Abstract
NOTCH signaling exerts essential roles in normal and malignant intestinal physiology and the homeostasis of cancer stem-like cells (CSC), but the basis for this latter role remains obscure. The signaling scaffold protein STRAP is upregulated in several cancers where it promotes tumorigenicity and metastasis. Here we report a novel oncogenic function for STRAP in maintaining CSC subpopulations in a heterogeneous mixture by antagonizing formation of the chromatin modifier PRC2 and by epigenetically activating NOTCH signals in human colorectal cancer (CRC). Silencing STRAP sensitized CRC cells to chemotherapeutic drugs in vitro and in vivo. STRAP depletion also contributed to a reduced stem-like phenotype of CRC cells, as indicated by reduced expression of the CSC signature and NOTCH signaling regulators in vitro and by diminished tumorigenesis in vivo. Genes encoding some upstream activators of NOTCH were highly enriched for H3K27me3, which form repressive chromatin domains upon STRAP silencing. Mechanistically, STRAP competitively disrupted association of the PRC2 subunits EZH2 and SUZ12, thereby inhibiting PRC2 assembly. Restoring the NOTCH pathway by lentiviral expression of NICD1 or HES1 in STRAP-depleted tumor cells reversed the CSC phenotype. In 90 CRC clinical specimens, a significant positive correlation was documented between the expression of STRAP and HES1. Overall, our findings illuminated a novel STRAP-NOTCH1-HES1 molecular axis as a CSC regulator in CRC, with potential implications to improve treatment of this disease.
Keywords: colorectal cancer, cancer stem-like cells, NOTCH signaling, epigenetic regulation, PRC2 complex
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in the United States (1). The 5-year relative survival rate is only 8% in patients with advanced CRC despite the development of treatment regimens (2). One of the major problems hindering the advancement of CRC treatment is the presence of cancer stem-like cells (CSCs), which are responsible for tumor initiation, growth, and metastasis (3). CRC CSCs show enhanced resistance to chemotherapy, a slow rate of cycling (4), and the capability of self-renewal, suggesting their potential role in drug resistance and tumor recurrence after initial response to chemotherapy. Therefore, therapies targeting the population of CRC CSCs can likely improve the therapeutic effectiveness of treatment (5–6).
Current findings support that NOTCH signaling plays a crucial role in the maintenance of CSCs found in solid tumors. Up-regulation of NOTCH1, JAG ligands and HES1 in CRC cell lines results in the increased expression of the EMT/stemness-associated proteins: CD44, SLUG, and SMAD3 (7,8). High expression of HES1 is significantly correlated with distal metastasis at diagnosis, and unfavorable prognostic outcome for CRC patients (9). Activation of NOTCH signaling also protects CSCs from apoptosis (10). HES1 induces stem-like properties in colon cancer cells by up-regulating stem cell markers at the transcriptional level (11). However, the upstream regulators of NOTCH signaling and the mechanism of their function remain unexplored.
The serine-threonine kinase receptor associated protein (STRAP) is highly expressed in several human cancers (12–14). STRAP consists of seven WD40 domains to serve as a scaffold protein (15). STRAP overexpression inhibits the anti-tumor effects of TGF-beta (16). Additionally, STRAP physically interacts with PDK1 and positively modulates its downstream targets such as PKB/Akt and Bad in vivo (17). Our recent studies showed that STRAP transcriptionally downregulates E-cadherin and p21Cip1 expressions (18), and it promotes the mesenchymal phenotype (19).
In this study, we demonstrate that STRAP epigenetically regulates the NOTCH pathway and maintains stem-like properties of CRC cells. Mechanistically, STRAP disassembles the PRC2 complex by disrupting the interaction between SUZ12 and EZH2, resulting in the activation of NOTCH signaling via epigenetic modification. Importantly, clinical data show that the expression of STRAP is significantly correlated with HES1 expression. Together, our results provide a novel mechanism for the regulation of CSCs via STRAP/NOTCH1/HES1 regulatory axis.
MATERIALS AND METHODS
Cell culture and reagents
Human colon adenocarcinoma cell lines: HCT116, LoVo, SW620, DLD-1, WiDr, HT29 and RKO, and HEK-293T cells were purchased from the American Type Culture Collection (Manassas, VA). Cell lines were cultured in McCoy’s 5A medium with 10% of FBS; and HEK-293T cells were maintained in DMEM medium with 10% of FBS. 5-FU (F6627) and Oxaliplatin (O9512) were obtained from Sigma Aldrich.
Adenoviral and lentiviral transduction
STRAP-expressing adenovirus was used as we described previously (13). Lentivirus constructs containing NICD1 and HES1 were obtained from Dr. Matthew Walters (Weill Cornell Medical College) and Dr. Linzhao Chang (Johns Hopkins University), respectively. STRAP shRNA lentivirus was produced as described (13). Lentivirus shRNA targeting SUZ12 was purchased from Sigma Aldrich.
AOM/DSS treatment of Strap+/− mice
Heterozygous knockout mice were generated using ZFN technology (20). Excision of exon 3 and 4 of the Strap gene in one allele knocks out the expression of the protein. Strap+/− mice were crossbred with wild-type mice in C57BL/6 background to achieve germline transmission. Details of AOM and DSS treatment are described in the corresponding figure legend and Supplementary material. All animal studies have been conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) at University of Alabama at Birmingham.
MTT assay, colony formation, and sphere formation assays
Cells were seeded in a 96-well plate and treated with the indicated compounds for 72 h. Cell viability was assessed using MTT assay (Millipore). For colony formation assay, cells were suspended in agarose containing 10% FBS medium and then plated on top of semi-solid agarose in 35-mm plates. Colonies were counted as described (13). For sphere formation assay, cell suspension in a serum-free conditioned medium was plated into an ultralow attachment 96-well plate. The conditioned medium contained DMEM/F-12 (1:1 ratio) supplemented with B27 supplement, N2 supplement, EGF, basic FGF, and insulin.
Drug resistance assay
Six to eight week-old male nude mice were injected subcutaneously with stable shCtrl or shSTRAP#1 clones from HCT116 or DLD-1 cells. When tumors reached a size of about 100 mm3, the mice (8 mice/group) were treated intraperitoneally with 5-FU (50 mg/kg) or Oxaliplatin (0.15mg/kg, LC Laboratories, # O-7111) 2 times/week for four consecutive weeks. Tumor volumes at indicated time points after treatment were calculated and plotted as we described (13).
Flow cytometric analysis
Apoptosis was evaluated using Annexin V-FITC and a PI staining kit (BD Transduction Labs). Flow cytometric analyses were used to detect CD133+/CD44+ cells. CD133/2(293C3)-PE (#130-090-853) and CD44-APC (#130-098-110) antibodies (Miltenyi Biotec Inc.) were utilized to label cells.
Luciferase assays
pHES1-luc and pHES5-luc were obtained from Addgene (#43806 and #26869, respectively ). All wells were also transfected with 25 ng of β-galactosidase (β-gal) as an internal control. Ratios of luciferase to β-gal readings were applied to plot the graph from triplicates values.
Immunofluorescence and immunohistochemical analyses
Cells were grown in chamber slides, fixed, and permeabilized, and then utilized to perform immunofluorescent staining with a rabbit anti-Sox2 antibody (#3579) and rabbit anti-Nanog antibody (#4903) (Cell Signaling Technology) overnight at 4°C, followed by goat anti-rabbit Alexa Fluor®488 antibody (A-11070, Life Technologies). Fluorescent cells were visualized and digital images were captured using an Olympus microscope.
For immunohistochemical (IHC) analyses, paraffin-embedded tissues were incubated with the indicated antibodies. The proportion score represents the estimated fraction of stained cells (0 = 0%, 1 = 1%–24%, 2 = 25%–49%, 3 = 50%–74%, and 4 = 75%–100%), while the intensity score represents their average staining intensity (0=no staining, 1=weak staining, 2=moderate staining, and 3=strong staining). The final staining score was determined by multiplying the intensity score by the proportion score. As a result, scoring was between 0 and 12.
qPCR and ChIP assays
qPCR analysis was performed as described previously (18). Purification of sonicated nuclear lysates and immunoprecipitation were performed using an EZ-ChIP assay kit (Upstate Biotechnology). The DNA samples recovered from the ChIP were analyzed by quantitative PCR using specific primers (Supplementary Table S1). Primers targeting 84 genes key to NOTCH signaling pathway were provided by the EpiTect ChIP qPCR Array kit (QIAGEN).
Co-immunoprecipitation (co-IP) and western blot
Co-IP and western blot assays were performed as described previously (18). Primary antibodies included: CD133 (18470-1-AP) (Proteintech Group); ABCG2 (sc-25822), HA (sc-805) (Santa Cruz Biotechnology); CD44 (#558739) and STRAP (#611346) (BD Transduction Labs); Cleaved Caspase 3 (#9661), Cleaved NOTCH1 (#4147), DLL1 (#2588), DLL4 (#2589), NUMB (#2756), TACE (#6978), JAG1 (#2620), JAG2 (#2210), HES1 (#11988), OCT4 (#2840), SNAIL (#3879), SLUG (#9585) and BMI1 (#5856) (Cell Signaling Technology); Tri-Methyl-Histone H3 (Lys27) (ab192985), SUZ12 (ab12073), EZH2 (ab3748), EED (ab169647) (Abcam); and β-actin (A5316) and Flag (F3165) (Sigma). More details are in Supplementary Information.
Statistical analyses
All values in the figures and text were derived from at least 3 independent experiments and expressed as means ±SD. Statistical analyses were completed using the SPSS statistical software package (SPSS/PC+, SPSS Inc.). Significant differences in mean values were evaluated by the student’s t-test. A two-sided P <0.05 was considered statistically significant.
Results
Reduced expression of STRAP enhances drug-induced apoptosis and sensitizes colorectal cancer cells to chemotherapy
Prompted by our previous studies pointing to a potential oncogenic role of STRAP (13,14), we hypothesized that STRAP may play an important role in drug responses in CRC. MTT assays were performed to evaluate the effect of 5-FU or Oxaliplatin on the survival of human CRC lines, HCT116 and DLD-1 at 72h post-treatment. We first knocked down (KD) STRAP expression with two independent STRAP shRNAs (shSTRAP#1 and shSTRAP#2) in these cell lines (Supplementary Fig. S1A). The shSTRAP#1 clone displayed more sensitivity to 5-FU and Oxaliplatin, indicated by the lower IC50 values as compared to those of the shCtrl clone (Supplementary Fig. S1B and C). After treatment, the percentage of apoptotic cells in STRAP-depleted HCT116 clone was dramatically increased in response to 5-FU or Oxaliplatin (10.0% to 23% or 20.5% to 32.2.5%, respectively) as compared to the shCtrl clone (Fig. 1A, upper panel). An increase in apoptosis was also observed in STRAP knockdown DLD-1 cells relative to control cells (8.5% to 18.3% with 5-FU; 14.4% to 25.8% with Oxaliplatin) (Fig. 1A, bottom panel). Interestingly, we did not observe much effect of STRAP knockdown in both cell lines without drug treatment. In an attempt to understand the mechanism, we observed an increase in cleaved caspase-3 activity in HCT116 and DLD-1 STRAP-depleted cells (Fig. 1B) with the same treatment. Taken together, these data suggest that loss of STRAP enhances 5-FU and Oxaliplatin-induced apoptosis and sensitizes CRC cells to these drugs.
Figure 1. STRAP knockdown increases the sensitivity of colorectal cancer (CRC) cells to drug treatment.
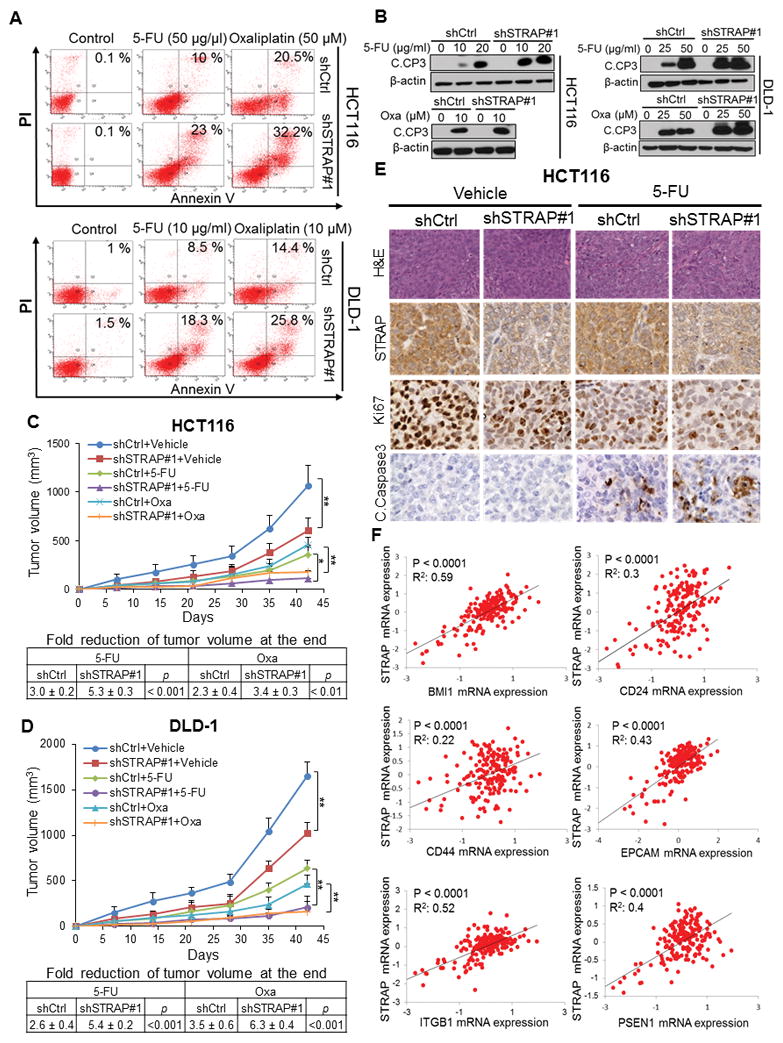
A. shSTRAP#1 and control clones were treated with 5-Fluorouracil (5-FU) or Oxaliplatin for 24 hours as indicated. The percentage of cells entering apoptosis was determined by flow cytometry using FITC-labeled annexin-V staining. B. Whole-cell lysates from control and STRAP knockdown clones treated with 5-FU or Oxaliplatin for 48 hours were subjected to western blot analyses using anti-cleaved Caspase-3 (C. CP3) antibody with β-actin as loading control. C and D. HCT116 and DLD-1 clones (STRAP-KD and control) were implanted subcutaneously into the flank regions of nude mice. When the tumors reached 100 mm3, drug treatment was initiated. Each group consisted of eight animals. After 4 weeks, mice were euthanized, tumor volumes were measured, and growth curves were plotted. The fold changes at the bottom represent the mean change in final tumor volume of the untreated group divided by the mean tumor volume of the treated group in the same category of animals. E. Representative Hematoxylin-Eosin (HE) stained images are shown. The expression of STRAP, Ki67 and cleaved Caspase-3 in the tumor from HCT116 clones was detected by IHC (magnification: X40). F. Scatter plots of mRNA expressions (log2) of STRAP versus mRNA expression levels of BMI1, CD24, CD44, EPCAM, ITGB1, and PSEN1 in colon adenocarcinoma (n=193) from TCGA dataset were plotted. P values and R2 are displayed.
To investigate the effect of downregulation of STRAP on drug sensitivity in vivo, we treated mice bearing subcutaneous tumors generated from shCtrl and shSTRAP#1 clonal cells with 5-FU or Oxaliplatin. Knockdown of STRAP alone had a significantly slower rate of growth of HCT116-derived tumors as compared to control (Fig. 1C). Mice bearing tumors from STRAP knockdown HCT116 clone showed significant sensitivity to 5-FU or Oxaliplatin when compared with those from shCtrl clone (Fig. 1C). Similarly, we observed that knockdown of STRAP in DLD-1 cells induced sensitization to 5-FU or Oxaliplatin treatment compared to the corresponding control clone (Fig. 1D). Apoptosis analyzed by active caspase-3 staining was significantly increased in 5-FU treated shSTRAP#1 tumors relative to its parallel group (Fig. 1E and Supplementary Figure S1D). In contrast, less staining of the proliferation marker Ki67 was observed in both untreated and treated shSTRAP#1 tumors relative to their corresponding controls (Fig. 1E and Supplementary Figure S1D). These results provide evidence that reduced expression of STRAP potentially promotes the sensitivity of CRC cells to 5-FU or Oxaliplatin-based chemotherapy.
Due to potential contributions of CSCs in drug resistance, we reasoned that STRAP may be involved in drug resistance through regulating CSCs. To assess this, we first analyzed the expressions of STRAP and CSC markers/regulators in 193 patient samples using TCGA mRNA-seq colon adenocarcinoma dataset. Linear regression analyses showed that STRAP expression is significantly positively correlated with the mRNA expressions of BMI1, CD24, CD44, EPCAM, ITGB1 (CD29), and PSEN1 (P<0.0001, Fig. 1F). These genes have been proposed for the identification of colorectal cancer stem-like cells (21). The upregulation of these markers is significantly correlated with invasion and metastases, and they are associated with poor prognosis (22–24). These analyses in conjunction with our above observations suggest that STRAP might play an important role in drug responses through regulating stemness of human CRC tumors.
STRAP is important in maintaining the homeostasis of colorectal cancer stem-like cell pool
To explore the potential effect of STRAP on CSCs, flow cytometry was employed to estimate the percentage of cells co-expressing CD44 and CD133 in six CRC cell lines. Expression of STRAP in each cell line after its knockdown was detected by western blot assays (Supplementary Fig. S1E). Despite differences in the genetic profiles of the cell lines, significant reduced expression of CD44/CD133 was observed in all shSTRAP clones (shSTRAP#1 and #2) compared to their control counterparts (Fig. 2A, Supplementary Fig.S2A). These data suggest that loss of STRAP could partly diminish the CD44+/CD133+ subpopulation in the heterogenous mixture of cells. The next set of experiments carried out were tumor sphere assays to determine whether STRAP might regulate the self-renewal behavior of CSCs. The number of spheres in STRAP KD HCT116 and DLD-1 cells was found to be >2-fold less than those formed in the control cells (P<0.01, Fig. 2B and C). Furthermore, the average diameter of spheres obtained from STRAP KD cells was much less than that of control cells. It is worth noting that downregulation of STRAP almost eliminated the spheres with size >200um in both cell lines, indicating that STRAP participates into the proliferation of CSCs. Colony formation assays showed that KD cells have less proliferation potency in the anchorage-independent condition (Fig. 2D). To further ascertain that STRAP could functionally mediate the stemness of CSCs, we increased the levels of STRAP by infecting cells with adenovirus-containing STRAP. Following the overexpression in two cell lines, there was a marked increase in sphere number and size, compared to the corresponding β-gal control cells (P<0.01, Fig. 2E).
Figure 2. STRAP maintains stem-like phenotype of CRC cells in vitro and in vivo.
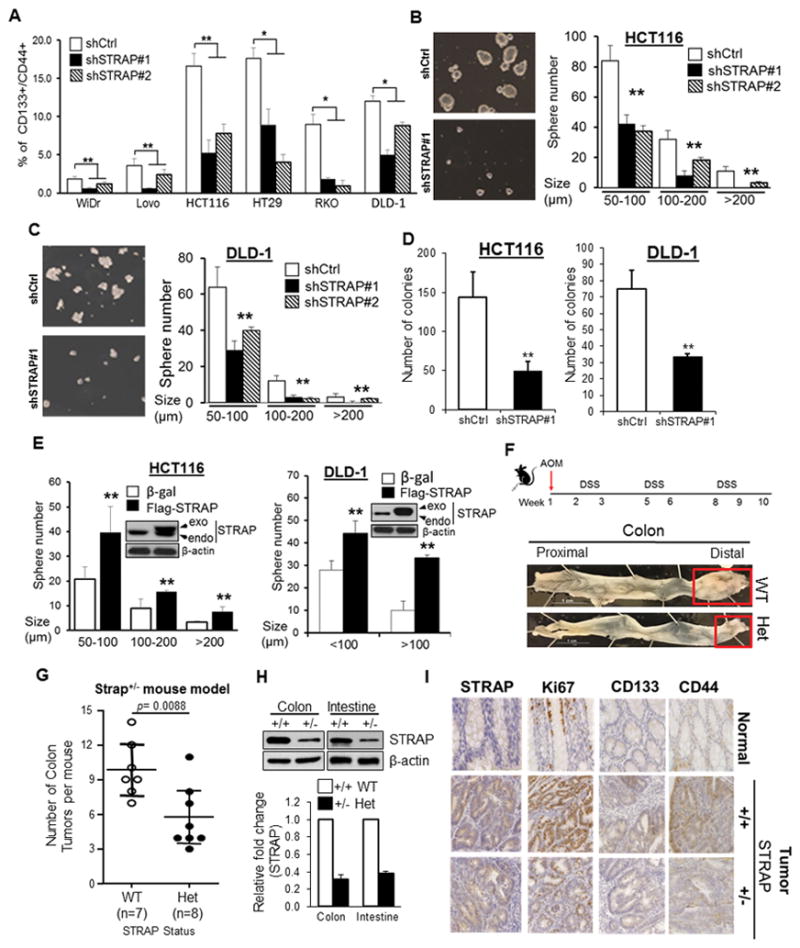
A. The fraction of CD133+/CD44+ cells in stable STRAP knockdown (KD) CRC clones (WiDR, LoVo, HCT116, HT-29, RKO and DLD-1) was analyzed by flow cytometry and plotted as % positive cells. Significance levels were determined by Student’s t test. n=3, *P<0.05, **P< 0.01, when compared with the control. B and C. Sphere-shaped cells were generated from HCT116 shSTRAP#1 and corresponding shCtrl cells after three days cultured in sphere-conditioned media (left panel). The number and size of spheres derived from STRAP KD cells were compared to those of control cells (right panel). Similar experiment was performed in STRAP KD DLD-1 cells. n=3, **P< 0.01 vs shCtrl. D. Colony formation assays were performed with HCT116 and DLD-1 cells. The number of colonies larger than 200 μm was counted, **P< 0.01, compared to control. E. The number and size of spheres derived from shSTRAP#1 HCT116 and DLD-1 clones after infection with Flag-tagged STRAP adenovirus were compared with those of β-gal adenovirus-infected cells. Endogenous and exogenous protein levels of STRAP were detected by western blot analyses, **P< 0.01 vs β-gal control. F. Schematic diagram of AOM and DSS administration to induce colon cancer development in mice (both wild type and Strap heterozygous mice) is presented. AOM (10 mg/kg) was injected on day 0. At the beginning of the second week, 2.5% DSS solution was administered to mice in their drinking water. Seven days of DSS treatment was followed by two weeks of water. Two more cycles of DSS and water were administered prior to sacrifice by the end of the tenth week (upper panel). As expected, the majority of tumors are located in the distal region (as indicated, bottom panel). G. Representative distribution of the number of tumors per mouse treated with AOM/DSS. Significance levels are determined by independent Student’s t-test. H. Expression of STRAP in colon and intestine tissues from indicated mice was examined by western blotting. β-actin was used as loading control (upper). Relative fold change of STRAP was determined by densitometry using ImageJ software (bottom). I. Immunohistochemical staining for the proliferation marker Ki67, CD133 and CD44 as well as for STRAP was analyzed in colon using tissue slides from AOM/DSS mouse model. Pictures are representative of both normal tissue and tumor segments from Strap+/+ and Strap+/− mice. Magnification, X40.
To evaluate the effect of STRAP on tumor stem-like cells in vivo, we employed Strap+/− mouse model and induced tumorigenicity in the colon with the carcinogen, azoxymethane (AOM) and with the inflammatory agent, dextran sodium sulphate (DSS). Heterozygous deletion of Strap markedly inhibited tumor growth in mice (Fig. 2F and G). We routinely observed a decrease in STRAP expression in the intestine and colon of heterozygous mice by 60–70% (Fig. 2H). As reported (25), histologically a majority of carcinomas were observed and no metastatic lesion was evident after 10 weeks of AOM/DSS treatment (Supplementary Fig. 2B). Immunohistochemical staining showed that nuclear staining of Ki67, a proliferative marker (26), was significantly decreased in colon tumors of Strap+/− mice compared to those in the wild type animals, indicating STRAP deficiency inhibits colon carcinogenesis in mice (Fig. 2I). Interestingly, we observed less expression of CD44 and CD133 in tumors generated from Strap+/− mice (Fig. 2I), further supporting our observations in human cell lines. Therefore, these findings suggest that STRAP is involved in CSC homeostasis and loss of STRAP results in lower proportion of CSCs.
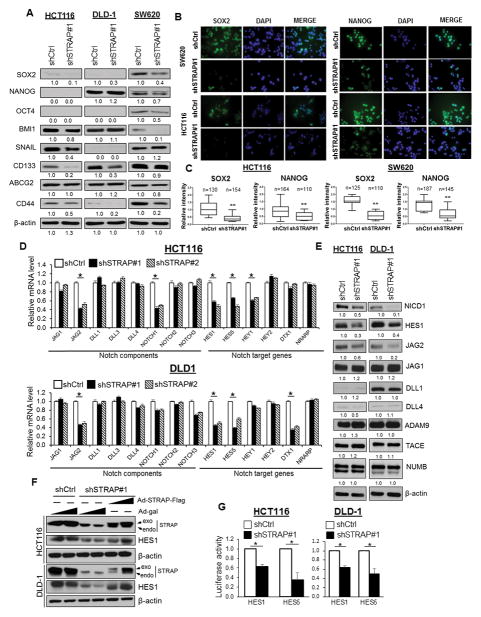
STRAP regulates the expression of a specific subset of NOTCH signaling effectors
To understand the mechanism of STRAP-induced regulation of cancer cell stemness, we assessed the expression of diverse sets of genes: common stemness regulators such as OCT4, SOX2, NANOG, and BMI1; Zinc-finger transcription factors including SNAIL, SLUG and TWIST; and CSC surface markers like CD44, CD133 and ABCG2. qPCR data revealed that SOX2, OCT4, BMI1, CD133, and CD44 were significantly reduced in different cell types after STRAP KD (Supplementary Fig S3A), suggesting the deregulation of stemness-related genes by STRAP in a context-dependent manner. Western blot assays confirmed that downregulation of STRAP was able to reduce the expression of the above markers in a cell-type specific manner (Fig. 3A). Immunofluorescence staining showed lower expression of SOX2 and NANOG in STRAP KD cells when compared to control cells (Fig. 3B and C). It has been reported that activated NOTCH signaling promotes the self-renewal capacity of colorectal CSCs (10). Here, we hypothesize that STRAP might participate in the activation of NOTCH signaling and modulates the molecular signature of CRC stem-like cells. To this end, we first detected a series of genes by qPCR, including four NOTCH receptors (NOTCH 1–4) and five ligands (DLL-1, -3, -4, JAG-1 and -2), as well as their well-documented target genes (HES1/5, HEY1/2, NRARP, and DTX) (27–29). mRNA levels of JAG2, HES1 and HES5 were significantly downregulated in both KD clones as compared to their control counterparts (Fig. 3D). In addition, shSTRAP HCT116 clones showed reduced NOTCH1 and HEY1, and DLD-1 clones showed reduced DTX1 expression (Fig. 3D). Notably, NOTCH1 intracellular domain (NICD1), HES1 and JAG2 proteins were lower in shSTRAP clones as compared to the corresponding control cells (Fig. 3E and Supplementary Fig. S3B). On the contrary, there were no changes in the expressions of NOTCH ligands (DLL1/4) and NOTCH upstream regulators (ADAM9, TACE and NUMB) in either cell line (Fig. 3E), indicating the specific effect of STRAP on selective regulation of NOTCH pathway components.
Figure 3. STRAP regulates the expression of CSC markers and NOTCH signaling genes.
A. Western blot was used to determine the expression of protein levels for indicated CSC markers in control and STRAP knockdown clones. B. Immunofluorescence staining of SOX2- and NANOG-positive cells in representative clones (Green). Cell nuclei were stained using DAPI stain (blue). C. Comparison of the relative expression (intensity) of SOX2 and NANOG from above staining in indicated KD cells and their control cells has been shown as box plots. D. Levels of mRNA in stable shSTRAP#1, shSTRAP#2, and shCtrl clones (HCT116 and DLD-1) as indicated were determined by qPCR and shown as the relative fold changes using GAPDH as a loading control. Significance levels were determined by Student’s t test. n=3, *P<0.05, when compared with the control. E. Protein expression of NOTCH components in shSTRAP#1 and shCtrl from HCT116 and DLD-1 cell lines was analyzed by western blot using antibodies against the corresponding proteins as indicated. F. Expression of HES1 was rescued by Flag-tagged STRAP adenovirus in shSTRAP#1 clone in a dose-dependent manner with β-gal adenovirus as a negative control. Endogenous and exogenous protein levels of STRAP were detected. G. Luciferase activities from HES1 and HES5 promoter reporters in STRAP knockdown and control cells normalized to β-gal activities were plotted. n=3, *P<0.05, when compared to the control.
To determine the specificity of the effect of STRAP on expression of endogenous HES1, an important transcriptional regulator of NOTCH pathway, shSTRAP#1 cells were infected with adenovirus-STRAP. HES1 protein level was reduced by knockdown of STRAP, which is reversed by re-expression of adenoviral STRAP in both cell lines (Fig. 3F). Next, we investigated whether intrinsic NOTCH signaling could be inhibited through the loss of endogenous STRAP expression. Luciferase assays showed the inhibition of HES1 and HES5 promoter activity mediated by reduced level of STRAP (Fig. 3G). These results suggest STRAP involvement in the control of stemness regulators/markers in CRC cells, likely through activation of NOTCH pathway.
STRAP helps to maintain the patterns of histone modification signature on NOTCH genes
Cumulating evidence suggests that histone modifications play a pivotal role in regulating transcriptional activity of NOTCH-related genes (30). We performed chromatin immunoprecipitation (ChIP) with anti-Histone H3K4me3 and H3K27me3 antibodies followed by qPCR analyses using shCtrl and shSTRAP#1 clones in HCT116 cells. The histone modification patterns for a set of genes involved in NOTCH signaling are represented as a heat map (Fig. 4A). We obtained 29 genes with low levels of H3K4me3 and 54 genes with H3K27me3 enrichment at the proximal promoter region (Fig. 4B). Bivalent domains bound by both H3K4me3 and H3K27me3 are found in some transcriptionally repressive genes (31). Therefore, we chose a total of 22 genes featured by increased H3K27me3 and reduced H3K4me3 levels with significant p-values (Supplementary Table 2), and we performed ChIP-qPCR (Supplementary Fig. 4). Upon depletion of STRAP, the transcripts of 14 and 11 genes were downregulated in HCT116 and DLD-1 cells, respectively (Supplementary Table 3). Collectively, these results indicate that the epigenetic status of NOTCH effectors and targets could be impaired by downregulation of STRAP.
Figure 4. Loss of STRAP leads to epigenetic alterations in the promoter regions of NOTCH genes, and STRAP impairs the association between EZH2 and SUZ12 by interacting with SUZ12.
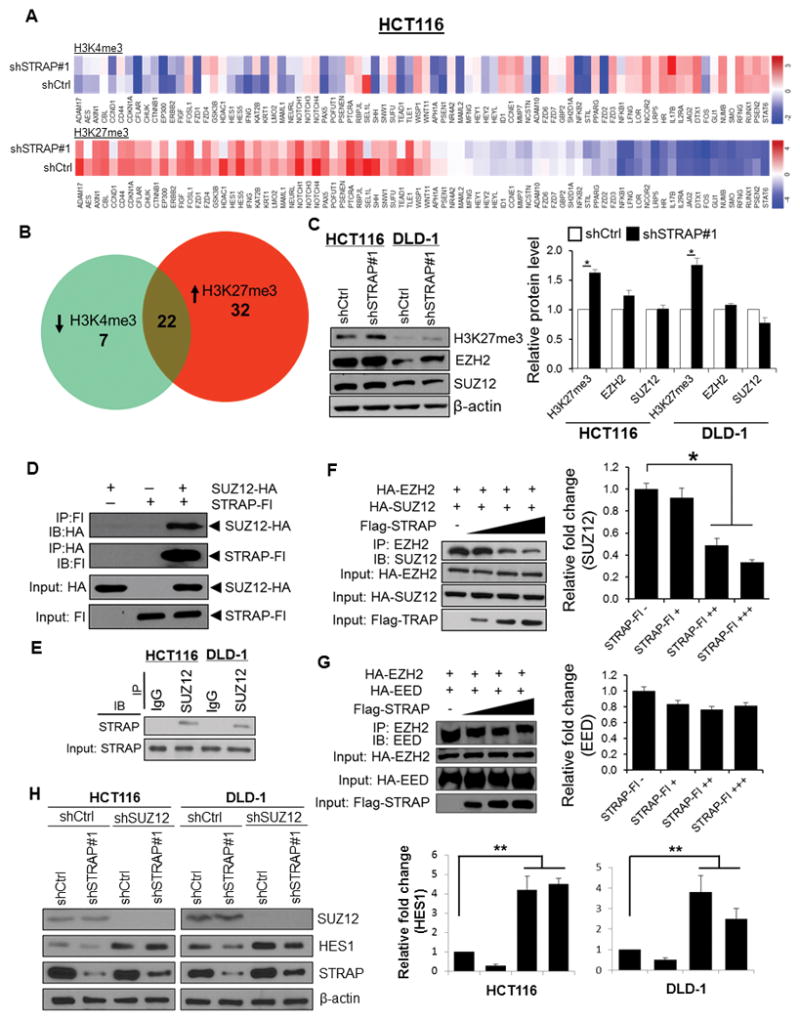
A. ChIP panel assays were performed to identify ChIP-enriched genomic DNA using antibodies against active chromatin marker H3K4me3 and repressive marker H3K27me3. Specifically designed qPCR primers focused on 1 kb genomic regions of individual promoter of 86 NOTCH signaling genes were used. Color bars in the heat maps indicate the altered fold from detected targets in shCtrl versus shSTRAP#1 HCT116 cells. B. Venn diagrams show the overlap of 22 genes with both low abundance of H3K4me3 (29 genes) and high abundance of H3K27me3 (54 genes) presented in KD cells relative to shCtrl parallel. C. Total cell lysates were analyzed for H3K27me3, EZH2, SUZ12, and β-actin expression by immunoblotting with their respective antibodies (left panel). Relative expressions of H3K27me3, EZH2 and SUZ12 normalized to β-actin were determined by densitometry using ImageJ software (right panel), *P<0.05, shSTRAP#1 vs. shCtrl. D. 293T cells were co-transfected with HA-tagged SUZ12 and Flag-tagged STRAP as indicated. Lysates were subjected to reciprocal immunoprecipitation with either anti-HA or anti-Flag antibody and analyzed by western blotting. Protein expression was also tested by immunoblotting. E. SUZ12 was immunoprecipitated with anti-SUZ12 antibody from lysates of indicated cell lines. Immune complexes were then analyzed by immunoblotting with anti-STRAP antibody. F and G. 293T cells were co-transfected with HA-tagged constructs of either SUZ12 or EED together with EZH2 as well as increased doses of Flag-tagged STRAP plasmid. Cell lysates were used for immunoprecipitation with the anti-EZH2 antibody. Co-precipitated SUZ12 (F) or EED (G) was detected by immunoblotting with the corresponding antibody (left panel). Protein expression was assessed by western blot. Relative fold changes of bound to SUZ12 (F) or EED (G) with increasing amounts of STRAP plasmid was determined by densitometry using ImageJ software (right panel). *P<0.05, when compared with no STRAP control. H. The indicated cells (shCtrl or shSTRAP#1) were infected with lentiviral shSUZ12RNA and scrambled shRNA. Lysates were subjected to western blotting with anti-SUZ12, anti-HES1, and anti-STRAP antibodies (left panel). β-actin was used as a loading control. Relative fold change of HES1 in each cell line was determined by densitometry using ImageJ software (right panel). **P<0.001, when compared with corresponding control.
STRAP dissociates polycomb repressive complex 2 (PRC2) via competitive binding with SUZ12
We observed that loss of STRAP led to a remarkable distribution of H3K27me3 on the loci of NOTCH genes. We then questioned whether the global levels of H3K27me3 could also be affected by STRAP, in addition to its bias on the accessibility of target gene loci. Western blot assay revealed that the total protein levels of H3K27me3 were modestly but significantly elevated in STRAP-depleted cells (Fig. 4C and Supplementary Figure S5A). In contrast, there is no effect of STRAP on the levels of two key subunits of PRC2 complex, SUZ12 and EZH2 (Fig. 4C and Supplementary Figure S5A), indicating that STRAP may structurally deregulate this complex function without disrupting its subunit expression. Therefore, we were prompted to examine the possible inhibitory effect of STRAP on PRC2 complex, which has an established role in trimethylating H3K27. Co-immunoprecipitation (co-IP) after co-expression of the tagged proteins in 293T cells showed that HA-tagged SUZ12 was detected in the anti-Flag immunoprecipitates. Reciprocally, Flag-tagged STRAP was also co-immunoprecipitated by the anti-HA antibody (Fig. 4D). In contrast, HA-EZH2 and HA-EED were not observed in the STRAP immunoprecipitates (Supplementary Figure S5B and C). These results suggest that there is a specific interaction between STRAP and SUZ12, despite STRAP not being incorporated into a complete PRC2 complex. Indeed, endogenous STRAP protein was also co-immunoprecipitated by anti-SUZ12 antibody from HCT116 and DLD-1 cells (Fig. 4E). However, we did not observe any endogenous interaction of STRAP with either EZH2 or EED (data not shown) as mentioned above. Together, the data suggest that STRAP selectively interacts with SUZ12, and not with EZH2 and EED.
The interaction between STRAP and SUZ12 led us to examine if STRAP affects the PRC2 complex assembly. We co-expressed proteins by co-transfecting HA tagged-EZH2 and -SUZ12 together with dose-dependent Flag-tagged STRAP. As expected, SUZ12 was co-immunoprecipitated by anti-EZH2 antibody. Interestingly, increasing STRAP overexpression greatly decreased the SUZ12 and EZH2 complex formation with no change in their protein levels (Fig. 4F). In contrast, there was no altered interaction between EZH2 and EED upon increased STRAP expression (Fig. 4G). To further assess whether depletion of STRAP-induced reduction in HES1 depends on SUZ12, endogenous expression of HES1 was tested by a specific shSUZ12 RNA. Western blot assay showed that SUZ12 KD had a positive effect on HES1, as evidenced by a significantly increased level of HES1 in shSUZ12 cells compared to that in scramble shRNA cells (Fig. 4H). Furthermore, restored expression of HES1 was observed in STRAP KD cells with concomitant loss of SUZ12 (Fig. 4H). Collectively, these data indicate that STRAP regulates the expression of HES1, in part through the presence of functional SUZ12, and HES1 might be a direct potential target of SUZ12 in colorectal cancer.
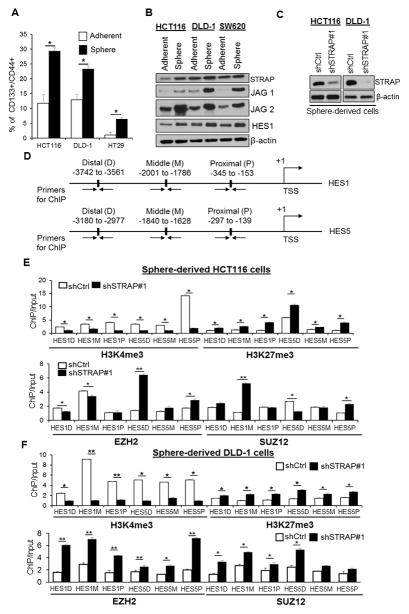
Loss of STRAP expression enhances PRC2 trimethyltransferase function on H3K27 at HES1 and HES5 loci
To address the functional regulation on local chromatin modification of target genes in CSCs by STRAP, we extended our studies to sphere-derived cells, which have more cancer stem-like properties than cells in adherent culture system (32). We first validated that sphere-derived cells are suitable for CSC model, based on significant enrichment of CD133+/CD44+ cells. Enrichment was accompanied by enhanced activation of NOTCH signaling, with increased expression of JAG receptors and HES1 (Fig. 5A and B). Surprisingly, increased expression of STRAP was observed in sphere culture condition, especially in HCT116 and DLD-1 cells, suggesting a potential involvement of STRAP in CSC maintenance (Fig. 5B). Next, we stably knocked-down STRAP using lentivirus-mediated shRNA#1 in HCT116 and DLD-1 sphere derived-cells (Fig. 5C). qPCR assay showed a significant decrease in CD133, CD44, HES1, and HES5 in KD HCT116 cells and CD133, CD44, and HES1 in KD DLD-1 cells (Supplementary Figure S6). To closely examine the chromatin signature of STRAP target genes in these cells, two transcription factors activated by NOTCH signaling, HES1 and HES5, were chosen to perform ChIP assay (Fig. 5D). H3K27me3 is highly distributed at several binding sites on the target genes along more than 3.0 kb upstream of TSS in KD sphere-derived cells; whereas H3K4me3 showed a decreased abundance at the same sites (Fig. 5D–F). These results indicate that the two markers compete with each other to occupy the loci of HES1 and HES5, and the inactive chromatin status is controlled by H3K27me3 upon STRAP downregulation. Of note, concurrence of EZH2 and SUZ12 enrichment with H3K27me3 abundances was also observed in most target binding regions (Fig. 5E and F), further evidencing the pattern of H3K27 trimethylation on its target cis-regulatory regions.
Figure 5. STRAP affects the local chromatin modification at HES1 and HES5 loci in sphere-derived cells.
A. The fraction of CD133+/CD44+ cells from the adherent and sphere cell population (HCT116, DLD-1, and HT-29) was analyzed by flow cytometry and plotted as % positive cells. *P< 0.05, vs. adherent group. B. Cell lysates from the adherent and sphere cells (HCT116, DLD-1, and SW620) were collected and subjected to immunoblot analyses with the indicated antibodies. C. Five-day old sphere-derived cells were used to establish STRAP knockdown clones and subsequent ChIP assay. Western blot analyzed the expression of STRAP in sphere-derived cells. D, E, and F. Anti-H3K4me3, -H3K27me3, -EZH2, and -SUZ12 antibodies were used for ChIP assays in sphere-derived cells. PCR amplification was done with a series of primers targeting distal, middle, and proximal sequences (see Supplementary Table S1 for primer sequences) in HES1 and HES5 loci. n=3, *P< 0.05, shSTRAP#1 vs. shCtrl in sphere-derived HCT116 and DLD-1 cells.
NICD1-HES1 signaling restores the stem-like phenotype of CSCs in STRAP KD cells
Next, we conducted gain-of-function assays either with NICD1- or with HES1-lentiviral constructs to re-activate NOTCH signaling in STRAP-KD cells. Restoration of NICD1 or HES1 resulted in significantly increased mRNA levels of NOTCH target genes (e.g. HES1, HES5, and TWIST), CD133, and CD44 in a cell-dependent manner (Fig. 6A and B), suggesting that NOTCH signaling is important in STRAP-mediated regulation of stemness-related genes. Western blot assays further confirmed the restored protein expressions of CD44, CD133, and HES1 by either NICD1 or HES1 in STRAP KD cells (Fig. 6C). The differential expression of CD44 and CD133 in different cell lines at mRNA and/or protein level regulated by STRAP may take place through different mechanisms. The percentage of CD44+/CD133+ cells was significantly increased in response to NICD1- or HES1-restoration in shSTRAP cells relative to β-gal control cells (Fig. 6D). To address the concurrent biological outcomes, we compared the proliferative ability of the rescued cells with corresponding STRAP-KD cells. Both NICD-1- and HES1-rescued cells showed the restoration in spheroid number and size compared to the STRAP-KD groups (Fig. 6E and F). Collectively, our data suggest that activation of STRAP-induced NOTCH signaling is involved in promoting stem-like phenotype of CRC cells.
Figure 6. Overexpression of NICD1 and HES1 can partially rescue stem-like phenotype of STRAP knock down cells.
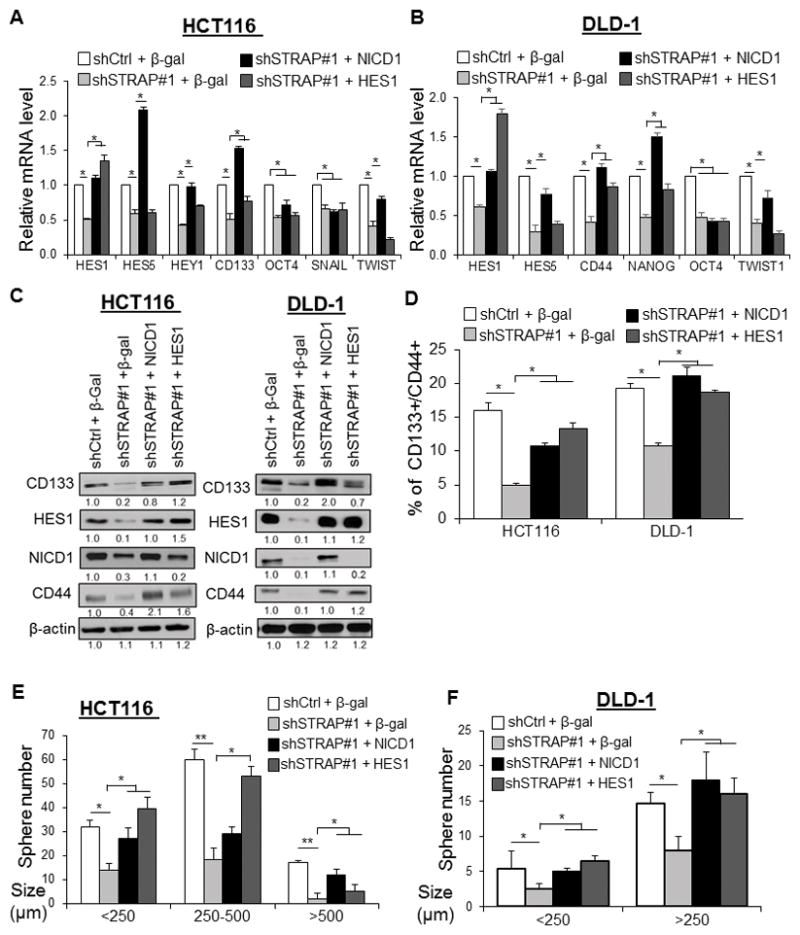
A and B. STRAP knockdown cells (HCT116, and DLD-1) were infected with lentivirus containing control vector (β-gal) or NICD1 or HES1 vector for 48 h. The relative mRNA levels of a subset of genes as indicated were determined by qRT-PCR. *P < 0.05, when compared with the corresponding control. C. Protein levels as indicated from HCT116 and DLD-1 rescued cell lines were analyzed by western blotting with the indicated antibodies. D. The fraction of CD133+/CD44+ cells from above rescued cell lines was analyzed by flow cytometry and plotted as %-positive cells. *P< 0.05, vs. the corresponding control. E and F. Stem-like spheres were generated from above rescued (NICD1 or HES1) shSTRAP or shCtrl cells after three days of culturing in sphere-conditioned medium. The number and size of spheres derived from STRAP KD cells (HCT116 and DLD-1) were compared to those of control cells. *P< 0.05, **P<0.01 relative to control.
STRAP positively correlates with the expression of HES1 in human CRC
To verify the biological significance of NOTCH signaling activation by STRAP, we examined the correlation of expression between STRAP and HES1 in human CRC. We performed immunohistochemical analyses for STRAP and HES1 in serial sections of colon tumor microarrays (TMA) containing 90 CRC patient specimens. The results showed that HES1 and STRAP are both upregulated in the colon cancer tissues compared with normal colon tissues with values 72.5% and 65.3%, respectively (Fig. 7A-D). We also observed that the higher score of HES1 is concomitant with that of STRAP (IHC score from 6 to 12) compared to the low expression group (IHC score from 0 to 4) (7.21 versus 3.24) (Fig. 7E, F and G). A statistically significant positive correlation was observed between the expressions of STRAP and HES1 in adenocarcinoma samples (Fig. 7H). Consistent with previous observations, STRAP and HES1 have been found to be present in both the nucleus and cytoplasm. Moreover, we found that the expression of both STRAP and HES1 was much higher in AJCC stage I than that in other stages (Fig. 7E and F), suggesting that STRAP and HES1 may coordinately function in CRC tumorigenesis. This observation is also consistent with previous studies showing that NOTCH signaling is high in early stages of human CRC (33). To further assess the clinical relevance of STRAP expression, TCGA data of 274 colorectal adenocarcinoma (COAD) patients and 41 controls were downloaded from UALCAN: http://ualcan.path.uab.edu/index.html (34). The mRNA level of STRAP was increased in all stages of colorectal cancer tissues compared to the normal tissues (Fig. 7I). The overall survival of CRC patients was significantly associated with STRAP expression (P<0.05) (Fig. 7J). The prognosis of patients with high STRAP expression was worse than cases with low expression. Taken together, these data indicate that STRAP-induced activation of NOTCH signaling is involved in colorectal cancer progression and in maintaining cancer stem-like subpopulation.
Figure 7. STRAP positively correlates with the expression of HES1 in colon tumors.
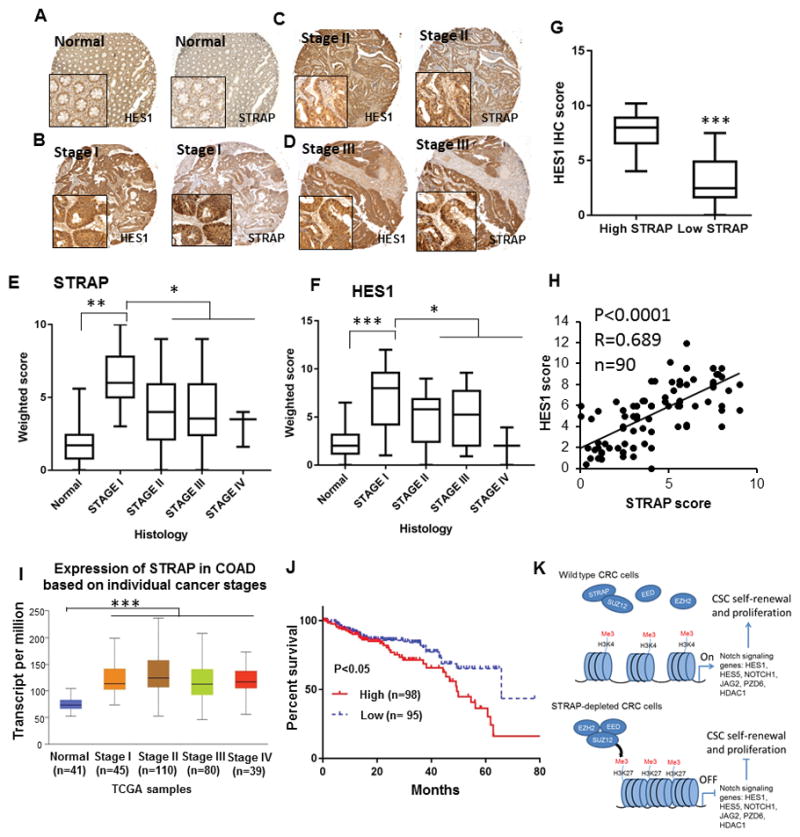
A–D. Representative images for STRAP and HES1 expressions in IHC using serial sections of tissue microarrays from normal human colon (A); adenocarcinoma stage I (B), stage II (C), stage III (D). Magnification, 100, and 400 for the insets. E and F. The weighted scores of the immunohistochemical staining for STRAP (E) or HES1 (F) plotted based on the histology *P< 0.05, **P< 0.01, ***P< 0.005. G. Tumors were divided into high STRAP IHC score (7–12) and low STRAP IHC score (0–4). The median IHC scores of HES1 were significantly different between the two groups using a two-tailed t-test. ***P< 0.001. H. The expression of STRAP and HES1 in CRC patient samples was analyzed for correlation by spearman rank correlation coefficient analysis. R = 0.689, P < 0.0001, n =90. I. Box plot analysis shows overexpression of STRAP in all stages of colorectal cancer vs. normal tissues by RNA-Seq derived expression data from TCGA. J. Kaplan-Meier graph to show the survival analyses of CRC patients based on STRAP expression. The median cutoff of STRAP expression is −0.0104 that is used to differentiate high and low expression groups for survival benefits. High STRAP represents patients with individual expression of STRAP ≥ cutoff, and low STRAP represents patients with individual expression < cutoff. K. Cartoon illustration of the underlying mechanisms by which STRAP regulates the NOTCH signaling pathway via PRC2 complex. The assembly of PRC2 complex is impaired in the presence of STRAP, resulting in the transcriptionally active chromatin marked with the high abundance of H3K4me3 at NOTCH signaling genes. Conversely, in the absence of STRAP, the expressions of these genes are epigenetically repressed by PRC2, as a result of the enrichment of H3K27me3.
Discussion
In this study, we observed the variation of CSC markers based on tumor cell types and this subpopulation constitutes 0.1–10% (35, 36). It is important to note that the best marker/s to identify CRC CSCs is not well-defined thus far. However, the expression of putative markers for CRC CSCs, like CD133 and CD44, are positively regulated by STRAP in a number cell types (Fig. 2A). This is supported by the result that knockdown of STRAP reduced the expression of some stem cell markers that are more frequently found in poorly differentiated tumors with poor clinical outcome (Fig. 3A–C) (37,38). Notably, mRNA of CD133 was rescued in HCT116 KD cells, and mRNA of CD44 was rescued in DLD-1 KD cells by NOTCH signaling. However, protein levels of CD133 and CD44 were up-regulated in both STRAP KD cell lines (Fig. 6A–C), suggesting the involvement of a different mechanism. The role of STRAP in regulating these stem-related markers is also supported by in vivo data using heterozygous mice (Fig. 2G–I).
JAG/NOTCH/HES signaling has been implicated in the regulation of cell proliferation, EMT, metastasis, stem-like phenotype, and chemoresistance. We have shown that STRAP-mediated activation of JAG2, NOTCH1, HES1, and HES5 plays an important role in CRC tumorigenicity and stemness (Fig. 3D, E, and Fig. 6). This is confirmed by the restoration of stemness markers and phenotype by the expresion of NICD1 or HES1 in STRAP knockdown HCT116 and DLD-1 clones (Fig. 6). As a result, STRAP KD cells are much less aggressive in terms of soft agar colony formation and sphere formation (Fig 2B–E). Our future work will determine whether other cytokines, chemokines, and environmental alterations functionally interact with the activation of intrinsic NOTCH signaling activated by STRAP.
Our data show that the protein level of STRAP is highest in stage I and its expression is lower but similar in the stages II/III/IV. However, all stage levels are significantly higher compared to levels in normal tissues (Fig 7E). The mRNA level of STRAP was increased in all stages of CRC tissues compared to normal tissues (Fig. 7I). Our data support the roles of STRAP in promoting cancer cell stem-like phenotype and in drug resistance. These observations are in agreement with a previous report that CRC patients with upregulation of STRAP had worse survival with adjuvant therapy, suggesting its vital role in chemoresistance (39). The level of STRAP did not increase in advanced stages (III/IV) from stage I, and it promotes cell proliferation in vivo (Fig. 1E and 2I). It has been shown that in advanced stages of CRC patients, tumor cells exhibit a decreased cell proliferation that might reduce the effectiveness of chemotherapeutics, such as 5-FU and Oxaliplatin, which primarily target rapidly-dividing cell populations (40). A medium level of STRAP expression (not high) in advanced stages may contribute to low level of cell proliferation, a factor important for resistance to cytotoxic drugs. In addition, the highest expression of STRAP in stage I may be involved in tumor progression. A similar expression pattern has been observed for another protein ZBP-89 that plays an important role in CRC progression and maintenance of cancer cell stemness (41). Together, high expression of STRAP in different stages of CRC may be linked to tumor progression and stemness phenotype.
Loss of PRC2 function contributes to oncogenesis in leukemia and lymphoma, indicating the component genes of PRC2 serve as tumor suppressors (42, 43). On the other hand, abnormal expressions of PRC2 genes correspond to human epithelial tumors (44, 45). Although the H3K27me3 level is not always sufficient to prevent transcriptional activation (46), PRC2-mediated H3K27 tri-methylation is found remarkably in the promoter regions of inactive transcripts of NOTCH genes in STRAP KD cells. These results point to a critical role of STRAP as a positive modulator of NOTCH signaling by effectively repressing the formation of PRC2 complex in CSCs (Fig. 7K). Therefore, our data provide evidence, for the first time, that 1) key components of PRC2 are necessary to selectively inactivate the transcription of NOTCH target genes in STRAP KD cells; 2) upregulation of STRAP in CRC impairs the association of the complex and influences H3K27me3, a hallmark for the repression of NOTCH related genes; and 3) EZH2 and SUZ12 might be the putative tumor suppressors in our models, depending on their activated function on target chromatin regions rather than the deregulated expression.
In conclusion, we show that STRAP exhibits oncogenic effects through mediating the epigenetic activation of NOTCH signaling, and is a regulator of tumor stemness behavior and drug response. In addition, the significant correlation of the expression of STRAP with HES1 and with stem markers in CRC patients highlights its clinical importance for novel therapeutic options in CRC treatment.
Supplementary Material
Acknowledgments
Financial support:
Supported by National Cancer Institute R01 CA95195, Veterans Affairs Merit Review Award, and a Faculty Development Award from UAB Comprehensive Cancer Center, P30 CA013148 (to PK Datta).
The authors thank Dr. Sejong Bae for statistical analyses and Dr. Tasha Smith for critically reading the manuscript.
Footnotes
CONFLICT OF INTEREST
The authors disclose no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Thomassen I, van Gestel YR, Lemmens VE, de Hingh IH. Incidence, prognosis, and treatment options for patients with synchronous peritoneal carcinomatosis and liver metastases from colorectal origin. Dis Colon Rectum. 2013;56:1373–80. doi: 10.1097/DCR.0b013e3182a62d9d. [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE, Lindeman GJ. Cancer stem cells: current statusand evolving complexities. Cell Stem Cell. 2012;10:717–28. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011;2011:1–11. doi: 10.1155/2011/396076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rich JN, Bao S. Chemotherapy and cancer stem cells. Cell Stem Cell. 2007;1:353–5. doi: 10.1016/j.stem.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Colak S, Medema JP. Cancer stem cells–important players in tumor therapy resistance. FEBS J. 2014;281:4779–91. doi: 10.1111/febs.13023. [DOI] [PubMed] [Google Scholar]
- 7.Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, Lockwood G, Gallinger S, Egan SE. Activation of NOTCH signaling in human colon adenocarcinoma. Int J Oncol. 2008;6:1223–9. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fender AW, Nutter JM, Fitzgerald TL, Bertrand FE, Sigounas G. NOTCH-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J Cell Biochem. 2015;11:2517–27. doi: 10.1002/jcb.25196. [DOI] [PubMed] [Google Scholar]
- 9.Yuan R, Ke J, Sun L, He Z, Zou Y, He X, Chen Y, Wu X, Cai Z, Wang L, Wang J, Fan X, Wu X, Lan P. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin Exp Metastasis. 2015;32:169–79. doi: 10.1007/s10585-015-9700-y. [DOI] [PubMed] [Google Scholar]
- 10.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, Lipkin SM. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer research. 2010;70:1469–75. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao F, Zhang Y, Wang S, Liu Y, Zheng L, Yang J, Huang W, Ye Y, Luo W, Xiao D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Sci Rep. 2014;4:4–10. doi: 10.1038/srep03963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda S, Katsumata R, Okuda T, Yamamoto T, Miyazaki K, Senga T, Machida K, Thant AA, Nakatsugawa S, Hamaguchi M. Molecular cloning and characterization of human MAWD, a novel protein containing WD-40 repeats frequently overexpressed in breast cancer. Cancer Res. 2000;60:13–7. [PubMed] [Google Scholar]
- 13.Halder SK, Anumanthan G, Maddula R, Mann J, Chytil A, Gonzalez AL, Washington MK, Moses HL, Beauchamp RD, Datta PK. Oncogenic function of a novel WD-domain protein, STRAP, in human carcinogenesis. Cancer Res. 2006;66:6156–66. doi: 10.1158/0008-5472.CAN-05-3261. [DOI] [PubMed] [Google Scholar]
- 14.Anumanthan G, Halder SK, Friedman DB, Datta PK. Oncogenic serine-threonine kinase receptor-associated protein modulates the function of Ewing sarcoma protein through a novel mechanism. Cancer Res. 2006;66:10824–32. doi: 10.1158/0008-5472.CAN-06-1599. [DOI] [PubMed] [Google Scholar]
- 15.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58:2085–97. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta PK, Chytil A, Gorska AE, Moses HL. Identification of STRAP, a novel WD domain protein in transforming growth factor-beta signaling. J Biol Chem. 1998;273:34671–4. doi: 10.1074/jbc.273.52.34671. [DOI] [PubMed] [Google Scholar]
- 17.Seong HA, Jung H, Choi HS, Kim KT, Ha H. Regulation of transforming growth factor-beta signaling and PDK1 kinase activity by physical interaction between PDK1 and serine-threonine kinase receptor-associated protein. J Biol Chem. 2005;280:42897–908. doi: 10.1074/jbc.M507539200. [DOI] [PubMed] [Google Scholar]
- 18.Jin L, Datta PK. Oncogenic STRAP functions as a novel negative regulator of E-cadherin and p21(Cip1) by modulating the transcription factor Sp1. Cell Cycle. 2014;13:3909–20. doi: 10.4161/15384101.2014.973310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashikar ND, Reiner J, Datta A, Datta PK. Serine threonine receptor-associated protein (STRAP) plays a role in the maintenance of mesenchymal morphology. Cell Signal. 2010;22:138–49. doi: 10.1016/j.cellsig.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433–40. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363–71. doi: 10.1002/stem.1031. [DOI] [PubMed] [Google Scholar]
- 22.Weichert W, Denkert C, Burkhardt M, Gansukh T, Bellach J, Altevogt P, Dietel M, Kristiansen G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res. 2005;11:6574–81. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 23.Choi D, Lee HW, Hur KY, Kim JJ, Park GS, Jang SH, Song YS, Jang KS, Paik SS. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009;15:2258–64. doi: 10.3748/wjg.15.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu QZ, Gao XH, Chang WJ, Gong HF, Fu CG, Zhang W, Cao GW. Expression of ITGB1 predicts prognosis in colorectal cancer: a large prospective study based on tissue. Int J Clin Exp Pathol. 2015;8:12802–10. [PMC free article] [PubMed] [Google Scholar]
- 25.Thaker AL, Shaker A, Rao MS, Ciorba MA. Modeling colitis-associted cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS) J Vis Exp. 2012;11:4100. doi: 10.3791/4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Azambuja E, Cardoso F, de Castro G, Jr, Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M. Ki-67 as a prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–13. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG. Nanog promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–45. doi: 10.1038/onc.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D, Pear WS. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing NOTCH1. Immunity. 2000;16:231–43. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 29.Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–93. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: molecular basis of cell context dependency. Eur J Cell Biol. 2011;90:572–81. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW, Yang MH. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nature cell biology. 2014;16:268–80. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 33.Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, Artavanis-Tsakonas S, Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106:6309–14. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrashekar DS, Bashel B, Balasubramanys SAH, Creighton CJ, Rodriguez IP, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasis. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 36.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;3:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller M, Hermann PC, Liebau S, Weidgang C, Seufferlein T, Kleger A, Perkhofer L. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 2016;16:349–57. doi: 10.1016/j.scr.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Buess M, Terracciano L, Reuter J, Ballabeni P, Boulay JL, Laffer U, Metzger U, Herrmann R, Rochlitz C. STRAP is a strong predictive marker of adjuvant chemotherapy benefit in colorectal cancer. Neoplasia. 2004;6:813–20. doi: 10.1593/neo.04307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anjomashoaa A, Nasri S, Human B, McCall JL, Chatterjee A, Yoon HS, McNoe L, Black MA, Reeve AE. Slow proliferation as abiological feature of colorectal cancer metastasis. Br J Cancer. 2009;101:822–28. doi: 10.1038/sj.bjc.6605229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essien BE, Sundaresan S, Ocadiz-Ruiz R, Chavis A, Tsao AC, Tessier AJ, Hayes MM, Photenhauer A, Saqui-Salces M, Kang AJ, Shah YM, Gyorffy B, Merchant JL. Transcription factor ZBP-89 drives a feedforward loop of beta-catenin expression in colorectal cancer. Caner Res. 2016;76:6877–87. doi: 10.1158/0008-5472.CAN-15-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28:44–9. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 43.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, Asp P, Hadler M, Rigo I, De Keersmaecker K, Patel J, Huynh T, Utro F, Poglio S, Samon JB, Paietta E, Racevskis J, Rowe JM, Rabadan R, Levine RL, Brown S, Pflumio F, Dominguez M, Ferrando A, Aifantis I. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, Tsao MS, Lam WL. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One. 2013;8:8–20. doi: 10.1371/journal.pone.0071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serresi M, Gargiulo G, Proost N, Siteur B, Cesaroni M, Koppens M, Xie H, Sutherland KD, Hulsman D, Citterio E, Orkin S, Berns A, van Lohuizen M. Polycomb Repressive Complex 2 Is a Barrier to KRAS-Driven Inflammation and Epithelial-Mesenchymal Transition in Non-Small-Cell Lung Cancer. Cancer Cell. 2016;29:17–31. doi: 10.1016/j.ccell.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.