaf | Chongqing, southwest China | KJ701004 | KM658112 | This study | ||
| NH 258 | Environment | Japan | AB498920 | – | Hamada & Abe (2010) | ||
| R70D1 | Leaf of living tree | Brazil, Bahia state, Saubara, Bahia state, Brazil | KC445295 | – | Research database | ||
| WM 04.477 | Environment | – | KU306361 | – | Research database | ||
| P. ellipsoidea, originally identified as P. verrucosa | CBS 286.47 (T) | ATCC 9541; MUCL 9768; UAMH 3635 | Human | Brazil | AF050282 | EU514715 | Untereiner & Naveau (1999), Untereiner et al. (2008) |
| CBS 224.97 | NIH 8701 | Mycetoma hand | Texas, USA | U31848 | KU306354 | Yan et al. (1995) | |
| P. expanda, originally identified as P. verrucosa | BMU 01245 | CBS 140322 | Chromoblastomycosis | China | KF881934 | KF971734 | This study |
| BMU 02323 (T) | CBS 140298 | Chromoblastomycosis | China | KF881937 | KF971737 | This study | |
| P. macrospora, originally identified as P. verrucosa | BMU 07676 | CBS 140320 | Facial phaeohyphomycosis (patient 4 case 5) | Wuhan, central China | KJ701006 | KM658113 | This study; Tong et al. (2013) Wang et al. (2014), |
| BMU 07163 | CBS 140293 | Phaeohyphomycosis skin; case 2 | Hebei, north China | KF360975 | KF971725 | This study; Zhang et al. (2015) | |
| BMU 04480 | CBS 140296 | Chromoblastomycosis face | North China | KF881927 | KF971726 | This study | |
| BMU 03356 | CBS 140295 | Chromoblastomycosis hand | East China | KF881928 | KF971727 | This study | |
| BMU 03082 | CBS 140321 | Chromoblastomycosis | East China | KF881938 | KF971738 | This study | |
| BMU 00849 | CBS 140297 | Chromoblastomycosis | East China | KF881945 | KF971746 | This study | |
| BMU 07066 | CBS 140294 | Chromoblastomycosis upper limb | Tianjin, north China | KF881933 | KF971759 | This study | |
| CBS 226.97 | NYS 303A | Human, facial burn | Tennessee, USA | U31846 | KU306349 | Yan et al. (1995) | |
| CBS 273.37 (T) | ATCC 10223; MUCL 9760; | Chromoblastomycosis | Brazil | AF050281 | EU514714 | Untereiner & Naveau (1999), Untereiner et al. (2008) | |
| IHEM 5639; UAMH 3964 | |||||||
| IMTSP.800 | Human | Uruguay | AF397135 | – | Heinrichs et al. (2012) | ||
| dH 12667 | Human | Mexico | KU317088 | – | Research database | ||
| dH 12665 | Human | Mexico | KU306363 | – | Research database | ||
| BMU 00106 | Soil | Xian, northwest China | KJ700948 | KM658124 | This study | ||
| BMU 00115 | Soil | Xian, northwest China | KJ700961 | KM658137 | This study | ||
| BMU 00149 | Soil | Xian, northwest China | KJ700952 | KM658128 | This study | ||
| CBS 839.69 | ATCC 34159; MUCL 15541 | Wood | Sweden | EU514701 | EU514716 | Untereiner et al. (2008) | |
| CBS 138.67 | LCP 971; dH 15384 | – | France | KU306356 | KU306355 | This study | |
| WM 08.287 | Environment | – | KU306357 | – | Research database | ||
| LY2 | – | – | KU306359 | – | Research database | ||
| CBS 273.57 | – | – | KU306360 | – | Research database | ||
| P. tarda, originally identified as P. verrucosa | CBS 111589 (T) | Invasive Chromoblastomycosis; case 13 | Libiya | KU306362 | KU306347 | This study | |
| BMU 07506 (ET) | CBS 140325 | Phaeohyphomycosis leg | Anhui, east China | KF881960 | KF971761 | This study; Hu et al. (2011), Wang et al. (2014) | |
| BMU 07678 | CBS 140299 | Phaeohyphomycosis skin | Jinan, east China | KJ701008 | KM658115 | This study; Xu et al. (2011), Wang et al. (2014) | |
| BMU 04928 | Phaeohyphomycosis back | Hebei, north China | KF881965 | KF971730 | This study; Gao et al. (2013), Wang et al. (2014) | ||
| BMU 05960 | CBS 140323 | Phaeohyphomycosis skin | Hebei, north China | KF881935 | KF971735 | This study; Gao et al. (2013), Wang et al. (2014) | |
| BMU 07712 | CBS 140324 | Chromoblastomycosis skin | Chengdu, southwest China | KJ700942 | KM658087 | This study | |
| CBS 115956 | Chromoblastomycosis | – | KU306364 | KU306352 | This study | ||
| Cladophialophora carrionii | CBS 160.54(T) | ATCC 16264 | Chromoblastomycosis | Australia | EU137266 | EU137201 | This study; De Hoog et al. (2007) |
| CBS 117906 | UNEFM 0014-96 = dH 14504 | Chromoblastomycosis hand | Venezuela | EU137288 | EU137171 | De Hoog et al. (2007) | |
| CBS 114402 | UNEFM 9902 = dH 13271 | Chromoblastomycosis arm | Venezuela | EU137275 | EU137158 | De Hoog et al. (2007) | |
| Cladophialophora yegresii | CBS 114406 | UNEFM SgSR1 = dH 13275 | Stenocereus griseus cactus | Venezuela | EU137323 | EU137208 | De Hoog et al. (2007) |
| CBS 114405(T) | UNEFM SgSR3 = dH 13274 (ex-T of C. yegresii) | Stenocereus griseus cactus | Venezuela | EU137322 | EU137209 | De Hoog et al. (2007) | |
1 CBS: Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; ATCC: American Type Culture Collection, Virginia, USA; MUCL: Mycotheque de l’Université de Louvain, Louvain-la-Neuve, Belgium; UAMH: Microfungus Herbarium and Collection, Edmonton, Canada; NIH: National Institutes of Health, Bethesda; WC: Wadsworth Center for Laboratory and Research, New York; NYS: New York State Department of Health, New York; CDC: Centers for Disease Control and Prevention, Atlanta, USA; AFTOL: Assembling the Fungal Tree of Life; BMU; Department of Dermatology, Beijing Medical University, Beijing, China; DCU: Department of Dermatology, School of Medicine, Chiba University, Chiba, Japan; IFM: Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University, Chiba, Japan; FMR: Facultat de Medicina i Ciencies de la Salut, Reus, Spain; FMC: Faculdade de Medicina, Caracas, Venezuela; UTHSC: University of Texas Health Science Center, San Antonio, TX, USA; IMTSP: Instituto de Medicina Tropical, São Paulo, Brazil; IHEM: The BCCM/IHEM Biomedical Fungi and Yeasts Collection, Brussels, Belgium; Conant: research collection of N.F. Conant; MR: research collection of M. Réblová; WUC: research collection of W.A. Untereiner; CJK Wang: research collection of C.J.K. Wang; dH: research collection of G.S. de Hoog.
2 ITS: internal transcribed spacer; BT2: β-tubulin; TEF1: translation elongation factor 1-α.
Morphology and physiology
For microscopy, small blocks were inoculated with three-point on slants of potato dextrose agar (PDA; Difco, Detroit, USA) at 30 °C for up to 7 d until rich sporulation was obtained. Observations were done with slide cultures using corn meal agar (CMA; Difco). Agar blocks of ~ 0.5 cm2 were placed on the agar plate and inoculated at the four sides. The block was subsequently covered with a sterile cover slip (~ 2 cm2). Plates were incubated at 30 °C for 7, 14 or 21 d in a closed plastic box with sterile gauze soaked with 5 mL sterile water to ovoid drying of the culture. Slides were made by Shear’s mounting medium without pigments. Micrographs were taken using a Nikon Eclipse 80i microscope and DS Camera Head DS-Fi1/DS-5 m/DS-2Mv/DS-2MBW using NIS-Element freeware package (Nikon Europe, Badhoevedorp, The Netherlands).
Cardinal growth temperatures were determined in triplicate on 2 % malt extract agar (MEA; Difco) by measuring colony diameters for a selection of 28 strains based on phylogenetic results. Plates were incubated in the dark for 3 wk at 21, 24, 27, 30, 33, 37 and 40 °C. In order to evaluate whether 37 °C and 40 °C was fungicidal, cultures were returned to 30 °C and incubated for two additional weeks. In addition, gross morphology was observed both on MEA and OA.
DNA extraction
Genomic DNA was extracted and purified from approximately 1 cm2 of fungal elements according to the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) with disruption of cells by glass beads (425–600 μm) (Sigma-Aldrich, Zwijndrecht, The Netherlands) and TissueLyser II (Qiagen). Extraction was according to the cetyltrimethylammonium bromide (CTAB) protocol according to Feng et al. (2012).
DNA amplification and sequencing
The following nuclear genes were amplified by PCR: ITS and partial TEF1, BT2, SSU and LSU. PCR amplifications and sequencing primers are shown in Table 2. Amplifications were done by the 2×EasyTaq PCR Super Mix protocol (TransGen Biotech, Beijing, China). Fifty to 100 ng of DNA template and a 0.2–0.4 μM concentration of forward and reverse primers were added in a total volume of 25 μL. Amplification was performed in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) and included initial denaturation at 94 °C for 5 min, followed by 30 cycles consisting of denaturation at 94 °C for 30 s, annealing for 30 s at 54 °C (ITS, BT2, SSU and LSU) or 52 °C (TEF1), and extension for 30 s (ITS, BT2 and TEF1) or 1 min (SSU and LSU) at 72 °C. A final extension step of 72 °C for 10 min was included. Reading was done with Gel Doc XR+ system (Biorad, Hercules, CA, USA) with Trans2K Plus DNA Marker (TransGen Biotech) as size and concentration marker. Purification was performed with Silica Bead DNA Gel Extraction Kit (Thermo Fisher Scientific, Vilnius, Lithuania), sequencing with an ABI 3730 automatic sequencer (Applied Biosystems, Foster City, CA, USA) and sequence data were adjusted by SeqMan Pro (DNAStar, Madison, WI, USA). GenBank accession numbers are given in Table 1 except for the TEF1 region because the sequence length was less than 200 bp.
Table 2.
Primers used for PCR amplification and sequencing.
| Gene region | Primer name | Primer sequence (5′- > 3′) | Reference |
|---|---|---|---|
| ITS | V9G | 5′-TTACGTCCCTGCCCTTTGTA-3′ | De Hoog & Gerrits van den Ende (1998) |
| LS266 | 5′-GCATTCCCAAACAACTCGACTC-3′ | Masclaux et al. (1995) | |
| ITS1 | 5′-TCCGTAGGTGAACCTGCGG-3′ | White et al. (1990) | |
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ | ||
| BT2 | Bt2a | 5′-GGTAACCAAATCGGTGCTGCTTTC-3′ | O’Donnell et al. (2000) |
| Bt2b | 5′-ACCCTCAGTGTAGTGACCCTTGGC-3′ | ||
| TEF1 | EF1-728F | 5′-CATCGAGAAGTTCGAGAAGG-3′ | Carbone & Kohn (1999) |
| EF1-986R | 5′-TACTTGAAGGAACCCTTACC-3′ | ||
| LSU | NL1 | 5′-GCATATCAATAAGCGGAGGAAAAG-3′ | O’Donnell (1993) |
| LR5 | 5′-TCCTGAGGGAAACTTCG-3′ | ||
| SSU | NS1 | 5′-GTAGTCATATGCTTGTCTC -3′ | White et al. (1990) |
| NS24 | 5′-AAACCTTGTTACGACTTTTA-3′ | Gargas & Taylor (1992) |
Alignment and phylogenetic reconstruction
Sequence data were aligned with Clustal W v. 1.6. Alignments were deposited in TreeBASE (number: 19135). Phylogenetic reconstructions were done for each locus and ITS-TEF1-BT2 combined using neighbour-joining (NJ), maximum likelihood (ML) and maximum parsimony (MP) implemented in MEGA v. 6.06 (Kimura 1980, Felsenstein 1985, Saitou & Nei 1987), and MrBayes trees were done by the CIPRES portal (http://www.phylo.org/). MEGA v. 6.06 selected the K2+G model was the most appropriate model of DNA substitution for NJ and ML analysis. Support for the internodes was assessed by bootstrap analysis from 1 000 replicates. MP heuristic search was performed for each dataset with 100 random taxon additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm. Partition Homogeneity Test (PHT) based on sequences of 3 loci (ITS, TEF1 and BT2) was done by PAUP v. 4.0 b10 with 1 000 replicates for the congruence of gene genealogies. Trees were viewed and edited with MEGA v. 6.06, FigTree v. 1.4.2 and Adobe Illustrator CS6.
RESULTS
Phenotypic data
Based on morphology test results of 50 strains, we found that the conidial system of the strains identified to belong to the Phialophora verrucosa complex invariably showed limited differentiation. Phialides were inserted directly on branched hyphae or sometimes occurring as intercalary adelophialides without basal septum and continuous with the supporting hypha. Sometimes phialides were inserted on a lateral supporting cell, or were part of a poorly branched system; conidiophores were not recognized. Discrete phialides were flask-shaped to subcylindrical. Collarettes were always discernible, but varied from short frills to funnel- or vase-shaped, occasionally with a thin-walled, extremely fragile balloon-like extension. Microscopic observation indicated that conidia showing a variety of forms. Conidia were produced in slimy heads with irregular arrangement, were one-celled, and varied in shape from subspherical, tear-shaped to short-cylindrical with rounded ends. Significant variation in shape could often be noted within a single strain. Growth at different temperatures indicated an optimum at 27–30 °C (Fig. 1) for most of the strains. No growth was observed at 40 °C. The following eight isolates were unable to grow at 37 °C: P. americana BMU 01246, BMU 04506, BMU 04541, CBS 220.97 and CBS 102234; P. verrucosa BMU 07506, BMU 07678; P. tarda CBS 111589. When returned at 30 °C for 3 wk all eight isolates grew well, and all except one (CBS 840.69) isolates originated from patients.
Fig. 1.
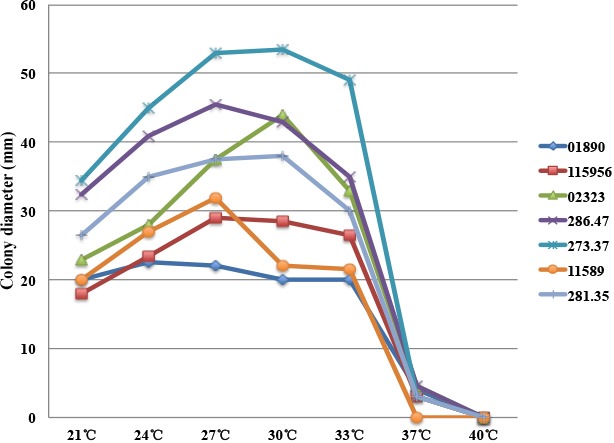
Colony diameters at various temperatures ranging from 21–40 °C, measured after 3 wk on 2 % MEA.
Molecular phylogeny
Phylogenetic reconstruction based on the ITS region and using NJ (Fig. 2), ML, MP and BI algorithms showed similar, more or less congruent topologies (data not shown), but generally with poor resolution. Three main aggregates of strains were recognizable, while seven branches were bootstrap-supported, with a single strain, CBS 111589 located in an isolated position. As a tendency, isolates from human infections were clustered. The preponderantly environmental clades also contained five clinical strains, while conversely, the mainly clinical clusters comprised four environmental isolates. Strains identified as Ca. semiimmersa (UAMH 10875, UAMH 10876, MUCL 40572 and MUCL 39979) and P. americana were preponderantly found in the environmental clades. The study set also contained the type strain of P. macrospora, CBS 273.37; it was located in a cluster that mainly contained strains from clinical samples. Strains of Ca. semiimmersa were indistinguishable from those of P. americana; a small group of strains denominated Ca. svrcekiana took an unresolved position paraphyletic to the P. americana / Ca. semiimmersa clade. Cladophialophora carrionii and Cl. yegresii, which are known to be phylogenetically close to P. verrucosa, were selected as out-groups and were clearly distinguishable by ITS (Fig. 2). Phylogenetic reconstruction based on SSU and LSU did not distinguish species of the P. verrucosa complex or related groups (data not shown).
Fig. 2.
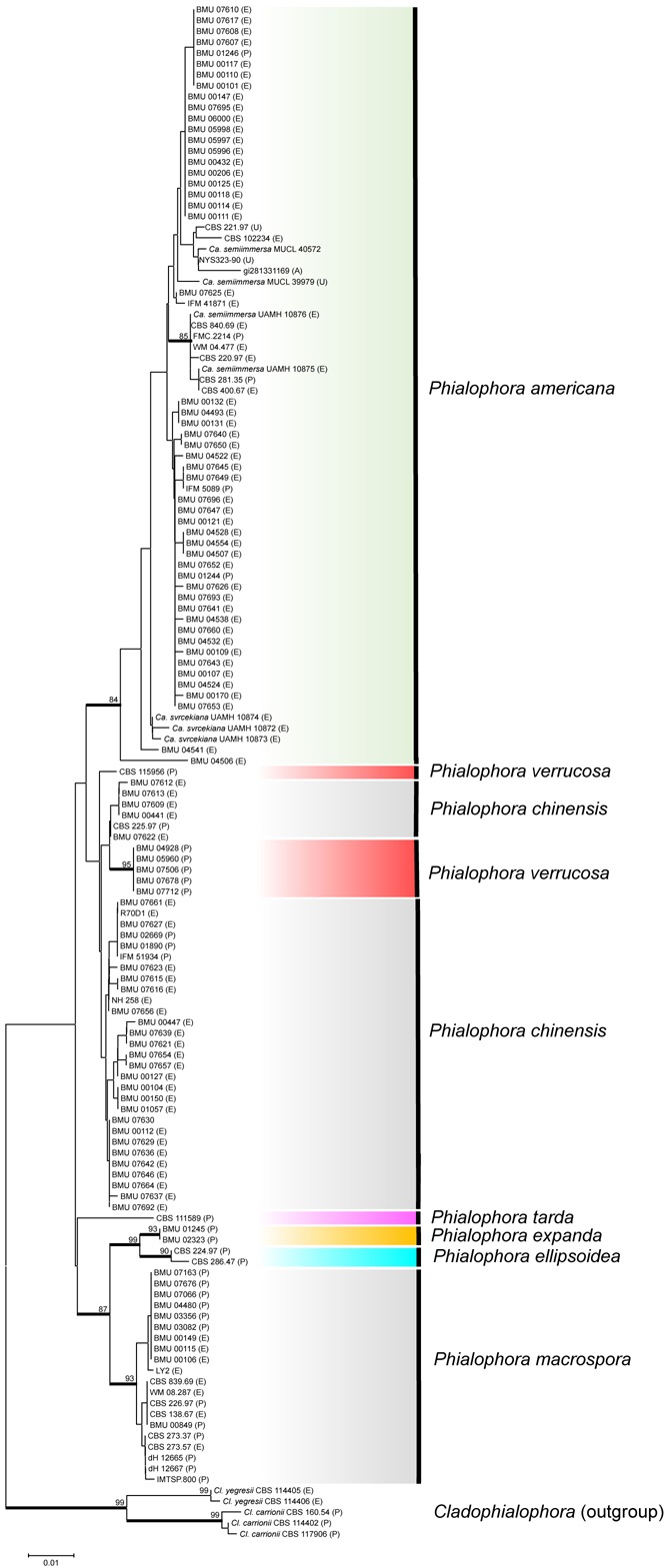
Neighbour-Joining tree obtained from the 141 ITS sequences data. Bootstrap values above 80 % are shown at the nodes. The carrionii-clade is selected as outgroup. P, E, A, U after strain number mean sources: patient, environment, animal and unknown.
To verify the ITS results and to explore a more detailed clustering, we analysed the BT2 and TEF1 regions of 118 strains phenotypically identified as P. verrucosa / P. americana, with the addition of CBS reference strains. Topologies were congruent with that of ITS, but at a higher level of resolution. Results of PHT showed that three gene lineages were congruent (P > 0.01). The tree of the combined 3-gene locus dataset (Fig. 3) revealed a topology similar to those of individual ITS, TEF1 and BT2 genes.
Fig. 3.

Maximum-Parsimony (MP) tree obtained from the combined DNA sequence data from three loci (ITS, BT2 and TEF1). Bootstrap values of Neighbour-Joining (NJ), Maximum-Likelihood (ML) and MP above 80 % / Bayesian (BS) posterior probability value above 0.80, are shown at the nodes (NJ/ML/MP/BS). Type strains and supported branches are drawn in bold. The carrionii-clade is selected as outgroup. Sources of isolation are mentioned at each strain.
The multilocus tree was used as a basis for a new taxonomic system for the P. verrucosa complex. The complex contained seven species, consistently separated with all partitions at high statistical support. Only a single cluster contained a type strain, i.e. CBS 273.37 of Phialophora macrospora. Strains generally identified and published in the literature with case reports as P. verrucosa comprised a small group of strains from five patients, two of which had been proven to have a CARD9 immunodeficiency, the two strains are BMU 07678 and BMU 07506, and this group kept the species name P. verrucosa (Gao et al. 2013, Tong et al. 2013, Wang et al. 2014, Zhang et al. 2015). A single, slow-growing isolate from a girl with a disseminated, severely mutilating chromoblastomycosis-like infection in Libya (Hofmann et al. 2005) took an isolated position in all analyses. One environmental cluster with 32 isolates contained strains collected in China from diverse environments such as soil, wood and plant debris, in addition to two isolates (BMU 01890 and BMU 02669) from human patients. Two small groups of strains with human-derived strains only were clearly separate from the main groups at high statistical support. A further, predominantly environmental group (4 clinical of 56 in total) contained strains that were identified in the literature (Untereiner & Naveau 1999) as P. americana and its sexual morph Ca. semiimmersa. For sequences deposited under the name Ca. svrcekiana no multi-locus data were available, but the position of these strains in the ITS tree, i.e. unresolved and adjacent to the P. americana group, suggested that the same taxonomic entity was concerned; for extended data see Untereiner et al. (2008).
TAXONOMY
Clade A
Phialophora verrucosa Medlar, Mycologia 7: 203. 1915 — MycoBank MBT203396; Fig. 4
Fig. 4.
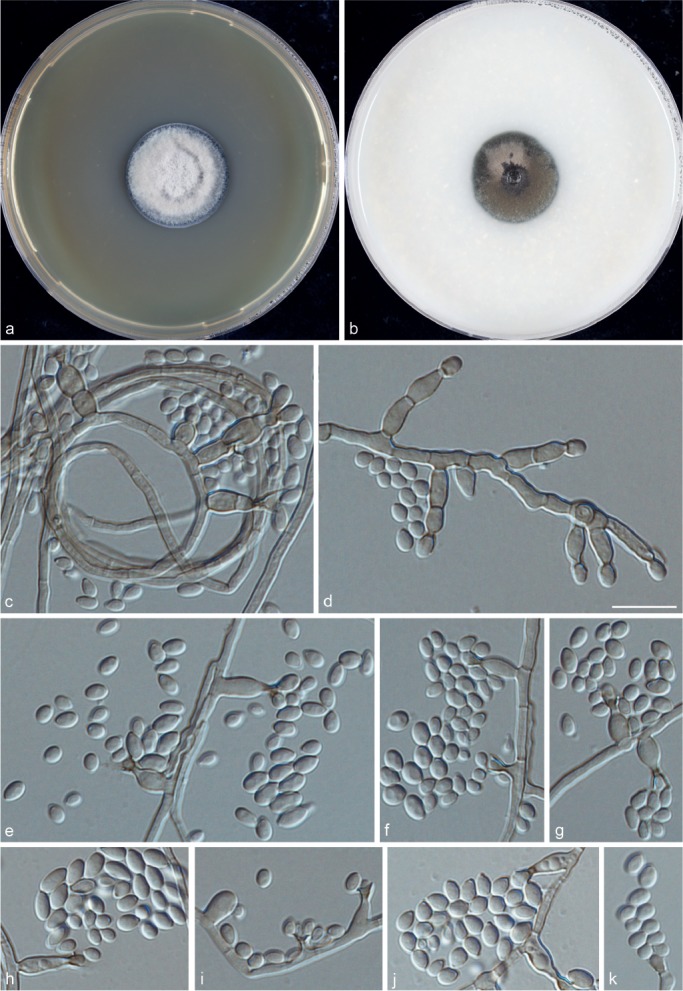
Phialophora verrucosa (CBS 140325). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–k. micromorphology showing phialides and conidia. — Scale bar = 10 μm.
Typus. Lectotype designated herewith f. 1 in Medlar (1915b: 201), an illustration of the fungus from a culture derived from a lesion in the buttock of a 22-yr-old Italian immigrant to Boston, USA. Whether original material of this strain has been preserved could not be ascertained. CHINA, from skin lesions of human disseminated phaeohyphomycosis patient with CARD9 deficiency, epitype designated here: CBS 140325 (preserved at CBS in metabolically inactive condition in liquid nitrogen). Living strain also deposited as BMU 07506.
Description of CBS 140325 after 3 wk incubation on OA, 30 °C: Colonies growing slowly, olivaceous brown, with black olivaceous in the centre and slightly pink margin. Reverse olivaceous black. On MEA, 30 °C: Colonies growing slowly, pale grey, woolly with smooth, moist margin; reverse olivaceous brown. No diffusible pigment produced. Hyphae olivaceous brown, irregularly separate, flexuous, 2.5 ± 0.5 (1.5–3.5) μm wide. Conidiophores absent. Phialides broadly flask-shaped to elongate, of variable length, often inserted on a subtending cell; adelophialides without basal septum are common. Collarettes large, funnel-shaped, sometimes small, darker brown than the supporting phialide, producing conidia in heads. Conidia hyaline, 4.5 ± 0.5 (3.0–5.5) × 2.5 ± 0.5 (2.0–3.5) μm, smooth-walled, teardrop-shaped with protruding beak on one end and remain aggregated around the phialides. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 30 °C, maximum 37 °C.
Additional material examined. Table 1.
Notes — The type isolate has been preserved at Research Center of Medical Mycology, Peking University and at CBS. Isolates belonging to this species were derived from five patients, including four from China, and two of them concerned cases of CARD9-related immunodeficiency phaeohyphomycosis reported by Wang et al. (2014).
Clade B
Phialophora chinensis Yali Li, de Hoog & R.Y. Li, sp. nov. — MycoBank MB815345; Fig. 5
Fig. 5.
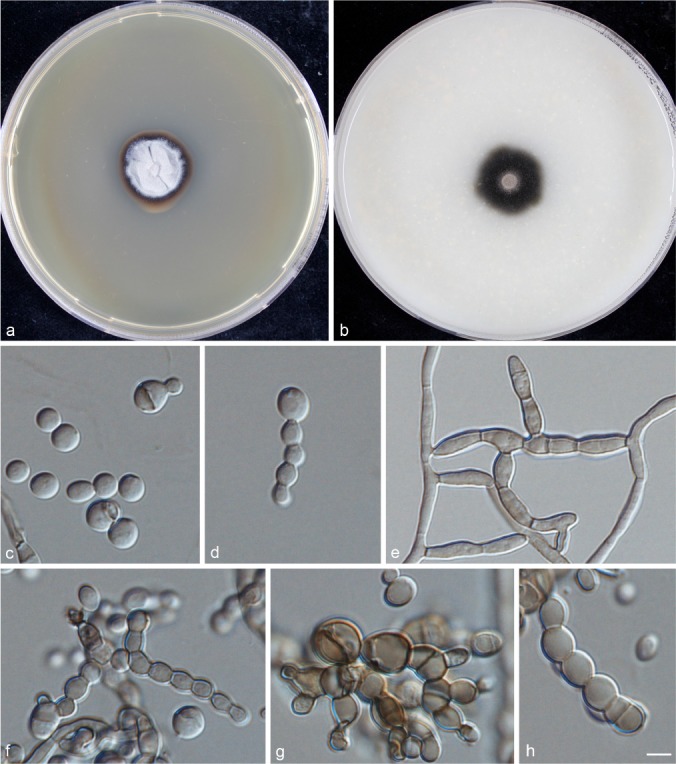
Phialophora chinensis (CBS 140326). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–h. micromorphology showing phialides, conidia, torulose hypha and muriform-like cells. — Scale bar = 10 μm.
Typus. CHINA, from skin lesions of human chromoblastomycosis patient, holotype CBS 140326 (preserved at CBS in metabolically inactive condition in liquid nitrogen). Living strain also deposited as BMU 01890.
Description of BMU 01890 after 3 wk incubation on OA, 30 °C: Colonies growing slowly, olivaceous black, with pale olivaceous centre. Reverse olivaceous black. On MEA, 30 °C: Colonies growing slowly, woolly, pale olivaceous grey with brown, smooth margin; reverse olivaceous brown. No diffusible pigment produced. Hyphae brown, regularly septate, 4.0 ± 0.5 (3.0–4.5) μm wide. Conidiophores absent. Phialides broadly flask-shaped. Conidia hyaline, smooth-walled, spherical to broadly ellipsoidal, 4.5 ± 0.5 (3.0–6.0) × 3.5 ± 0.5 (2.0–5.5) μm, some larger conidia developing a median septum resembling muriform cells, or show budding. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 24 °C, maximum 40 °C.
Additional material examined. Table 1.
Notes — The species grows with short cells producing thick-walled, swollen cells with median septa strongly resembling muriform cells on routine media. Nevertheless, nearly all strains known of P. chinensis are environmental, mostly being isolated from soil and plant debris. Two of the strains examined (Table 1) were derived from human patients, causing chromoblastomycosis.
Clade C
Phialophora americana (Nannf.) S. Hughes, Canad. J. Bot. 36: 795. 1958 — MycoBank MB203397; Fig. 6
Fig. 6.
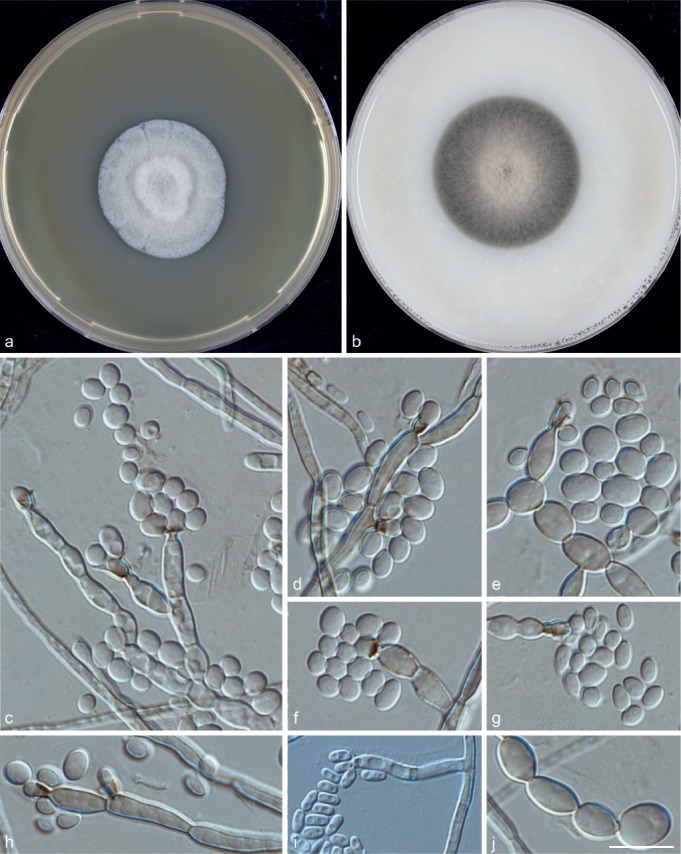
Phialophora americana (CBS 281.35). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–j. micromorphology showing phialides, conidia and torulose hypha. — Scale bar = 10 μm.
Basionym. Cadophora americana Nannf., in Melin & Nannf., Svensk Skogsvårdsförening Tidskr. 3–4: 412. 1934.
= Dictyotrichiella semiimmersa Cand. & Sulmont, Rev. Mycol. 36: 242. 1972.
≡ Capronia semiimmersa (Cand. & Sulmont) Unter. & F.A. Naveau, Mycologia 91: 73. 1999.
= Capronia svrcekiana Réblová, Czech Mycol. 49: 82. 1996.
Typus. USA, Wisconsin, woodpulp, A. Richards, holotype of P. americana slide 6320-2 (UPS). Living strain also deposited as UAMH 10875 = CDC 10.
Description of CBS 281.35 after 3 wk incubation on OA, 30 °C: Colonies growing moderately rapidly, olivaceous brown and pale at the centre. Reverse olivaceous black. On MEA, 30 °C: woolly, olivaceous grey; reverse olivaceous black. No diffusible pigment produced. Hyphae irregular, 2.5 ± 0.5 (1–3) μm wide. Distinct conidiophores absent. Phialides variable, flask-shaped to cylindrical or elongated, with darker, vase-shaped or tubular collarettes, which may also be sessile directly on undifferentiated hyphae. Conidia hyaline, 5.0 ± 0.5 (3.5–7.0) × 3.0 ± 0.5 (2–4) μm, subspherical to broadly ellipsoidal, occasionally subcylindrical, of variable size, mostly adhering in loose clumps at the collarette openings, rarely arranged in loose strings. Cardinal temperatures: minimum below 21 °C, optimum 30 °C, maximum 37 °C.
Additional material examined. Table 1.
Notes — The strain taken by several authors as representative for the species, CBS 281.35 was derived from a verrucous dermatosis of the legs of a human chromoblastomycosis patient, USA. The isolate was first described as Phialophora verrucosa by Schol-Schwarz (1970) as representative of that species, but later it was redescribed as P. americana by Yamagishi (in Yamagishi et al. 1997), Untereiner (in Untereiner et al. 2008) and Takizawa (in Takizawa et al. 2011). The species was also reported as Capronia semiimmersa from a herbarium specimen by Candousseau & Sulmont (1971). Untereiner & Naveau (1999) judged living strain MUCL 40572, parasitizing a lichen on Populus wood in France, identical to the type specimen and provided an illustration of its monomorphic Phialophora asexual morph with deep, vase-shaped phialidic collarettes. Strains UAMH 10872, 10873, 10874 are representative of Ca. svrcekiana and are also identical to P. americana in the ITS tree (Fig. 2), confirming conclusions of Untereiner et al. (2008).
Clade D
Phialophora tarda Yali Li, de Hoog & R.Y. Li, sp. nov. — MycoBank MB815349; Fig. 7
Fig. 7.
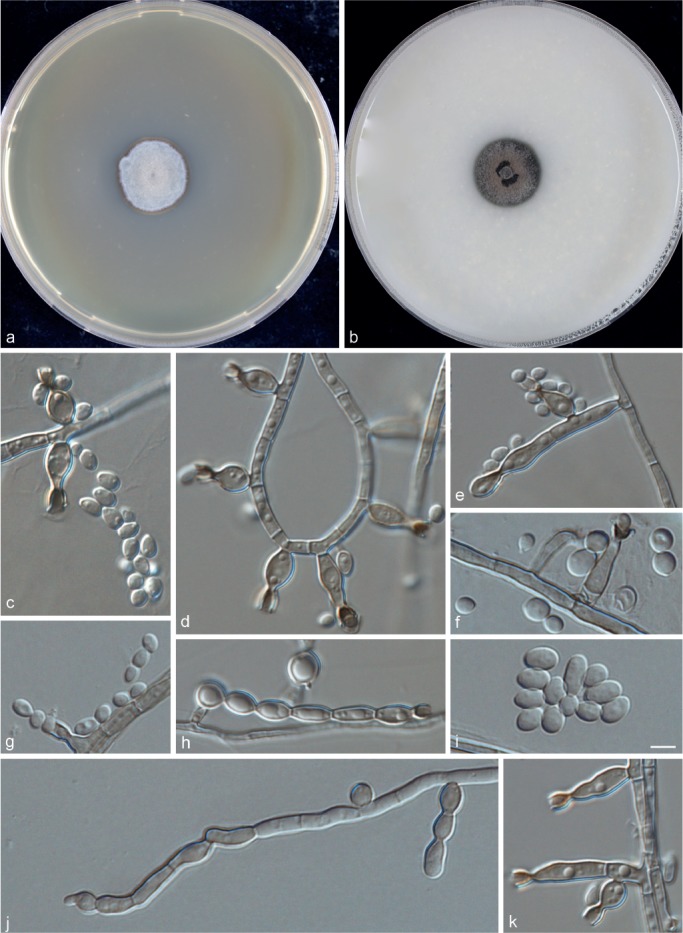
Phialophora tarda (CBS 111589). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–k. micromorphology showing phialides and conidia. — Scale bar = 10 μm.
Typus. LIBYA, from tissue of disseminated chromoblastomycosis-like infection in human patient (Hofmann et al. 2005), holotype CBS 111589 (preserved at CBS in metabolically inactive condition in liquid nitrogen).
Description of CBS 111589 after 3 wk incubation on OA, 30 °C: Colonies growing slowly, olivaceous brown, with black olivaceous centre. Reverse olivaceous black. On MEA, 30 °C: Colonies growing slowly, pale olivaceous grey, woolly, with narrow smooth margin; reverse olivaceous brown. No diffusible pigment produced. Hyphae olivaceous brown, flexuous, 2.0 ± 0.5 (1.5–2.5) μm wide. Conidiophores absent. Phialides regularly flask-shaped to elongate; adelophialides uncommon. Collarettes slightly darker than the rest of the phialide, narrow funnel-shaped to almost cylindrical, up to 5.6 μm long. Conidia hyaline, variable in shape, mostly broadly ellipsoidal, 3.5 ± 1.0 (2.0–5.5) × 2.5 ± 0.5 (1.5–3.5) μm, smooth-walled. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 27 °C, maximum 40 °C.
Notes — The species is known from a single strain causing a severely mutilating, disseminated infection in a girl from Libya, initially identified as P. verrucosa (Hofmann et al. 2005). The patient was judged to be immunocompetent, but at that time the existence of CARD9- or STAT1-based or other rare inherited genetic immune defects was not known. Muriform cells in tissue had a variable appearance without typical cruciate septation.
Clade E
Phialophora expanda Yali Li, de Hoog & R.Y. Li, sp. nov. — MycoBank MB815350; Fig. 8
Fig. 8.
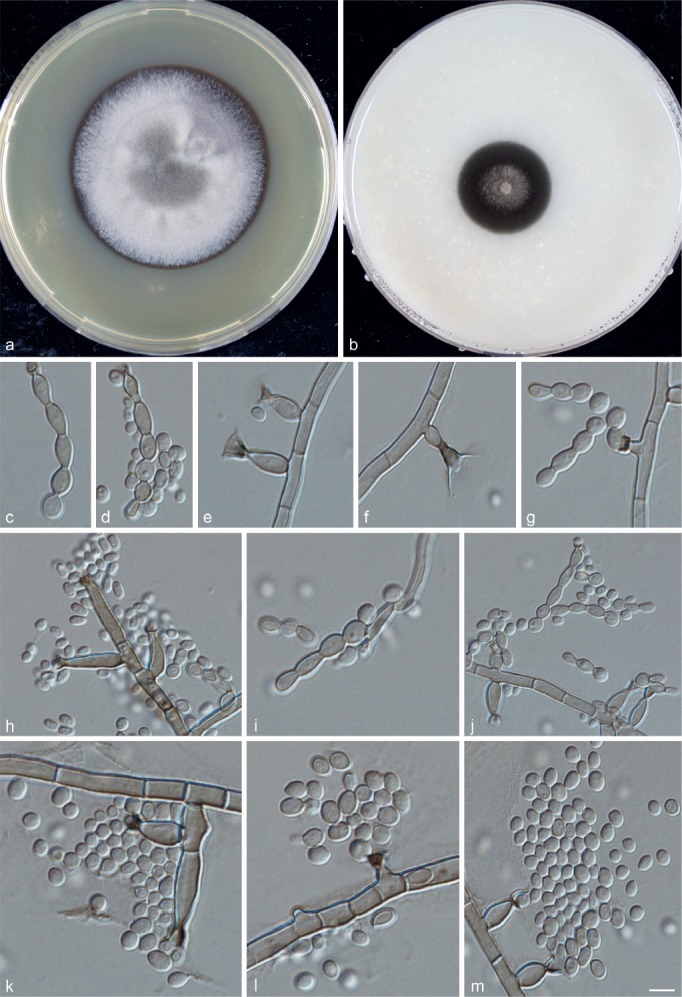
Phialophora expanda (CBS 140298). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–l. micromorphology showing phialides, conidia and torulose hypha. — Scale bar = 10 μm.
Typus. CHINA, from skin lesions of chromoblastomycosis patient, holotype CBS 140298 (preserved at CBS in metabolically inactive condition in liquid nitrogen). Also deposited as living strain CBS 140298 = BMU 02323.
Description of BMU 02323 after 3 wk incubation on OA, 30 °C: Colonies growing slowly, olivaceous black, with brown, woolly hyphae near the centre. Reverse olivaceous black. On MEA, 30 °C: Colonies growing moderately rapidly, woolly, pale olivaceous grey, with smooth margin; reverse olivaceous brown. No diffusible pigment produced. Hyphae olivaceous brown, often emerging from torulose hyphae and flexuous, 2.0 ± 0.5 (2.0–3.5) μm wide. Conidiophores absent or poorly differentiated. Most phialides flask-shaped to elongate, narrowed towards the tip; short adelophialides without basal septa frequently present. Collarettes darker than the rest of the phialide, funnel-shaped to almost cylindrical, often with a large, less intensely pigmented and very fragile apical portion, which is widely open. Conidia hyaline, ellipsoidal, 3.5 ± 0.5 (2.0–5.0) × 2.5 ± 0.5 (1.5–3.5) μm, smooth-walled, occasionally budding, aggregated in heads, sometimes in short chains. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 30 °C, maximum 40 °C.
Additional material examined. Table 1.
Notes — This isolate was collected by Peking University First Hospital from a chromoblastomycosis patient in 2000. It always clustered with the isolate BMU 01245 that was collected in 1999 from another patient.
Clade F
Phialophora ellipsoidea Yali Li, de Hoog & R.Y. Li, sp. nov. — MycoBank MB815351; Fig. 9
Fig. 9.
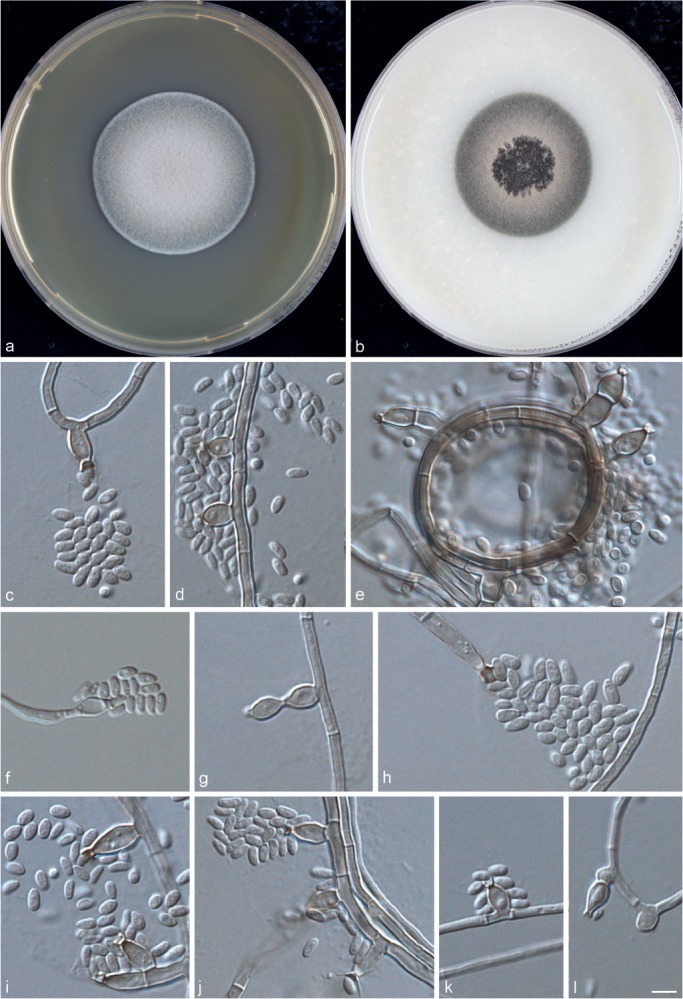
Phialophora ellipsoidea (CBS 286.47). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–k. micromorphology showing phialides and conidia. — Scale bar = 10 μm.
Typus. BRAZIL, from human patient, holotype CBS 286.47 (preserved at CBS in metabolically inactive condition in liquid nitrogen). Living strain CBS 286.47 = ATCC 9541 = MUCL 9768 = UAMH 3635.
Description of CBS 286.47 after 3 wk incubation on OA, 30 °C: Colonies growing moderately rapidly, olivaceous brown, with black and purple granules at the centre. Reverse olivaceous black. On MEA, 30 °C: woolly, olivaceous grey; reverse olivaceous brown. No diffusible pigment produced. Hyphae pigmented with slightly brown, separate uniform with 2 ± 0.5 (1.5–2.5) μm wide. Distinct conidiophores absent. Part of the phialides flask-shaped, later enlarge to become subellipsoidal; some of the phialides give rise to a second phialide. Collarettes mostly small, sometimes longer, 1.5 ± 0.5 (0.5–2.0) μm. Conidia hyaline, ellipsoidal, 3.0 ± 0.5 (2.0–4.5) × 1.5 ± 0.5 (1.5–2.0) μm. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 27 °C, maximum 37 °C.
Additional material examined. Table 1.
Notes — This isolate had been identified as P. verrucosa all the time. Now, according to the ITS, BT2 and TEF1 gene analyses, it always clustered together with CBS 224.97 with high support value, and they are from human patients.
Clade G
Phialophora macrospora M. Moore & F.P. Almeida, Ann. Mo. Bot. Gdn 23: 545. 1936. — MycoBank MB270192; Fig. 10
Fig. 10.
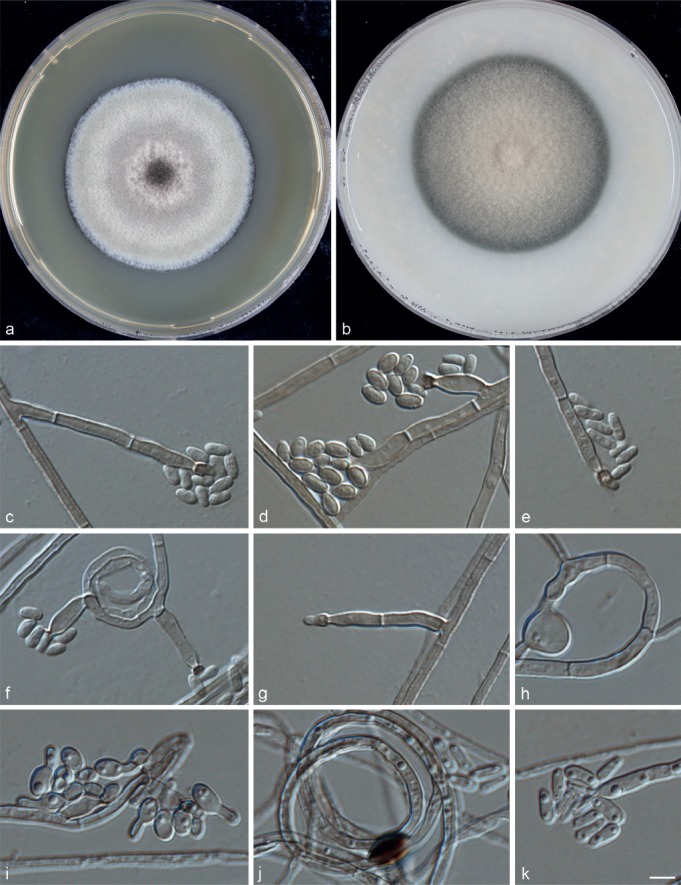
Phialophora macrospora (CBS 273.37). a. Colonies grown on MEA for 3 wk; b. colonies grown on OA for 3 wk; c–k. micromorphology showing phialides and conidia. — Scale bar = 10 μm.
Typus. BRAZIL, São Paulo, from human chromoblastomycosis-like infection, M. Moore, holotype CBS 273.37 = ATCC 10223 = MUCL 9760.
= Fonsecaea pedrosoi (Brumpt) Negroni var. phialophora Carrión, Mycologia 34: 432. 1942.
Description of CBS 273.37 after 3 wk incubation on OA, 30 °C: Colonies growing rapidly, olivaceous brown, pale at the centre. Reverse olivaceous black. On MEA, 30 °C: Colonies growing rapidly, pale grey, brown at the centre, woolly with smooth, moist margin; reverse olivaceous brown. No diffusible pigment produced. Hyphae brownish, regularly septate, 2.0 ± 0.3 (1.5–2.5) μm wide, flexuous. Phialides inserted directly on hyphae, flask-shaped. Collarettes small and short, vase- to funnel-shaped, part of them darker than the rest of the phialide. Conidia hyaline, sometimes showing some budding, 4.0 ± 0.5 (3.0–5.5) × 2.0 ± 0.5 (1.5–2.5) μm. Sexual morph unknown. Cardinal temperatures: minimum below 21 °C, optimum 30 °C, maximum 40 °C.
Notes — This isolate had been identified as P. verrucosa (Untereiner & Naveau 1999, Untereiner et al. 2008, Heinrichs et al. 2012), or P. americana (Yamagishi et al. 1997).
DISCUSSION
The present study aims to investigate the biodiversity and taxonomy of Phialophora verrucosa, which has been reported in older literature as one of the uncommon agents of human chromoblastomycosis (Guerriero et al. 1998). However, also other types of infection have been ascribed to this species, among which are mycetoma (Turiansky et al. 1995), disseminated (Hofmann et al. 2005, Tong et al. 2013) and particularly different kinds of subcutaneous infection, often with cystic encapsulation (Iwatsu & Miyaji 1978, Schnadig et al. 1986, Kimura et al. 2003). Most infections were noted in patients with apparently good health; the share of immunocompromised patients, such as transplant recipients (Lundstrom et al. 1997), those with AIDS (Duggan et al. 1995) or with chronic use of antibiotics (Hochfelder & Fetto 2013) are relatively limited. In addition to human infection, the species has also been isolated from the environment, by enrichment in a mammal vector (Gezuele et al. 1972) but also with methods that are standard for direct black yeast isolation (Iwatsu et al. 1981). The majority of these isolates have, however, not been preserved, and their identity thus can no longer be verified.
Our data provide evidence that separate species are concerned, with different predilection and possibly causing different disorders. The combined ITS-TEF1-BT2 tree showed seven clades, six of which were supported by high bootstrap values and the seventh took an isolated position in all partitions. It was concluded that seven putative phylogenetic species exist in the P. verrucosa complex. Most of the recognised phylogenetic species exhibited a high degree of origin specificity, with species significantly differing in apparent pathogenicity. Two of the main environmental clusters contained 6.3–7.1 % strains from human patients, whereas in the combined ‘pathogenic’ clusters this ratio was 83.3 %.
Species identification for black yeasts in general (Zeng et al. 2013) including members of the genus Phialophora s.str. (Chowdhary et al. 2014) is possible by ITS sequencing. Our study shows the taxonomy of P. verrucosa in more detail. The phylogenetic trees of both ITS and BT2 distinguish the P. verrucosa complex unambiguously from its close the relatives Cl. carrioni and Cl. yegresii. Within the complex, rDNA ITS provides insufficient resolution in that the seven species-clusters have statistical support due to strains in paraphyletic position. Nevertheless, characteristic ITS-profiles are recognizable for each species, so that ITS can be used as barcode for routine identification (Schoch et al. 2012).
Only a single sexual morph of P. americana, Ca. semiimmersa, is known in the P. verrucosa complex (Untereiner et al. 2008). Sexual connections (Ca. semiimmersa including Ca. svrcekiana) were made by isolation of ascospores from natural samples, and sequences of cultures invariably clustered in P. americana (Untereiner et al. 2008, Réblová 1996). Phialophora americana is a preponderantly environmental species and is predicted to have low human pathogenicity judging from isolation sources.
Pathogenicity and virulence is known to differ significantly between closely related species of black fungi (Chowdhary et al. 2014). Virulence factors listed thus far include melanin and carotene, thick cell walls, muriform cells, yeast-like phases, thermo- and perhaps also osmotolerance, adhesion, hydrophobicity, aromatic hydrocarbon assimilation, and production of siderophores, factors exerting variable influence upon location and severity of the infection (Seyedmousavi et al. 2014). These are general factors attributed to the entire family Herpotrichiellaceae; significant differences between species as yet have not been found. It remains difficult to explain why closely related species, as in P. verrucosa and its allies, differ significantly in this respect, while on the other hand agents of a highly specific disease as chromoblastomycosis are scattered over the family. Infections caused by members of the P. verrucosa complex can be destructive and highly refractory to therapy. Clinical isolates collected in the course of our study mostly were derived from patients with chromoblastomycosis or phaeohyphomycosis, while treatment outcomes of those patients were quite different (Tong et al. 2013, Wang et al. 2014). Remarkably, two patients were ultimately proven to have a mutation in the CARD9 signalling pathway interfering with Dectin-1 immunity. Phialophora verrucosa isolates caused recalcitrant infections, and a species named P. tarda was collected from an invasive disseminated mycosis in Libya (Hofmann et al. 2005) in a patient that may also have had a CARD9 immune defect. It is not understood why such patients acquire just a single mycotic infection, and why black fungi are relatively frequent in these hosts. Infections by P. americana and P. chinensis are environmental fungi with opportunistic behaviour after local trauma.
Isolates used in this study had been recovered from diverse environmental sources across the world such as plant debris, soil and rotten wood. These environmental isolates tended to aggregate in a limited number of clusters, different from the subgroups with preponderantly human sources of isolation according to the phylogenetic trees. The overabundance of Chinese strains probably is a sampling effect; we expect that all environmental species have a global distribution. The most enigmatic species in the complex is P. tarda, originating from a severe human infection and without known environmental source. Notably, despite extensive environmental sampling, P. verrucosa (s.str.), P. expanda and P. ellipsoidea were not encountered either.
CONCLUSIONS
Distinction of six clades described here and summarised in Fig. 1 was achieved with molecular characters, phenotype and ecology. Optimum temperatures differ between strains and are therefore compared below at an average of 27 °C after 3 wk. Phialophora chinensis (B), nearly exclusively derived from environmental sources, in culture nevertheless has a strong tendency to production of isodiametric cells resembling muriform cells of chromoblastomycosis, and shows some yeast-like cells; hyphae are scant and conidiophores are absent. Growth is moderately slow (19–42 mm). Phialophora verrucosa s.str. (A) contains clinical strains only. Phialides have a wide base and a dark, funnel-shaped collarette. Growth 22–31 mm. Phialophora tarda (D) is known only from a moderately slow-growing (32 mm) clinical strain with well-differentiated, flask-shaped phialides. Phialophora expanda (E), with slow or fast-growing colonies (15–44 mm), has more slender phialides and very dark collarettes which have a huge expansion when young. Phialophora macrospora (G) has expanding, woolly colonies; phialides are slender, nearly cylindrical, with ellipsoidal conidia. Phialophora ellipsoidea (F), known from two clinical cases grows moderately rapidly (22–45 mm), has flask-shaped phialides but with short collarettes. Phialophora americana (C) is an environmental species with moderate growth (26–37 mm), differentiated conidiogenous cells with dark, funnel- to vase-shaped collarettes and broadly ellipsoidal conidia. Several species need further study when additional material becomes available.
Acknowledgments
We express our gratitude to all colleagues who provided us with fungal strains. This work was supported by the grants from the National Natural Science Foundation and the Ministry of Science and Technology of China (81520108026 and 2013ZX10004612-002). We declare that we have no relevant conflicts of interest.
REFERENCES
- Attili-Angelis D, Duarte APM, Pagnocca FC, et al. 2014. Novel Phialophora species from leaf-cutting ants (tribe Attini). Fungal Diversity 65: 65–75. [Google Scholar]
- Badali H, Prenafeta-Boldú FX, Guarro J, et al. 2011. Cladophialophora psammophila, a novel species of Chaetothyriales with a potential use in the bioremediation of volatile aromatic hydrocarbons. Fungal Biology 115: 1019–1029. [DOI] [PubMed] [Google Scholar]
- Candoussau F, Sulmont P. 1971. Dictyotrichiella semiimmersa nov. sp. Revue de Mycologie 36: 238–242. [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Caretta G, Tosi S, Piontelli E, et al. 2006. Phialophora sessilis, a lithobiont fungus. Mycotaxon 95: 281–284. [Google Scholar]
- Chowdhary A, Perfect J, De Hoog GS. 2014. Black molds and melanized yeasts pathogenic to humans. Cold Spring Harbor Perspectives in Medicine 5: a019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Wingfield MJ, et al. 2012. Fungal Planet description sheets: 128–153: Phialophora livistonae Crous & Summerell, sp. nov. Fungal Planet 138. Persoonia 29: 146–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Gerrits van den Ende AHG. 1998. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183–189. [DOI] [PubMed] [Google Scholar]
- De Hoog GS, Nishikaku AS, Fernandez-Zeppenfeldt G, et al. 2007. Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Studies in Mycology 58: 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Vicente VA, Najafzadeh MJ, et al. 2011. Waterborne Exophiala species causing disease in cold-blooded animals. Persoonia 27: 46–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Weenink XO, Gerrits van den Ende AHG. 1999. Taxonomy of the Phialophora verrucosa complex with the description of two new species. Studies in Mycology 43: 107–122. [Google Scholar]
- Duggan JM, Wolf MD, Kauffman CA. 1995. Phialophora verrucosa infection in an AIDS patient. Mycoses 38: 215–218. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Feng PY, Lu QY, Najafzadeh MJ, et al. 2012. Cyphellophora and its relatives in Phialophora: biodiversity and possible role in human infection. Fungal Diversity 65: 17–45. [Google Scholar]
- Gams W. 2000. Phialophora and some similar morphologically little-differentiated anamorphs of divergent ascomycetes. Studies in Mycology 45: 187–199. [Google Scholar]
- Gao LJ, Yu J, Wang DL, et al. 2013. Recalcitrant primary subcutaneous phaeohyphomycosis due to Phialophora verrucosa. Mycopathologia 175: 165–170. [DOI] [PubMed] [Google Scholar]
- Gargas A, Taylor JW. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenised fungi. Mycologia 84: 589–592. [Google Scholar]
- Gezuele E, Mackinnon JE, Conti-Diaz IA. 1972. The frequent isolation of Phialophora verrucosa and Phialophora pedrosoi from natural sources. Sabouraudia 10: 266–273. [PubMed] [Google Scholar]
- Guerriero C, De Simone C, Tulli A. 1998. A case of chromoblastomycosis due to Phialophora verrucosa responding to treatment with fluconazole. European Journal of Dermatology 8: 167–168. [PubMed] [Google Scholar]
- Hamada N, Abe N. 2010. Growth characteristics of four fungal species in bathrooms. Biocontrol Science 15: 111–115. [DOI] [PubMed] [Google Scholar]
- Heinrichs G, De Hoog GS, Haase G. 2012. Barcode identifiers as a practical tool for reliable species assignment of medically important black yeast species. Journal of Clinical Microbiology 50: 3023–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochfelder J, Fetto J. 2013. Phialophora verrucosa as a cause of deep infection following total knee arthroplasty. American Journal of Orthopedics 42: 515–518. [PubMed] [Google Scholar]
- Hofmann H, Choi SM, Wilsmann-Theis D, et al. 2005. Phialophora verrucosa causing invasive chromoblastomycosis and sinusitis in a child from northern Africa. Mycoses 48: 456–461. [DOI] [PubMed] [Google Scholar]
- Hu SQ, Li XF, Lv GX, et al. 2011. A case report of facial phaeohyphomycosis caused by Phialophora verrucosa. Chinese Journal of Dermatology 44: 564–566. [In Chinese.] [Google Scholar]
- Iwatsu T, Miyaji M. 1978. Subcutaneous cystic granuloma caused by Phialophora verrucosa. Mycopathologia 64: 165–168. [DOI] [PubMed] [Google Scholar]
- Iwatsu T, Miyaji M, Okamoto S. 1981. Isolation of Phialophora verrucosa and Fonsecaea pedrosoi from nature in Japan. Mycopathologia 75: 149–158. [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kimura M, Goto A, Furuta T, et al. 2003. Multifocal subcutaneous phaeohyphomycosis caused by Phialophora verrucosa. Archives of Pathology & Laboratory Medicine 127: 91–93. [DOI] [PubMed] [Google Scholar]
- Lane C. 1915. A cutaneous lesion caused by a new fungus (Phialophora verrucosa). Journal of Cutanous Disease 33: 840–846. [Google Scholar]
- Lopez Martinez R, Mendez Tovar LJ. 2007. Chromoblastomycosis. Clinical Dermatology 25: 188–194. [DOI] [PubMed] [Google Scholar]
- Lundstrom TS, Fairfax MR, Dugan MC, et al. 1997. Phialophora verrucosa infection in a BMT patient. Bone Marrow Transplantation 20: 789–791. [DOI] [PubMed] [Google Scholar]
- Marimón R, Cano J, Gené J, et al. 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. Journal of Clinical Microbiology 45: 3198–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimón R, Gené J, Cano J, et al. 2006. Molecular phylogeny of Sporothrix schenckii. Journal of Clinical Microbiology 44: 3251–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux F, Guého E, De Hoog GS, et al. 1995. Phylogenetic relationships of human-pathogenic Cladosporium (Xylohypha) species inferred from partial LS rRNA sequences. Journal of Medical and Veterinary Mycology 33: 327–338. [DOI] [PubMed] [Google Scholar]
- McGinnis MR. 1983. Chromoblastomycosis and phaeohyphomycosis: new concepts, diagnosis, and mycology. Journal of the American Academy of Dermatology 8: 1–16. [DOI] [PubMed] [Google Scholar]
- Medlar EM. 1915a. A cutaneous infection caused by a new fungus, Phialophora verrucosa, with a study of the fungus. The Journal of Medical Research 32: 507–522. [PMC free article] [PubMed] [Google Scholar]
- Medlar EM. 1915b. A new fungus, Phialophora verrucosa, pathogenic for man. Mycologia 7: 200–203. [Google Scholar]
- O’Donnell K. 1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, Wallingford. [Google Scholar]
- O’Donnell K, Nirenberg H, Aoki T, et al. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78. [Google Scholar]
- Réblová M. 1996. Two new Capronia species from the Czech Republic. Czech Mycology 49: 77–83. [Google Scholar]
- Réblová M, Untereiner WA, Réblová K. 2013. Novel evolutionary lineages revealed in the Chaetothyriales (fungi) based on multigene phylogenetic analyses and comparison of its secondary structure. PLoS One 8, 5: e63547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Saunte DM, Tarazooie B, Arendrup MC, et al. 2012. Black yeast-like fungi in skin and nail: it probably matters. Mycoses 55: 161–167. [DOI] [PubMed] [Google Scholar]
- Schnadig VJ, Long EG, Washington JM, et al. 1986. Phialophora verrucosa-induced subcutaneous phaeohyphomycosis. Fine needle aspiration findings. Acta Cytologica 30: 425–429. [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences of the USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schol-Schwarz MB. 1970. Revision of the genus Phialophora (Moniliales). Persoonia 6: 59–94. [Google Scholar]
- Seyedmousavi S, Netea MG, Mouton JW, et al. 2014. Black yeasts and their filamentous relatives: principles of pathogenesis and host defence. Clinical Microbiology Reviews 27: 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa K, Hashizume T, Kamei K. 2011. Occurrence and characteristics of group 1 introns found at three different positions within the 28S ribosomal RNA gene of the dematiaceous Phialophora verrucosa: phylogenetic and secondary structural implications. BMC Microbiology 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZS, Chen SC, Chen L, et al. 2013. Generalized subcutaneous phaeohyphomycosis caused by Phialophora verrucosa: report of a case and review of literature. Mycopathologia 175: 301–306. [DOI] [PubMed] [Google Scholar]
- Turiansky GW, Benson PM, Sperling LC, et al. 1995. Phialophora verrucosa: a new cause of mycetoma. Journal of the American Academy of Dermatology 32: 311–315. [DOI] [PubMed] [Google Scholar]
- Untereiner WA, Angus A, Réblová M, et al. 2008. Systematics of the Phialophora verrucosa complex: new insights from analyses of β-tubulin, large subunit nuclear rDNA and ITS sequences. Botany 86: 742–750. [Google Scholar]
- Untereiner WA, Naveau FA. 1999. Molecular systematics of the Herpotrichiellaceae with an assessment of the phylogenetic positions of Exophiala dermatitidis and Phialophora americana. Mycologia 91: 67–83. [Google Scholar]
- Wang XW, Wang WY, Lin ZM, et al. 2014. CARD9 mutations linked to subcutaneous phaeohyphomycosis and TH17 cell deficiencies. Journal of Allergy and Clinical Immunology 133: 905–908. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR Protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California. [Google Scholar]
- Xu YH, Li CY, Zhao J, et al. 2011. A case of phaeohyphomycosis caused by Phialophora verrucosa. Chinese Journal of Dermatology 44: 809–811. [In Chinese.] [Google Scholar]
- Yamagishi Y, Kawasaki K, Ishizaki H. 1997. Mitochondrial DNA analysis of Phialophora verrucosa. Mycoses 40: 329–334. [DOI] [PubMed] [Google Scholar]
- Yan ZH, Rogers SO, Wang CJK. 1995. Assessment of Phialophora species based on ribosomal DNA internal transcribed spacers and morphology. Mycologia 87: 72–83. [Google Scholar]
- Zeng JS, De Hoog GS. 2008. Exophiala spinifera and its allies: diagnostics from morphology to DNA barcoding. Medical Mycology 46: 193–208. [DOI] [PubMed] [Google Scholar]
- Zeng JS, Feng PY, Gerrits van den Ende AHG, et al. 2013. Multilocus analysis of the Exophiala jeanselmei clade containing black yeasts involved in opportunistic disease in humans. Fungal Diversity 65: 3–16. [Google Scholar]
- Zhang Y, Wang XW, Li RY, et al. 2015. Facial subcutaneous phaeohyphomycosis caused by Phialophora verrucosa: successful treatment with itraconazole and local resection. Medical Mycology Case Reports 2: e000010. [Google Scholar]
- Zhuang JL, Zhu MQ, Zhang R, et al. 2010. Phialophora sessilis, a species causing flyspeck signs on bamboo in China. Mycotaxon 113: 405–413. [Google Scholar]


