Abstract
While microglia and astrocytes are known to produce key inflammatory and anti-viral mediators following infection with replicative DNA viruses, the mechanisms by which these cell types perceive such threats are poorly understood. Recently, cyclic GMP-AMP synthase (cGAS) has been identified as an important cytosolic sensor for DNA viruses and retroviruses in peripheral leukocytes. Here we confirm the ability of human microglial and astrocytic cell lines and primary human glia to respond to foreign intracellular double stranded DNA. Importantly, we provide the first demonstration that human microglia and astrocytes show robust levels of cGAS protein expression at rest and following activation. Furthermore, we show these cell types also constitutively express the critical downstream cGAS adaptor protein, stimulator of interferon genes (STING). The present finding that human glia express the principle components of the cGAS-STING pathway provides a foundation for future studies to investigate the relative importance of these molecules in clinically relevant viral CNS infections.
Keywords: human, microglia, astrocytes, cyclic GMP-AMP synthase, nucleic acid sensing
INTRODUCTION
Glia cells such as microglia and astrocytes are known to contribute to both damaging and protective immune responses during viral infections of the central nervous system (CNS). However, the mechanisms by which microglia and astrocytes respond to replicative viruses have been poorly understood, and it has only recently become apparent that cytosolic sensors for viral nucleic acids play a key role in glial cell responses [1]. For example, we have shown that the cytosolic RNA sensor retinoic acid-inducible gene I (RIG-I) is important for the recognition of productive infections with the RNA virus vesicular stomatitis virus (VSV) in murine glia and primary human astrocytes [2, 3]. Additionally, we have demonstrated that the DNA sensor, DNA-dependent activator of interferon-regulatory factors (DAI), is expressed by murine glial cells and plays a significant role in the initiation of inflammatory cytokine production by these cells in response to HSV-1 infection [4]. However, several other cytosolic DNA sensors have recently been identified in peripheral leukocytes that may also contribute to the innate immune responses of glia to replicative DNA viruses [5, 6], some of which have been shown to be expressed in murine glial cells [7]. However, far less is known about the expression and role of such sensors in human glial cells.
Of particular interest is the recently discovered cytosolic DNA sensor, cyclic GMP-AMP synthase (cGAS) [8]. Upon binding to double stranded DNA (dsDNA), cGAS produces a unique secondary metabolite 2′3′ cyclic GMP-AMP that then binds to the critical adaptor protein, stimulator of interferon genes (STING). STING, in turn, recruits TANK binding kinase 1 (TBK1), which phosphorylates interferon regulatory factor 3 (IRF-3) that then dimerizes and enters the nucleus to initiate transcription of interferon-stimulated genes including interferon-beta (IFN-β) [9–12]. The significance of cGAS is underscored by its apparent ability to recognize clinically important DNA viruses and retroviruses such as herpes simplex virus-1 (HSV-1) and human immunodeficiency virus-1 (HIV-1) that infect the human CNS [13, 14]. An antiviral role for cGAS in the CNS is supported by recent work in mice demonstrating that the microglial responses mediated by this sensor limit productive HSV-1 infection [15]. To date, the expression of cGAS and the critical adaptor protein STING has not been demonstrated in human glial cells. Here we show that human microglia and astrocytes functionally respond to cytosolic B form DNA and that these cell types constitutively express robust levels of cGAS and STING.
METHODS
Source and propagation of human glial cell lines and primary cells
U87 MG, an immortalized human astrocytic cell line, was obtained from the ATCC (HTB-14). Cells were maintained in Eagle’s Minimum Essential Medium supplemented with 10% FBS and penicillin/streptomycin. The human microglial cell line, hμglia, was a kind gift from Dr. Jonathan Karn (Case Western Reserve University). These cells were derived from primary human cells transformed with lentiviral vectors expressing SV40 T antigen and hTERT, and have been classified as microglia due to their microglia-like morphology, migratory and phagocytic activity, presence of the microglial cell surface markers CD11b, TGFβR, and P2RY12, and characteristic microglial RNA expression profile [16]. This cell line was maintained in Dulbecco’s Modified Eagle Medium supplemented with 5% FBS and penicillin/streptomycin. Primary human astrocytes and microglia were purchased from ScienCell Research Laboratories (Carlsbad, CA) and were cultured in medium supplied by the vendor.
In vitro stimulation of human microglia and astrocytes with B-DNA
Synthetic double stranded B-DNA analog poly(deoxyadenylic-deoxythymidylic) acid sodium salt (Poly(dA:dt); Invivogen, San Diego, CA), a reported cGAS ligand [17], was directly introduced into microglial or astrocytic cells at concentrations of 0.01 and 0.1 μg/ml using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. For comparison purposes, cells were exposed to transfection reagent alone or were untreated. At the indicated time points following transfection, whole cell protein isolates were collected and RNA was isolated for immunoblot analysis and semi-quantitative RT-PCR, respectively.
Immunoblot analysis for phospho-IRF3 (pIRF3), cGAS, and STING
Whole cell protein isolates were collected from microglial and astrocytic cells using Triton lysis buffer (10 mM Tris-HCl pH 10.5, 5 mM MgCl2, and 1% (v/v) Triton X-100) and analyzed by immunoblot analysis. Samples were electrophoresed on a 12% SDS-polyacrylamide gel and transferred to Immobilon-P transfer membranes (Millipore). Membranes were blocked with either 5% milk (cGAS and STING) or 5% BSA (pIRF3) for one hour and then incubated overnight at 4°C with a rabbit polyclonal antibody directed against human cGAS (Sigma Aldrich; 1:1000 dilution) or STING (Abcam; 1:1000 dilution), or a rabbit monoclonal antibody directed against pIRF3 (Ser396) (Cell Signaling Technology; clone 4D4G; 1:1000 dilution). Blots were then washed and incubated in the presence of a horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody. Bound enzyme was detected with the Super Signal system (Thermo Fisher Scientific). To assess total protein loading in each well, immunoblots were re-probed with a goat anti-mouse β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoblots shown are representative of at least three separate experiments. Protein bands were detected using a Bio-Rad ChemiDoc imaging system and quantification analysis was performed using ImageLab software (Bio-Rad). Results are presented as fold increases in the number of arbitrary densitometric units over untreated cells corrected for background intensity and normalized to the expression of β-actin.
RNA extraction and semi-quantitative reverse transcription PCR (RT-PCR)
Total RNA was isolated from cultured glial cells using Trizol Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions and quantified using a Nanodrop ND-1000 spectrophotometer. Prior to reverse transcription, RNA was treated with amplification grade DNase (Sigma Aldrich) to remove genomic DNA. All RNA samples were diluted to the same concentration and reverse transcribed in the presence of random hexamers using 200 U of RNase H minus Moloney leukemia virus reverse transcriptase (Promega, Madison, WI) in the buffer supplied by the manufacturer. Semi-quantitative RT-PCR was performed on 5% of the reverse-transcribed cDNA product to assess the relative levels of expression of mRNA-encoding IFN-β and the housekeeping gene product glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Positive and negative strand PCR primers used, respectively, were GACGCCGCATTGACCATCTA and CCTTAGGATTTCCACTCTGACT to amplify mRNA encoding IFN-β, and CCATCACCATCTTCCAGGAGCGAG and CACAGTCTTCTGGGTGGCAGTGAT to amplify mRNA encoding GAPDH.
Quantification of IFN-β in glial cell culture supernatants
A specific capture ELISA was performed to quantify concentrations of human IFN-β using a polyclonal rabbit anti-human IFN-β capture antibody and a biotinylated polyclonal rabbit anti-human IFN-β detection antibody (Abcam, Cambridge, MA). Bound antibody was detected by addition of streptavidin-horseradish peroxidase (BD Biosciences). After addition of TMB substrate and H2SO4 stop solution, absorbances were measured at 450 nm. A standard curve was constructed using varying dilutions of recombinant human IFN-β (Abcam) and the cytokine content of culture supernatants determined by extrapolation of absorbances to the standard curve.
Statistical analysis
Data is presented as the mean +/− standard error of the mean (SEM). Statistical analyses were performed using Student’s t-test or one-way analysis of variance (ANOVA) with Tukey’s post hoc test using commercially available software (GraphPad Prism, GraphPad Software, La Jolla, CA). In all experiments, results were considered statistically significant when a P-value of less than 0.05 was obtained.
RESULTS
Human microglia and astrocytes functionally respond to B-form DNA
To determine if primary human astrocytes and microglia functionally respond to foreign cytosolic dsDNA, we transfected these cells with BDNA and assessed the expression of pIRF3 protein at 1, 2, and 3 hours post administration. The human microglial cell line, hμglia, and the astrocytic cell line, U87 MG, were transfected with BDNA (0.1 μg/mL) or left untreated, and whole cell lysates collected at the indicated time points. The presence of pIRF3 was determined by immunoblot analysis and was observed in hμglia and U87-MG cells as early as an hour following challenge (Figure 1) with maximal increases seen at 3 hrs post transfection for hμglia (4.75 +/− 0.96 arbitrary densitometric units versus 0.10 +/− 0.40 in untransfected control cells: p < 0.05, n = 3) and at 2 hrs post transfection for U87-MG cells (1.40 +/− 0.15 arbitrary densitometric units versus 0.14 +/− 0.50 in untransfected control cells: p < 0.05, n = 3). In agreement with glial cell lines, primary human microglia and astrocytes similarly demonstrated rapid elevations in pIRF3 levels following challenge with BDNA (Figure 1) with expression seen at 2 hrs post transfection in primary microglia from undetectable levels in untransfected control cells and significant increases in expression at 3 hrs post BDNA administration in primary astrocytes (0.79 +/− 0.16 arbitrary densitometric units versus 0.15 +/− 0.07 in untransfected control cells: p < 0.05, n = 3).
FIGURE 1.
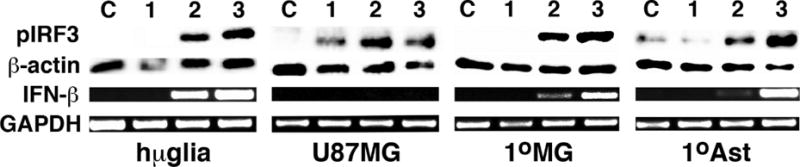
Human microglia and astrocyte cell lines and primary cells functionally respond to foreign intracellular double stranded DNA. Immortalized human microglial (hμglia) and an astrocytic cell line (U87MG) or primary human microglia (1°MG) and astrocytes (1°Ast) (3 × 105 cell per well) were treated with transfection reagent alone for 3 hrs (C) or exposed to intracellular B-form DNA (BDNA; 0.1 μg/mL) for 1, 2, or 3 hours (1, 2, 3). At these time points, whole cell lysates were collected and analyzed for the expression of phosphorylated IRF3 (pIRF3) by immunoblot analysis, and RNA was isolated and the expression of mRNA encoding IFN-β was determined by semi-quantitative RT-PCR. Expression of β-actin protein and GAPDH mRNA housekeeping gene products is shown. Results shown are representatives of at least three independent experiments and in some cases sections of the blots have been repositioned for presentation purposes.
To determine the functional significance of changes in pIRF3 levels, we investigated the expression of IFN-β mRNA following BDNA challenge. Human astrocytes, microglia, and cell lines, were transfected with BDNA (0.1 μg/mL), or left untreated, prior to RNA isolation at 1, 2, and 3 hours post challenge. As shown in Figure 1, expression of IFN-β was significantly elevated in hμglia microglia-like cells with a maximal increase seen at 3 hrs post transfection (1.69 +/− 0.06 arbitrary densitometric units versus 0.08 +/− 0.06 in untransfected control cells: p < 0.05, n = 3), and 24 hour exposure to BDNA elicited significant (p < 0.05, n = 3) IFN-β protein production by these cells at in a dose dependent manner (0.90 +/− 0.23 and 1.29 +/− 0.33 ng/ml with 1 and 5 ug/ml BDNA, respectively, from undetectable levels in untransfected cells and cells treated with transfection reagent alone). IFN-β mRNA was similarly detectable in primary human microglia and astrocytes within two hours of challenge. However, intracellular BDNA administration failed to induce demonstrable IFN-β mRNA expression in the U87 MG astrocytic cell line despite inducing IRF3 activation (Figure 1).
Human microglia and astrocytes constitutively express robust levels of the viral DNA sensor cGAS
To begin to determine the mechanisms by which glial cells respond to BDNA, we investigated the expression of the recently identified cytosolic DNA sensor, cGAS, in human glia. Hμglia and U87 MG cell lines were transfected with BDNA (0.01 and 0.1 μg/mL), treated with transfection reagent alone (L2K), or left untreated (Con). After 24 hours, whole cell lysates were collected and the level of cGAS protein expression was determined by immunoblot analysis. Both hμglia and U87 MG cell lines at rest showed robust expression levels of a 50 kDa protein corresponding to the predicted size of one of the two cGAS isoforms, and in agreement with the size previously reported for cGAS in hepatic and tonsil tissue samples detected using this antibody as reported by the vendor (Sigma Aldrich). Such expression could not be elevated significantly by exposure to BDNA (Figure 2). The expression of cGAS protein was confirmed in resting primary human microglia and astrocytes (Figure 2). Interestingly, primary human microglia showed a decrease in cGAS protein expression when challenged with the higher concentration of BDNA (Figure 2) that could not be attributed to changes in cell viability as assessed by MTT assay (data not shown).
FIGURE 2.
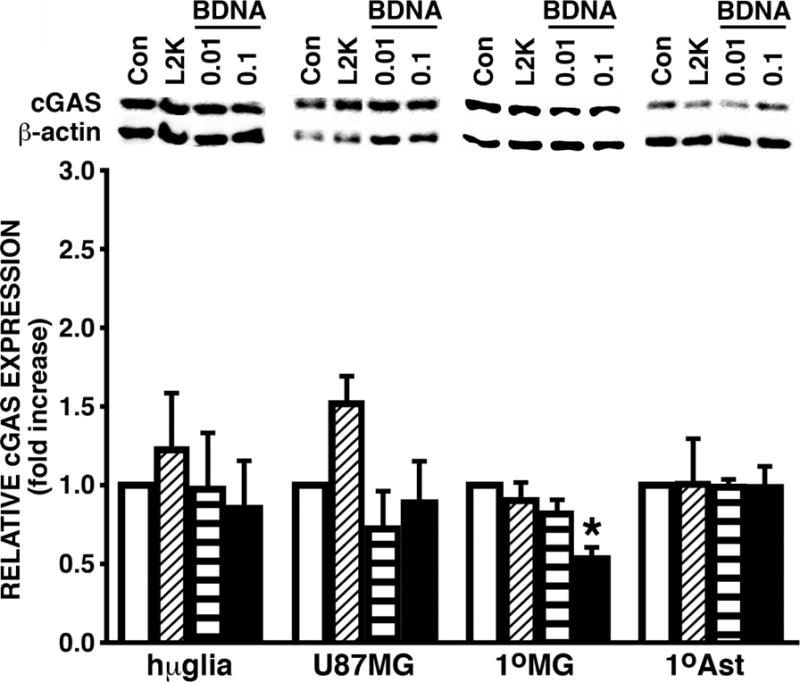
Human microglia and astrocyte cell lines and primary cells constitutively express robust levels of the viral DNA sensor cGAS. Immortalized human microglial (hμglia) and an astrocytic cell line (U87MG) or primary human microglia (1°MG) and astrocytes (1°Ast) (3 × 105 cell per well), were untreated (Con), treated with transfection reagent alone (L2K) or transfected with B-form DNA (BDNA; 0.01 or 0.1 μg/mL). At 24 hours post challenge, whole cell lysates were collected and analyzed for the expression of cGAS or the housekeeping gene product β-actin by immunoblot analysis. Blots shown are representatives of at least three independent experiments and in some cases sections of the blots have been repositioned for presentation purposes. The relative cGAS expression was determined by densitometric analysis and normalized to untreated cells. Data is expressed as the mean +/− the standard error of the mean (SEM) of at least 3 independent experiments and an asterisk indicates a statistically significant difference from transfection reagent only treated cells.
Human microglia and astrocytes constitutively express robust levels of the critical downstream adaptor protein STING
We next sought to determine the presence of the cGAS downstream adaptor protein STING in human glial cells. Hμglia and U87 MG cell lines were transfected with BDNA (0.01 and 0.1 μg/mL), treated with transfection reagent alone (L2K), or left untreated (Cells). At 24 hours following challenge, whole cell lysates were collected and the level of STING protein expression was determined by immunoblot. Both hμglia and U87 MG cell lines demonstrated robust constitutive expression of STING in unstimulated cells, and such expression was not elevated significantly by exposure to BDNA (Figure 3). These findings were confirmed in primary human astrocytes and microglia, which also demonstrated robust constitutive expression of STING protein that was not elevated further with increasing concentrations of BDNA (Figure 3).
FIGURE 3.
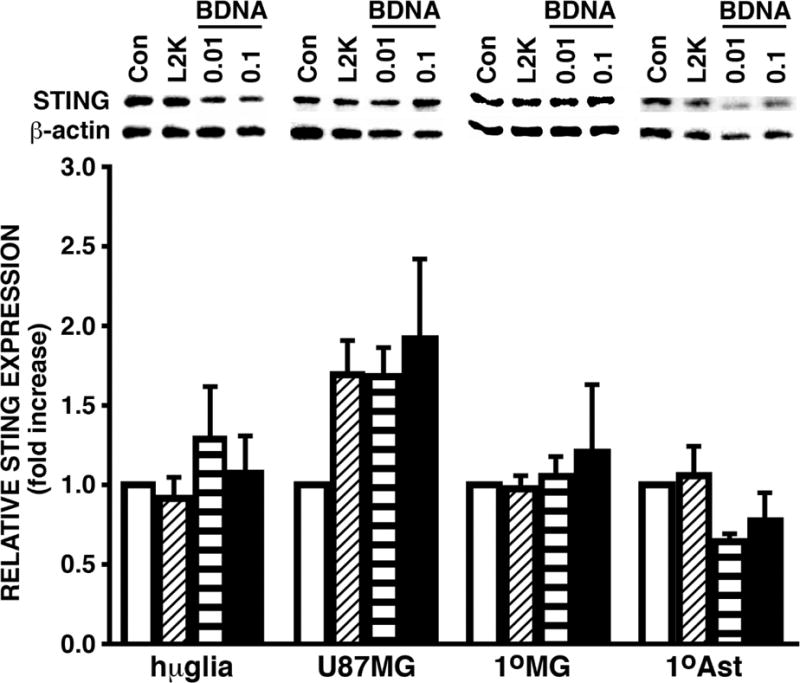
Human microglia and astrocyte cell lines and primary cells constitutively express robust levels of the cGAS downstream adaptor molecule STING. Immortalized human microglial (hμglia) and an astrocytic cell line (U87MG) or primary human microglia (1°MG) and astrocytes (1°Ast) (3 × 105 cell per well) were untreated (Cells), treated with transfection reagent alone (L2K) or transfected with B-form DNA (BDNA; 0.01 or 0.1 μg/mL). At 24 hours post challenge, whole cell lysates were collected and analyzed for the expression of STING or the housekeeping gene product β-actin by immunoblot analysis. Blots shown are representatives of at least three independent experiments and in some cases sections of the blots have been repositioned for presentation purposes. The relative STING expression was determined by densitometric analysis and normalized to untreated cells. Data is expressed as the mean +/− the standard error of the mean (SEM) of at least 3 independent experiments and no statistically significant differences were observed from transfection reagent only treated cells.
DISCUSSION
In the present study, we show that human microglia and astrocyte-like cell lines and primary human glial cells functionally respond to transfection with BDNA with the activation of the key transcription factor IRF3 and the expression of mRNA encoding the type I interferon IFN-β. This is in agreement with recent studies from our laboratory and others demonstrating the ability of primary murine glia to express IFN-β mRNA in response to BDNA [7, 2]. As such, both human glial cell types possess the means to perceive foreign cytosolic DNA. Importantly, we also provide the first demonstration that human microglia and astrocyte-like cell lines and authentic primary human glial cells express the novel cytosolic DNA sensor cGAS and its critical downstream adaptor molecule STING. We have determined that cultured human microglia and astrocytes constitutively show robust levels of cGAS and STING protein expression, levels that could not be elevated further following activation with foreign cytosolic DNA. Indeed, levels of cGAS protein expression actually showed a significant decrease in primary microglia at 24 hrs following transfection with the highest dose of BDNA, an effect that could not be attributed to a decrease in cell viability. The finding that human glial cells express components of the cGAS-STING DNA sensor pathway are in agreement with the recent study from Reinert and co-workers showing that murine microglia express cGAS [15], and the demonstration by this group [15] and our own [4] that murine glia possess STING.
It is interesting to note that while IFN-β mRNA expression was induced in primary human astrocytes following cytosolic BDNA administration, the human astrocytic cell line U87-MG failed to show such expression despite exhibiting BDNA-induced IRF3 activation. This observation perhaps reflects the limitations of the use of immortalized cell lines in such studies. Finally, it is also important to note that BDNA could be recognized via other cytosolic DNA sensors in human glial cells, or may even be perceived using RNA sensors if the foreign DNA is transcribed by host cytosolic RNA polymerase III. As such, the present demonstration that human microglia and astrocytes express the principle components of the cGAS-STING pathway sets the stage for further studies to determine the relative importance of these molecules in human glial responses to clinically relevant viral pathogens.
HIGHLIGHTS.
Primary and immortalized human microglia and astrocytes are responsive to foreign intracellular double stranded DNA.
Human microglia and astrocytes show robust expression of the recently identified cytosolic DNA sensor, cyclic GMP-AMP synthase (cGAS), at rest and following activation.
Human microglia and astrocytes constitutively express the critical downstream cGAS adaptor protein, stimulator of interferon genes (STING).
Acknowledgments
Not applicable.
Funding: This work was supported by grant NS050325 and NS097840 to IM from the National Institutes of Health.
ABBREVIATIONS
- AMP
adenosine monophosphate
LIST OF ATCC: American type tissue collection
- ANOVA
analysis of variance
- BDNA
B-form DNA
- CNS
central nervous system
- cDNA
complementary DNA
- cGAS
cyclic GMP-AMP synthase
- DAI
DNA-dependent activator of interferon-regulatory factors
- DNA
deoxyribonucleic acid
- dsDNA
double stranded DNA
- ELISA
enzyme linked immunosorbent assay
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- GMP
guanine monophosphate
- HSV-1
herpes simplex virus-1
- IFN
interferon
- IRF3
interferon regulatory factor 3
- L2K
Lipofectamine 2000
- mRNA
messenger ribonucleic acid
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- RIG-I
retinoic acid-inducible gene-I
- RNA
ribonucleic acid
- RT-PCR
reverse transcribed polymerase chain reaction
- SEM
standard error of the mean
- STING
stimulator of interferon genes
- TERT
telomerase reverse transcriptase
- TBK1
TANK binding kinase 1
- VSV
vesicular stomatitis virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics approval: Not applicable.
Availability of data and material: The data used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests: The authors declare that they have no competing interests.
Author Contributions: AMJ carried out the in vitro experiments, performed the semi-quantitative RT-PCR and immunoblot analyses, and performed data analysis. IM conceived the study, contributed to the experimental design, and drafted the manuscript. All authors read and approved the final version of the manuscript.
References
- 1.Furr SR, Marriott I. Viral CNS infections: role of glial pattern recognition receptors in neuroinflammation. Front Microbiol. 2012:201. doi: 10.3389/fmicb.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crill EK, Furr-Rogers SR, Marriott I. RIG-I is required for VSV-induced cytokine production by murine glia and acts in combination with DAI to initiate responses to HSV-1. Glia. 2015:2168–2180. doi: 10.1002/glia.22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furr SR, Moerdyk-Schauwecker M, Grdzelishvili VZ, Marriott I. RIG-I mediates nonsegmented negative-sense RNA virus-induced inflammatory immune responses of primary human astrocytes. Glia. 2010:1620–1629. doi: 10.1002/glia.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furr SR, Chauhan VS, Moerdyk-Schauwecker MJ, Marriott I. A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation. 2011:99. doi: 10.1186/1742-2094-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology. 2013:1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Dempsey A, Bowie AG. Innate Immune Recognition of DNA: a recent history. Virology. 2015:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox DJ, Field RH, Williams DG, Baran M, Bowie AG, Cunningham C, Dunne A. DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia. 2015:812–825. doi: 10.1002/glia.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′–5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzner AM, Hagmann CA, Goldeck M, Wolter S, Kubler K, Wittmann S, Gramberg T, Andreeva L, Hopfner KP, Mertens C, Zillinger T, Jin T, Xiao TS, Bartok E, Coch C, Ackermann D, Hornung V, Ludwig J, Barchet W, Hartmann G, Schlee M. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol. 2015:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinert LS, Lopusna K, Winther H, Sun C, Thomsen MK, Nandakumar R, Mogensen TH, Meyer M, Vaegter C, Nyengaard JR, Fitzgerald KA, Paludan SR. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun. 2016:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Mesa Y, Jay TR, Checkley MA, Luttge B, Dobrowolski C, Valadkhan S, Landreth GE, Karn J, Alvarez-Carbonell D. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol. 2017:47–66. doi: 10.1007/s13365-016-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]


