Abstract
Aims
The onset of clinical type 1 diabetes (T1D) is preceded by the occurrence of disease-specific autoantibodies. The level of autoantibody titers is known to be associated with progression time from the first emergence of autoantibodies to the onset of clinical symptoms, but detailed analyses of this complex relationship are lacking. We aimed to fill this gap by applying advanced statistical models.
Methods
We investigated data of 613 children from the prospective TEDDY study who were persistent positive for IAA, GADA and/or IA2A autoantibodies. We used a novel approach of Bayesian joint modeling of longitudinal and survival data to assess the potentially time- and covariate dependent association between the longitudinal autoantibody titers and progression time to T1D.
Results
For all autoantibodies we observed a positive association between the titers and the T1D progression risk. This association was estimated as time constant for IA2A, but decreased over time for IAA and GADA. For example the hazard ratio [95% credibility interval] for IAA (per transformed unit) was 3.38 [2.66, 4.38] at 6 months after seroconversion, and 2.02 [1.55, 2.68] at 36 months after seroconversion.
Conclusions
These findings indicate that T1D progression risk stratification based on autoantibody titers should focus on time points early after seroconversion. Joint modeling techniques allow for new insights into these associations.
Keywords: autoantibodies, joint modeling, type 1 diabetes
INTRODUCTION
Type 1 diabetes (T1D) is one of the most common chronic diseases in childhood, with worldwide increasing incidence [1]. The disease is preceded by a preclinical period of islet autoimmunity, which most commonly develops in early infancy [2,3]. The presence of islet autoantibodies is associated with the progression to clinical diabetes [4]. However, the time from the first emergence of autoantibodies, called seroconversion, to the onset of clinical symptoms varies considerably between individuals, ranging from weeks to decades [4]. It is also known that the combination of different autoantibodies as well as the autoantibody titer is associated with progression time [5]. For insulin autoantibodies (IAA), both their titers around seroconversion and their mean levels over time have been found to be associated with progression to T1D [2,6], and similar findings have been recently reported for other islet autoantibodies [7–9]. Nevertheless detailed analyses of autoantibody titers over time are lacking.
Here, we investigated data of more than 600 islet-autoantibody positive children followed up within the prospective The Environmental Determinants of Diabetes in the Young (TEDDY) study [10,11]. In contrast to previous analyses, we used joint models of longitudinal and survival data. This class of models has the advantage to avoid potential bias due to characteristics of the longitudinal markers (here autoantibodies), such as random biological fluctuations, informative censoring and discrete measurement time points [12]. By applying a novel approach of joint modeling, we gained further insights into the potentially complex relationship between longitudinal islet autoantibody measures and the time to T1D progression, particularly with respect to time-varying associations of both.
METHODS
TEDDY is an ongoing prospective cohort study funded by the National Institutes of Health with the primary goal to identify environmental causes of T1D. The TEDDY study enrolled 8,676 children with increased genetic risk for T1D who were recruited in six clinical research centers located in the USA, Finland, Germany, and Sweden between 2004 and 2010 shortly after birth. Detailed information on study design, eligibility and methods has been previously published [11,13,14]. Written informed consents were obtained for all participants from a parent or primary caretaker, separately, for genetic screening and for participation in prospective follow-up before inclusion in the study. The study was approved by local Institutional Review or Ethics Boards and is monitored by the External Advisory Board formed by the National Institutes of Health. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). For this analysis, we used the data of all children who had developed one or more persistent islet autoantibodies by the time of our data access (31 December 2014). At that timepoint the median age of the children analyzed at their last visit was 6.5 years with a range from 0.75 to 10.2 years.
Definition of islet autoimmunity
Development of persistent islet autoimmunity was assessed every 3 months and defined by the presence of at least one islet autoantibody among autoantibodies to insulin (IAA), glutamic acid decarboxylase (GADA), and insulinoma-associated protein 2 (IA2A) on two or more consecutive visits confirmed by two laboratories. Date of persistent autoimmunity to an autoantibody was defined as the draw date of the first sample of the two consecutive samples which deemed the child persistent confirmed positive for this autoantibody. As described in more detail elsewhere [7], the respective autoantibody titers were standardized to be comparable across study laboratories (University of Bristol, UK; and University of Colorado, Denver, US) by subtracting the laboratory- and antibody-specific threshold and dividing by the laboratory- and autoantibody-specific standard deviation, and were log-transformed afterwards.
Study outcome
The main outcome of this analysis was the time to development of T1D after seroconversion in months. T1D diagnosis was based on American Diabetes Association criteria [15].
Statistical analyses
Of the 8,676 children enrolled, 613 had developed one or more autoantibodies at the time of our data access. We created three subsets of the data where we restricted the data to children who had seroconverted to IAA (n=442), GADA (n=466) or IA2A (n=288), respectively. These subsets were not mutually exclusive, as children had potentially seroconverted to multiple autoantibodies. Children were assigned to each subset irrespectively of whether the specific autoantibody was amongst the first islet autoantibodies to appear or appeared at a later time during follow-up. For example, if a child developed autoantibodies to IAA first and autoantibodies to GADA later, the child would be assigned to both the IAA and GADA subset.
We used a novel shared parameter joint model approach to assess the association between the longitudinal autoantibody titers from seroconversion with the time to T1D. Joint models allow the incorporation of longitudinal titers as time-varying covariates into the survival model of progression to T1D by estimating a longitudinal model and a proportional hazards model, using a joint likelihood for both submodels [16]. We further extended this model to a more flexible joint model, where we were able to assess heterogeneous and nonlinear individual biomarker trajectories and to explore complex associations between the biomarkers and the time to event [17]. We refer to the Appendix for further details. Using this novel approach we specified the autoantibody titers over time as smooth, nonlinear, subject-specific trajectories in the longitudinal model. Furthermore we allowed the association between the modeled trajectories and the time to T1D to be time-varying in our main analysis. In additional explorative analyses we allowed the association to differ between subjects with different characteristics, and to differ over time between subjects with different characteristics.
We fitted these models for each of the three autoantibodies IAA, GADA and IA2A, separately, within each autoantibody-specific subset. In the longitudinal submodels, we assessed the associations of each autoantibody titer with a) age at seroconversion of the respective autoantibody, b) a binary variable indicating whether the autoantibody was among the first autoantibodies to appear, and c) two time-varying binary variables indicating which of the other two autoantibodies were present at each observed time point. In each proportional hazards submodel, we assessed the associations of the smooth subject-specific autoantibody trajectories from the longitudinal model with progression time from seroconversion of the respective autoantibody to T1D. Baseline covariates were a) the age at seroconversion of the respective autoantibody and b) whether the autoantibody was among the first autoantibodies to appear. We further assessed whether the association between the autoantibody trajectories and the time to T1D differed over time between subjects with and without a first-degree relative with T1D or between girls and boys. Additionally we checked for differences in the association between HLA genotypes. Due to the limited size of certain HLA subgroups we modelled this association as time-constant.
All models were estimated within a Bayesian framework using the R-package bamlss [18]. Weakly informative normal priors were used for all coefficients. We report the posterior mean estimates/hazard ratios and 95% credibility intervals (CI) for all modeled parameters. Bayesian CIs can be interpreted as the interval in which the population parameter lies with a given probability (here 95%). We assessed convergence of the Markov chains by visual inspection of traceplots and conducted sensitivity analyses with regards to prior specification. All calculations were carried out with R version 3.2.5 [19].
RESULTS
Table 1 shows the study characteristics in each subset, i.e. the subsets of children who developed IAA, GADA or IA2A autoantibodies, respectively, at any time during follow-up. In most cases, either IAA, GADA, or both, were present at the time of the first seroconversion, whereas IA2A occurred at a later time point. The children seroconverted to the different autoantibodies at different median ages (p < 0.001, Kruskal-Wallis Test) with IAA seroconversion taking place at a lower median age. Apart from that, children with different autoantibodies were similar regarding the progression time to T1D and other variables.
Table 1.
Description of the Study Population by Type of Persistent Autoantibody. Values are Reported as n (% of Non-Missing Observations) for Categorical Variables and Median (Interquartile Range) for Continuous Variables.
| Variable | Total | Type of persistent autoantibody | ||
|---|---|---|---|---|
|
| ||||
| IAA | GADA | IA2A | ||
| Total number of children | 613 | 442 | 466 | 288 |
| Age at respective seroconversion (years) | 2.2 (1.2, 3.8) | 2.0 (1.1, 3.5) | 2.7 (1.6, 4.2) | 2.8 (1.9, 4.5) |
| Girls | 268 (44%) | 200 (45%) | 212 (45%) | 112 (39%) |
| Country | ||||
| US | 206 (34%) | 136 (31%) | 166 (36%) | 94 (33%) |
| Finland | 153 (25%) | 125 (28%) | 109 (23%) | 85 (30%) |
| Germany | 47 (8%) | 40 (9%) | 32 (7%) | 22 (8%) |
| Sweden | 207 (34%) | 141 (32%) | 159 (34%) | 87 (30%) |
| Child having a first degree relative with T1D | 128 (21%) | 105 (24%) | 97 (21%) | 71 (25%) |
| HLA-DR genotype | ||||
| DR3/4 | 311 (51%) | 241 (55%) | 251 (54%) | 148 (51%) |
| DR4/4 | 106 (17%) | 74 (17%) | 81 (17%) | 64 (22%) |
| DR4/8 | 92 (15%) | 71 (16%) | 51 (11%) | 46 (16%) |
| DR3/3 | 76 (12%) | 30 (7%) | 64 (14%) | 16 (6%) |
| other | 28 (5%) | 26 (6%) | 19 (4%) | 14 (5%) |
| Additionally autoantibody positive for | ||||
| IAA | 302 (65%) | 252 (88%) | ||
| GADA | 302 (68%) | 237 (83%) | ||
| IA2A | 252 (57%) | 237 (51%) | ||
| Autoantibody present at first seroconversion | 353 (80%) | 344 (74%) | 40 (14%) | |
| Number of children who developed T1D | 175 (29%) | 162 (37%) | 134 (29%) | 127 (44%) |
The individual autoantibody patterns over time after seroconversion were heterogeneous, but on average IAA titers declined after an initial increase, and GADA and IA2A titers increased shortly after seroconversion and remained relatively stable thereafter (Supplementary figure 1).
In the joint modeling of autoantibody titers over time and the time to T1D, we observed for all autoantibodies a positive association between the titer and the risk of progression to T1D. Titers over time were lower for subjects who seroconverted at an older age for the respective autoantibody, and higher if the respective autoantibody appeared at the initial seroconversion, and if other autoantibodies were present (Table 2). For each autoantibody, a higher age at the respective seroconversion was also associated with lower risk of progression to clinical T1D. For example, children had a hazard ratio [95% CI] of 0.84 [0.72, 0.98] if they seroconverted one year later for IAA.
Table 2.
Posterior Mean Estimates of Coefficients (β) and Hazard Ratios with Corresponding 95% Credibility Intervals from Joint Models of Autoantibody Trajectories (IAA, GADA and IA2A, as Estimated in Longitudinal Submodels) and Progression to T1D (Survival Submodels).
| Autoantibodies | Covariate | Longitudinal models | Survival models | ||
|---|---|---|---|---|---|
|
| |||||
| β | 95% CI | HR | 95% CI | ||
| IAA | IAA present at first seroconversion | 0.35 | 0.20, 0.51 | 0.66 | 0.42, 1.02 |
| IAA seroconversion age (years) | −0.10 | −0.13, −0.07 | 0.84 | 0.72, 0.98 | |
| GADA positive (time-varying variable) | 0.31 | 0.24, 0.37 | a | a | |
| IA2A positive (time-varying variable) | 0.17 | 0.12, 0.22 | a | a | |
| GADA | GADA present at first seroconversion | 0.34 | 0.19, 0.47 | 0.72 | 0.51, 1.03 |
| GADA seroconversion age (years) | −0.06 | −0.09, −0.03 | 0.61 | 0.52, 0.72 | |
| IAA positive (time-varying variable) | 0.25 | 0.20, 0.32 | a | a | |
| IA2A positive (time-varying variable) | 0.06 | 0.02, 0.11 | a | a | |
| IA2A | IA2A present at first seroconversion | 0.31 | 0.11, 0.50 | 1.07 | 0.61, 1.80 |
| IA2A seroconversion age (years) | −0.05 | −0.08, −0.01 | 0.66 | 0.56, 0.78 | |
| IAA positive (time-varying variable) | 0.22 | 0.09, 0.36 | a | a | |
| GADA positive (time-varying variable) | 0.29 | 0.18, 0.41 | a | a | |
CI Credibility Interval; HR Hazard Ratio
Covariate only included in the longitudinal submodel.
Bold font indicates that the 95% CI does not include 0 (for β) or 1 (for HR).
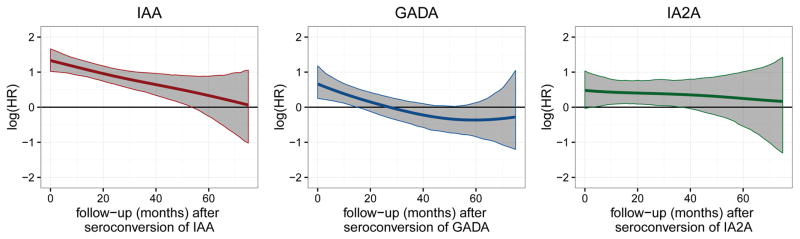
We further investigated whether the association between the estimated trajectories of autoantibodies and the progression to T1D was time-varying or constant. By using our approach, we observed that the association was time-varying for IAA and GADA with the association being highest early after seroconversion and decreasing over time (Figure 1) and stronger for IAA than GADA: The hazard ratio for IAA (per transformed unit) was 3.38 [2.66; 4.38] at 6 months after seroconversion, 3.02 [2.44, 3.81] at 12 months after seroconversion, and 2.02 [1.55, 2.68] at 36 months after seroconversion (Table 3) with an average decrease in the hazard ratio of 10% [95% CI; 2%, 18%] every 6 months. The hazard ratio for GADA (per transformed unit) was 1.63 [1.20, 2.30] at 6 months after seroconversion, 1.40 [1.07, 1.85] at 12 months after seroconversion, and 0.85 [0.61, 1.17] at 36 months after seroconversion with an average decrease of 9% [1%, 15%] every 6 months. For IA2A, the positive association between autoantibody titer and T1D progression was estimated as time-constant: The hazard ratios for IA2A (per transformed unit) were 1.56 [1.04, 2.42] at 6 months after seroconversion, 1.53 [1.10, 2.16] at 12 months after seroconversion, and 1.44 [1.005, 2.16] at 36 months after seroconversion with a negligible average decrease of 2% [−8%, 13%] every 6 months. As indicated by the credibility intervals in Figure 1, positive associations with T1D progression were observed for IAA up to 54 months after seroconversion, for GADA up to 18 months after seroconversion and for IA2A between 6 and 36 months after seroconversion. The traceplots indicated satisfactory convergence of the Markov chains (Supplementary Figures 2–4) and sensitivity analyses showed robustness against different prior specifications (Supplementary Figure 5).
Figure 1.
Posterior mean estimates (lines) and 95% credibility intervals (shaded areas) of ηα(t) the time-varying log hazard ratio (HR) of the association between longitudinal autoantibody trajectories and type 1 diabetes.
Table 3.
Posterior Mean Hazard Ratios (HR) with Corresponding 95% Credibility Intervals (CI) at Different Time-Points After Seroconversion of each Autoantibody for the Association Between Autoantibody Trajectories From the Longitudinal Model and Progression Time to Type 1 Diabetes.
| Autoantibodies | Time point | HR | 95% CI |
|---|---|---|---|
| IAA | 0 months | 3.78 | 2.78, 5.28 |
| 6 months | 3.38 | 2.66, 4.38 | |
| 12 months | 3.02 | 2.44, 3.81 | |
| 24 months | 2.43 | 1.94, 3.02 | |
| 36 months | 2.02 | 1.55, 2.68 | |
| 48 months | 1.69 | 1.17, 2.50 | |
| 60 months | 1.39 | 0.77, 2.42 | |
|
| |||
| GADA | 0 months | 1.94 | 1.28, 3.25 |
| 6 months | 1.63 | 1.20, 2.30 | |
| 12 months | 1.40 | 1.07, 1.85 | |
| 24 months | 1.07 | 0.80, 1.41 | |
| 36 months | 0.85 | 0.61, 1.17 | |
| 48 months | 0.73 | 0.50, 1.04 | |
| 60 months | 0.69 | 0.43, 1.14 | |
|
| |||
| IA2A | 0 months | 1.62 | 0.96, 2.81 |
| 6 months | 1.56 | 1.04, 2.42 | |
| 12 months | 1.53 | 1.10, 2.16 | |
| 24 months | 1.49 | 1.08, 2.13 | |
| 36 months | 1.44 | 1.005, 2.16 | |
| 48 months | 1.37 | 0.82, 2.33 | |
| 60 months | 1.28 | 0.58, 2.74 | |
Bold font indicates that the 95% CI of the respective association does not include the 1.
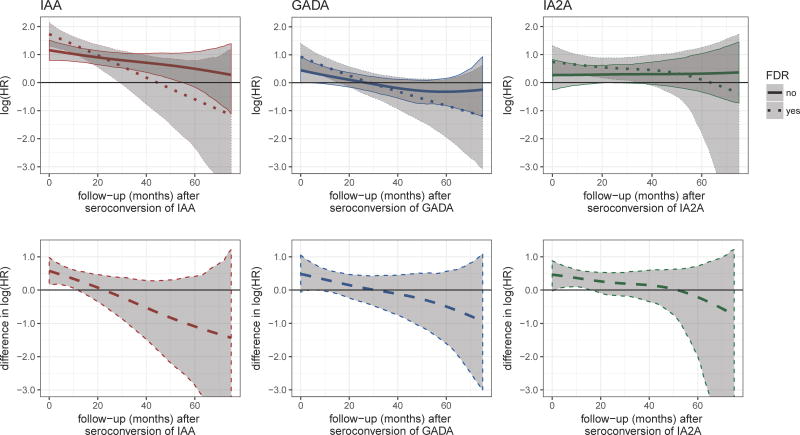
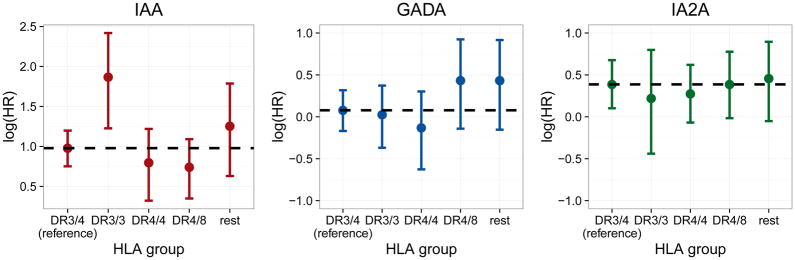
We further observed differences in the time-varying association of autoantibodies with progression to T1D between children with and without a first-degree relative with T1D. For all autoantibodies the associations were higher amongst children with a first degree relative at early time points and decreased more strongly within this group (Figure 2, upper panel). For IAA, the associations between the two groups differed from seroconversion until about 12 months thereafter, as indicated by the credibility bands of the differences (Figure 2, lower panel), but only from 4 to 6 months after seroconversion for GADA and from 1 to 16 months after seroconversion for IA2A. For all autoantibodies HLA subgroups were similar in the association between autoantibody trajectories and the time to T1D (Figure 3). An exception was a higher association for subjects with IAA autoantibodies and the DR3/3 genotype, a genotype which is less prevalent among IAA positive children (n = 30, 7%). In accordance with the difference in the hazard, the mean titer levels between progressors and non-progressors differed more strongly within the small subgroup of DR3/3 than within other HLA genotypes with non-progressors showing an especially low level (Supplementary Figure 6). We did not observe consistent differences in the association over time between girls and boys (Supplementary Figure 7).
Figure 2.
Posterior mean estimates (lines/dots) and 95% credibility intervals (shaded areas) of ηα(t, FDR), the time-varying log hazard ratio (HR) of the association between longitudinal autoantibody trajectories and type 1 diabetes (T1D) progression stratified for children that had a first-degree relative (FDR) with T1D or not (upper panel) and of the difference of the association between the groups over time, ηα(t, FDR=1)- ηα(t, FDR=0) (lower panel).
Figure 3.
Posterior mean estimates and 95% credibility intervals of ηα(HLA), the time-constant log hazard ratio (HR) of the association between longitudinal autoantibody trajectories and type 1 diabetes progression, per HLA genotype. The dashed line represents the estimated log hazard ratio of the reference group
DISCUSSION
In the present study the complex relationship between longitudinally measured autoantibodies and the risk of progression to T1D diabetes was explored using a novel joint modeling approach. We observed potentially time-varying positive associations between the autoantibody titers of IAA and GADA, and the risk of T1D progression, indicating that the T1D progression risk associated with autoantibody titers was highest shortly after seroconversion of the respective autoantibody. The hazard ratio was highest for IAA, especially at early time points. Additionally, we observed that the associations of the autoantibody titer and the T1D risk early after seroconversion were more pronounced in children with first-degree relatives with T1D.
These results were in line with earlier results from other cohorts, where initial and mean IAA and IA2A titers were shown to be associated with the risk of progression [20,2,6] as well as from a more recent and methodologically advanced study based on the TEDDY data. In this study the relationship between titers of the same autoantibodies over time and the risk of progression to T1D was modeled assuming a time constant association [7]. By using mean levels of the respective autoantibodies as time-varying predictors in a Cox model, the authors could show a positive association between autoantibody titers and the time to T1D progression for IAA and IA2A in their analyses.
Potential limitations of this previous approach are however that (a) only subject’s mean titers until a certain time point are taken into account and not all observed values over time, (b) in a time-varying Cox model the time-varying predictor is assumed constant between observations, and (c) the association between autoantibodies and the risk of progression is assumed to be time-constant. These limitations were addressed by our joint modeling approach. Here, we flexibly modeled the trajectories of all three autoantibodies in each subject as a smooth function of time, i.e. obtaining predictions for the autoantibody titers between the measurements at discrete time points, and could use all this information as a time-varying covariate in the survival model. Additionally, we allowed their association with the risk of T1D progression to vary over time and between groups of subjects (children with and without first degree relatives with T1D as well as boys and girls). In consequence, we were able to explore the association between autoantibodies and the risk of T1D beyond the previous results. For example, we observed that increased GADA titers may predict T1D progression within the first 1.5 years after seroconversion, but not thereafter. As this association averages to 0 over the whole time range this association was potentially not captured in the simpler modeling from the previous analysis. Furthermore our modeling approach revealed that the time-varying associations appear to be more pronounced in children with first degree relatives with T1D compared to children without.
The modeling of autoantibodies as longitudinal biomarkers and the time to clinical T1D poses a challenge due to the nature of the data beyond the aspects mentioned above. Longitudinal biomarkers usually contain potential random variation both due to the laboratory measurement process as well as short and long term biological fluctuations, and are only observed until an event occurs. Whereas not accounting for the random fluctuations in a time-varying Cox model might result in an underestimation of the hazard ratio [12], ignoring the latter might distort the estimation of covariate effects in the longitudinal model. By jointly analyzing the longitudinal and survival model we could address these issues and gained further insights as to how covariates affected both the autoantibody titers over time, and the risk of T1D progression. We found that earlier seroconversion for the respective autoantibody, if the respective seroconversion was the initial one, as well as the presence of other autoantibodies was associated with higher autoantibody titers. The age at the respective seroconversion was also inversely related to the risk of T1D progression for every autoantibody. While joint modeling approaches allow for detailed and unbiased estimations, they demand a high number of subjects, especially when complex associations are modeled in the survival part. TEDDY is the largest prospective study on the determinants of T1D worldwide and thus offers a unique opportunity to explore the application of joint modeling techniques on these complex relationships due to the high number of subjects and the detailed measurement schedule.
Currently, the presented flexible joint model only allows the assessment of one longitudinal biomarker at a time. In consequence, one limitation is that we were not able to combine all three markers into one joint model. We partly addressed this issue by including information on the presence of other autoantibodies and the order of their occurrence in our model. While they provide insights into the mechanisms of disease progression, a drawback of our results is that they cannot easily be translated from a cohort setting with frequent measurements into clinical practice, as the age at the respective seroconversion plays a crucial role in the prediction of T1D progression risk, but is not readily available in practice.
In conclusion, by using state of the art joint modeling techniques we were able to give insights into the complex relationship between longitudinal autoantibody titers and the risk of progression to clinical T1D. Risk stratification basing on autoantibody titers should focus on time points early after seroconversion.
Supplementary Material
Supplementary Figure 1. Individual and mean transformed titers of IAA, GADA and IA2A autoantibodies after seroconversion to the respective autoantibody.
Supplementary Figure 2. Convergence plots for exemplary coefficients from the joint model of IAA trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 3. Convergence plots for exemplary coefficients from the joint model of GADA trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 4. Convergence plots for exemplary coefficients from the joint model of IA2A trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 5. Results from the sensitivity analysis for the joint models of autoantibody trajectories (IAA, GADA and IA2A) and progression to T1D data. Estimated coefficients from model fits based on (i) the main model as presented in the Application section with IG(0.0001, 0.00011) as prior distribution for the variance parameters and N(0, 10002) as weakly informative prior for the parametric terms, (ii) with IG(0.001, 1) as prior distribution for the variance parameters, and (iii) N(0, 502) as prior for the parametric terms. Left panel: Posterior mean estimates (lines) and 95% pointwise credibility intervals (shaded areas) of ηα(t). Right panel: Posterior mean estimates of coefficients and hazard ratios with corresponding 95% credibility intervals.
Association denotes the intercept of ηα(t), long the coefficients of the longitudinal submodel and survival the coefficients (i.e. the log hazard ratio) of the survival model.
Supplementary Figure 6. Individual and mean transformed titers of IAA autoantibodies after seroconversion per HLA genotype, stratified for progression to T1D.
Supplementary Figure 7. Posterior mean estimates (lines/dots) and 95% credibility intervals (shaded areas) of ηα(t, SEX) the time-varying log hazard ratio (HR) of the association between longitudinal autoantibody trajectories and T1D progression stratified for girls and boys (upper panel) and of the difference of the association between the groups over time, ηα(t, boys) – ηα(t, girls) (lower panel).
Acknowledgments
Funding
The TEDDY study was supported by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Juvenile Diabetes Research Foundation (JDRF), and Centers for Disease Control and Prevention (CDC). Additionally this work was supported by funds from the Helmholtz International Research Group [HIRG-0018] and the Juvenile Diabetes Research Foundation (JDRF) [grant number 2-SRA-2015-13-Q-R] to AGZ, and the German Research Foundation DFG [Emmy Noether grant GR 3793/1-1] to SG. The funders had no impact on the design, implementation, analysis and interpretation of the data.
APPENDIX
The Bayesian flexible additive joint model allows for a high variety of effect specifications in the longitudinal submodel, the survival submodel, and the association between both. All parts of the model are parameterized as structured additive models. We refer to the original methodological paper [1] and the documentation of the package bamlss [2] for further details. In our estimated models the longitudinal model per autoantibody y for subject i at the observed measurement times tij was specified as a mixed model allowing smooth, nonlinear, and subject-specific trajectories over time as well as linear and nonlinear effects of the covariates xk
where the error ε(tij) was assumed independently and identically (iid) normally distributed around 0. We assumed ημi(tij) to be the true underlying trajectories, modeled by a smooth function of time f1(tij), a random intercept f2(i), smooth subject-specific functions of time f3(tij, i) as well as a sum of linear and nonlinear effects of covariates xk
The proportional hazards model for the log-hazard is
where ηλ(t) denotes the log-baseline hazard, explicitly modeled by Bayesian P-Splines [3], ηγi is the sum of the linear and nonlinear effects of baseline covariates, ημi(t) are the true subject-specific autoantibody trajectories from the mixed model as continuous-time time-varying predictors and ηαi(t) denotes the association between this autoantibody and the log-hazard. As a novelty in this joint model this association between longitudinal trajectories and the log-hazard can be a function of time and subject-specific covariates. The full model was estimated using a derivative-based Metropolis-Hastings algorithm. We used vague normal priors for all regression coefficients in the model with mean 0 and a standard deviation of 1000. Smooth and random effects terms were regularized using suitable multivariate normal priors on the coefficients. Inverse Gamma hyperpriors IG(0.0001, 0.0001) were used for the variance parameters. For details on the priors and the model estimation we refer to the package bamlss. For each model we sampled 33.000 iterations with a burnin of 3000 and a thinning of 30 obtaining 1000 samples. Satisfactory convergence and mixing was assessed by visual inspection of traceplots. Additionally sensitivity analyses were conducted for the main models using (a) IG(0.001, 1) for the variance parameters and (b) more informative normal priors N(0, 502) for all regression coefficients.
In order to assess the time-varying nature of the association between longitudinal autoantibody titers and the time to T1D, we computed the average slope of the estimated association over time. This estimate and corresponding credibility intervals can be easily obtained from the samples of the posterior.
In more detail, for every posterior sample we first numerically approximated the first derivative of the association at every observed event and follow-up time, and afterwards computed their mean. In consequence we obtained an average first derivative for every sample. From this empirical distribution we computed the posterior mean estimate and corresponding credibility intervals of the first derivative, which was used as a measure of the average decrease of the hazard ratio over time.
- 1.Köhler M, Umlauf N, Beyerlein A, Winkler C, Ziegler A-G, Greven S. Flexible Bayesian additive joint models with an application to type 1 diabetes research. Biometrical Journal. 2017 doi: 10.1002/bimj.201600224. (To appear) [DOI] [PubMed] [Google Scholar]
- 2.Umlauf N, Klein N, Zeileis A. BAMLSS: Bayesian Additive Models for Location, Scale and Shape (and Beyond). Working Paper 2017-05. Working Papers in Economics and Statistics, Research Platform Empirical and Experimental Economics, Universität Innsbruck. 2017 URL http://EconPapers.RePEc.org/RePEc:inn:wpaper:2017-05.
- 3.Lang S, Brezger A. Bayesian P-Splines. Journal of Computational and Graphical Statistics. 2004;13(1):183–212. doi: 10.1198/1061860043010. [DOI] [Google Scholar]
The Teddy Study Group
Colorado Clinical Center
Marian Rewers, M.D., Ph.D., PI1,4,5,6,10,11, Kimberly Bautista12, Judith Baxter9,10,12,15, Ruth Bedoy2, Daniel Felipe-Morales, Kimberly Driscoll, Ph.D.9, Brigitte I. Frohnert, M.D.2,14, Patricia Gesualdo2,6,12,14,15, Michelle Hoffman12,13,14, Rachel Karban12, Edwin Liu, M.D.13, Jill Norris, Ph.D.2,3,12, Adela Samper-Imaz, Andrea Steck, M.D.3,14, Kathleen Waugh6,7,12,15, Hali Wright12. University of Colorado, Anschutz Medical Campus, Barbara Davis Center for Childhood Diabetes.
Finland Clinical Center
Jorma Toppari, M.D., Ph.D., PI¥,^,1,4,11,14, Olli G. Simell, M.D., Ph.D.¥,^,1,4,11,13, Annika Adamsson, Ph.D.^,12, Suvi Ahonen*,±,§, Heikki Hyöty, M.D., Ph.D.*,±,6, Jorma Ilonen, M.D., Ph.D.¥,¶,3, Sanna Jokipuu^, Tiina Kallio^, Leena Karlsson^, Miia Kähönenμ,¤, Mikael Knip, M.D., Ph.D.*,±,5, Lea Kovanen*,±,§, Mirva Koreasalo*,±,§,2, Kalle Kurppa, M.D., Ph.D.*,±,13, Tiina Latva-ahoμ,¤, Maria Lönnrot, M.D., Ph.D.*,±,6, Elina Mäntymäki^, Katja Multasuoμ,¤, Juha Mykkänen, Ph.D.¥,3, Tiina Niininen±,*,12, Sari Niinistö±,§, Mia Nyblom*,±, Petra Rajala^, Jenna Rautanen±,§, Anne Riikonen*,±,§, Mika Riikonen^, Jenni Rouhiainen^, Minna Romo^, Tuula Simell, Ph.D., Ville Simell^,¥,13, Maija Sjöberg¥,^,12,14, Aino Steniusμ,¤,12, Maria Leppänen^, Sini Vainionpää^, Eeva Varjonen¥,^,12, Riitta Veijola, M.D., Ph.D.μ,¤,14, Suvi M. Virtanen, M.D., Ph.D.*,±,§2, Mari Vähä-Mäkilä^, Mari Åkerlund*,±,§, Katri Lindfors, Ph.D.*,13 ¥University of Turku, *University of Tampere, μUniversity of Oulu, ^Turku University Hospital, Hospital District of Southwest Finland, ±Tampere University Hospital, ¤Oulu University Hospital, §National Institute for Health and Welfare, Finland, ¶University of Kuopio.
Georgia/Florida Clinical Center
Jin-Xiong She, Ph.D., PI1,3,4,11, Desmond Schatz, M.D.*,4,5,7,8, Diane Hopkins12, Leigh Steed12,13,14,15, Jamie Thomas*,6,12, Janey Adams*,12, Katherine Silvis2, Michael Haller, M.D.*,14, Melissa Gardiner, Richard McIndoe, Ph.D., Ashok Sharma, Joshua Williams, Gabriela Young, Stephen W. Anderson, M.D.^, Laura Jacobsen, M.D.*,14 Center for Biotechnology and Genomic Medicine, Augusta University. *University of Florida, ^Pediatric Endocrine Associates, Atlanta.
Germany Clinical Center
Anette G. Ziegler, M.D., PI1,3,4,11, Andreas Beyerlein, Ph.D.2, Ezio Bonifacio Ph.D.*,5, Michael Hummel, M.D.13, Sandra Hummel, Ph.D.2, Kristina Foterek¥,2, Nicole Janz, Mathilde Kersting, Ph.D.¥,2, Annette Knopff7, Sibylle Koletzko, M.D.¶,13, Claudia Peplow12, Roswith Roth, Ph.D.9, Marlon Scholz, Joanna Stock9,12,14, Katharina Warncke, M.D.14, Lorena Wendel, Christiane Winkler, Ph.D.2,12,15. Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, and Klinikum rechts der Isar, Technische Universität München. *Center for Regenerative Therapies, TU Dresden, ¶Dr. von Hauner Children’s Hospital, Department of Gastroenterology, Ludwig Maximillians University Munich, ¥Research Institute for Child Nutrition, Dortmund.
Sweden Clinical Center
Åke Lernmark, Ph.D., PI1,3,4,5,6,8,10,11,15, Daniel Agardh, M.D., Ph.D.13, Carin Andrén Aronsson2,12,13, Maria Ask, Jenny Bremer, Ulla-Marie Carlsson, Corrado Cilio, Ph.D., M.D.5, Emelie Ericson-Hallström, Lina Fransson, Thomas Gard, Joanna Gerardsson, Rasmus Bennet, Monica Hansen, Gertie Hansson, Susanne Hyberg, Fredrik Johansen, Berglind Jonsdottir, M.D., Helena Elding Larsson, M.D., Ph.D. 6,14, Marielle Lindström, Markus Lundgren, M.D.14, Maria Månsson-Martinez, Maria Markan, Jessica Melin12, Zeliha Mestan, Karin Ottosson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Sara Sibthorpe, Birgitta Sjöberg, Ulrica Swartling, Ph.D.9,12, Evelyn Tekum Amboh, Carina Törn, Ph.D. 3,15, Anne Wallin, Åsa Wimar12,14, Sofie Åberg. Lund University.
Washington Clinical Center
William A. Hagopian, M.D., Ph.D., PI1,3,4, 5, 6,7,11,13, 14, Michael Killian6,7,12,13, Claire Cowen Crouch12,14,15, Jennifer Skidmore2, Josephine Carson, Maria Dalzell, Kayleen Dunson, Rachel Hervey, Corbin Johnson, Rachel Lyons, Arlene Meyer, Denise Mulenga, Alexander Tarr, Morgan Uland, John Willis. Pacific Northwest Diabetes Research Institute.
Pennsylvania Satellite Center
Dorothy Becker, M.D., Margaret Franciscus, MaryEllen Dalmagro-Elias Smith2, Ashi Daftary, M.D., Mary Beth Klein, Chrystal Yates. Children’s Hospital of Pittsburgh of UPMC.
Data Coordinating Center
Jeffrey P. Krischer, Ph.D.,PI1,4,5,10,11, Michael Abbondondolo, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Rasheedah Brown12,15, Brant Burkhardt, Ph.D.5,6, Martha Butterworth2, Joanna Clasen, David Cuthbertson, Christopher Eberhard, Steven Fiske9, Dena Garcia, Jennifer Garmeson, Veena Gowda, Kathleen Heyman, Francisco Perez Laras, Hye-Seung Lee, Ph.D.1,2,13,15, Shu Liu, Xiang Liu, Ph.D.2,3,9,14, Kristian Lynch, Ph.D. 5,6,9,15, Jamie Malloy, Cristina McCarthy12,15, Steven Meulemans, Hemang Parikh, Ph.D.3, Chris Shaffer, Laura Smith, Ph.D.9,12, Susan Smith12,15, Noah Sulman, Ph.D., Roy Tamura, Ph.D.1,2,13, Ulla Uusitalo, Ph.D.2,15, Kendra Vehik, Ph.D.4,5,6,14,15, Ponni Vijayakandipan, Keith Wood, Jimin Yang, Ph.D., R.D.2,15. Past staff: Lori Ballard, David Hadley, Ph.D., Wendy McLeod. University of South Florida.
Project scientist
Beena Akolkar, Ph.D.1,3,4,5,6,7,10,11. National Institutes of Diabetes and Digestive and Kidney Diseases.
Autoantibody Reference Laboratories
Liping Yu, M.D.^,5, Dongmei Miao, M.D.^, Polly Bingley, M.D., FRCP*,5, Alistair Williams*, Kyla Chandler*, Saba Rokni*, Claire Williams*, Rebecca Wyatt*, Gifty George*, Sian Grace*. ^Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, *School of Clinical Sciences, University of Bristol UK.
HLA Reference Laboratory
Henry Erlich, Ph.D.3, Steven J. Mack, Ph.D., Anna Lisa Fear. Center for Genetics, Children’s Hospital Oakland Research Institute.
Repository
Sandra Ke, Niveen Mulholland, Ph.D. NIDDK Biosample Repository at Fisher BioServices.
Other contributors
Kasia Bourcier, Ph.D.5, National Institutes of Allergy and Infectious Diseases. Thomas Briese, Ph.D.6,15, Columbia University. Suzanne Bennett Johnson, Ph.D.9,12, Florida State University. Eric Triplett, Ph.D.6, University of Florida.
Committees
1Ancillary Studies, 2Diet, 3Genetics, 4Human Subjects/Publicity/Publications, 5Immune Markers, 6Infectious Agents, 7Laboratory Implementation, 8Maternal Studies, 9Psychosocial, 10Quality Assurance, 11Steering, 12Study Coordinators, 13Celiac Disease, 14Clinical Implementation, 15Quality Assurance Subcommittee on Data Quality.
Footnotes
Statement of Human and Animal Rights
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5).
Conflicts of interest
The authors declare that they have no conflict of interest.
Statement of Informed Consent
Informed consent was obtained from all patients for being included in the study.
Author contributions
AGZ and EB designed this manuscript proposal. MK analyzed the data and wrote the first and final draft of the manuscript together with AB, SG and AGZ. EB and KV contributed to the interpretation of the results and to subsequent drafts of the manuscript. NU contributed to specific programming aspects. MR, WAH, JXS, AL, JT, BA, AGZ, and JPK are principal investigators of the TEDDY study, contributed to conception and design of the TEDDY study, data acquisition, and funding for TEDDY, reviewed the manuscript and contributed to subsequent drafts. This work is part of MK’s PhD thesis within the graduate school HELENA at the Helmholtz Zentrum München in collaboration with the Ludwig-Maximilians-Universität München, Germany.
Contributor Information
Meike Köhler, Institute of Diabetes Research, Helmholtz Zentrum München, Munich, Germany. Forschergruppe Diabetes, Klinikum rechts der Isar, Technische Universität München, Neuherberg, Germany. Forschergruppe Diabetes e.V., Neuherberg, Germany.
Andreas Beyerlein, Institute of Diabetes Research, Helmholtz Zentrum München, Munich, Germany. Forschergruppe Diabetes, Klinikum rechts der Isar, Technische Universität München, Neuherberg, Germany. Forschergruppe Diabetes e.V., Neuherberg, Germany.
Kendra Vehik, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa, FL, USA.
Sonja Greven, Department of Statistics, Ludwig-Maximilians-Universität München, Munich, Germany.
Nikolaus Umlauf, Department of Statistics, University of Innsbruck, Innsbruck, Austria.
Åke Lernmark, Department of Clinical Sciences, Lund University/CRC, Skåne University Hospital SUS, Malmö, Sweden.
William A. Hagopian, Pacific Northwest Diabetes Research Institute, Seattle, WA, USA
Marian Rewers, Barbara Davis Center for Childhood Diabetes, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
Jin-Xiong She, Center for Biotechnology and Genomic Medicine, Medical College of Georgia, Georgia Regents University, Augusta, GA, USA.
Jorma Toppari, Department of Physiology, Institute of Biomedicine, University of Turku, and Department of Pediatrics, Turku University Hospital, Turku, Finland.
Beena Akolkar, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, USA.
Jeffrey P. Krischer, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa, FL, USA
Ezio Bonifacio, Center for Regenerative Therapies Dresden and Paul Langerhans Institute Dresden, Technische Universität Dresden, Germany.
Anette-G. Ziegler, Institute of Diabetes Research, Helmholtz Zentrum München, Munich, Germany. Forschergruppe Diabetes, Klinikum rechts der Isar, Technische Universität München, Neuherberg, Germany. Forschergruppe Diabetes e.V., Neuherberg, Germany.
References
- 1.Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G Eurodiab Study Group. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373(9680):2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 2.Parikka V, Nanto-Salonen K, Saarinen M, Simell T, Ilonen J, Hyoty H, Veijola R, Knip M, Simell O. Early seroconversion and rapidly increasing autoantibody concentrations predict prepubertal manifestation of type 1 diabetes in children at genetic risk. Diabetologia. 2012;55(7):1926–1936. doi: 10.1007/s00125-012-2523-3. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler AG, Bonifacio E Babydiab-Babydiet Study Group. Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia. 2012;55(7):1937–1943. doi: 10.1007/s00125-012-2472-x. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, Winkler C, Ilonen J, Veijola R, Knip M, Bonifacio E, Eisenbarth GS. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA: the journal of the American Medical Association. 2013;309(23):2473–2479. doi: 10.1001/jama.2013.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achenbach P, Warncke K, Reiter J, Naserke HE, Williams AJ, Bingley PJ, Bonifacio E, Ziegler AG. Stratification of type 1 diabetes risk on the basis of islet autoantibody characteristics. Diabetes. 2004;53(2):384–392. doi: 10.2337/diabetes.53.2.384. [DOI] [PubMed] [Google Scholar]
- 6.Steck AK, Johnson K, Barriga KJ, Miao D, Yu L, Hutton JC, Eisenbarth GS, Rewers MJ. Age of islet autoantibody appearance and mean levels of insulin, but not GAD or IA-2 autoantibodies, predict age of diagnosis of type 1 diabetes: diabetes autoimmunity study in the young. Diabetes care. 2011;34(6):1397–1399. doi: 10.2337/dc10-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steck AK, Vehik K, Bonifacio E, Lernmark A, Ziegler AG, Hagopian WA, She J, Simell O, Akolkar B, Krischer J, Schatz D, Rewers MJ TEDDY Study Group. Predictors of Progression From the Appearance of Islet Autoantibodies to Early Childhood Diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY) Diabetes Care. 2015;38(5):808–813. doi: 10.2337/dc14-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steck AK, Dong F, Waugh K, Frohnert BI, Yu L, Norris JM, Rewers MJ. Predictors of slow progression to diabetes in children with multiple islet autoantibodies. Journal of autoimmunity. 2016;72:113–117. doi: 10.1016/j.jaut.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endesfelder D, Hagen M, Winkler C, Haupt F, Zillmer S, Knopff A, Bonifacio E, Ziegler AG, Zu Castell W, Achenbach P. A novel approach for the analysis of longitudinal profiles reveals delayed progression to type 1 diabetes in a subgroup of multiple-islet-autoantibody-positive children. Diabetologia. 2016 doi: 10.1007/s00125-016-4050-0. [DOI] [PubMed] [Google Scholar]
- 10.Krischer JP, Lynch KF, Schatz DA, Ilonen J, Lernmark Å, Hagopian WA, Rewers MJ, She J-X, Simell OG, Toppari J, Ziegler A-G, Akolkar B, Bonifacio E Group tTS. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia. 2015;58:980–987. doi: 10.1007/s00125-015-3514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teddy Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatric diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 12.Asar O, Ritchie J, Kalra PA, Diggle PJ. Joint modelling of repeated measurement and time-to-event data: an introductory tutorial. International journal of epidemiology. 2015;44(1):334–344. doi: 10.1093/ije/dyu262. [DOI] [PubMed] [Google Scholar]
- 13.Teddy Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Annals of the New York Academy of Sciences. 2008;1150:1–13. doi: 10.1196/annals.1447.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagopian WA, Erlich H, Lernmark A, Rewers M, Ziegler AG, Simell O, Akolkar B, Vogt R, Jr, Blair A, Ilonen J, Krischer J, She J, Group TS. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatric diabetes. 2011;12(8):733–743. doi: 10.1111/j.1399-5448.2011.00774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Diabetes Association. Executive summary: standards of medical care in diabetes--2011. Diabetes Care. 2011;34(Suppl 1):S4–10. doi: 10.2337/dc11-S004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizopoulos D. Joint models for longitudinal and time-to-event data, with applications in R. Chapman and Hall/CRC; Boca Raton, Florida: 2012. [Google Scholar]
- 17.Köhler M, Umlauf N, Beyerlein A, Winkler C, Ziegler A-G, Greven S. Flexible Bayesian additive joint models with an application to type 1 diabetes research. Biometrical Journal. 2017 doi: 10.1002/bimj.201600224. (To appear) [DOI] [PubMed] [Google Scholar]
- 18.Umlauf N, Klein N, Zeileis A, Koehler M. bamlss: Bayesian Additive Models for Location Scale and Shape (and Beyond) 2016. [Google Scholar]
- 19.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 20.Mrena S, Virtanen SM, Laippala P, Kulmala P, Hannila M-L, Åkerblom HK, Knip M Group tCDiFS. Models for Predicting Type 1 Diabetes in Siblings of Affected Children. Diabetes Care. 2006;29(3):662–667. doi: 10.2337/diacare.29.03.06.dc05-0774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Individual and mean transformed titers of IAA, GADA and IA2A autoantibodies after seroconversion to the respective autoantibody.
Supplementary Figure 2. Convergence plots for exemplary coefficients from the joint model of IAA trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 3. Convergence plots for exemplary coefficients from the joint model of GADA trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 4. Convergence plots for exemplary coefficients from the joint model of IA2A trajectories and progression to T1D. Coefficients with index μ represent the longitudinal submodel and the γ survival submodel (see also Appendix).
Supplementary Figure 5. Results from the sensitivity analysis for the joint models of autoantibody trajectories (IAA, GADA and IA2A) and progression to T1D data. Estimated coefficients from model fits based on (i) the main model as presented in the Application section with IG(0.0001, 0.00011) as prior distribution for the variance parameters and N(0, 10002) as weakly informative prior for the parametric terms, (ii) with IG(0.001, 1) as prior distribution for the variance parameters, and (iii) N(0, 502) as prior for the parametric terms. Left panel: Posterior mean estimates (lines) and 95% pointwise credibility intervals (shaded areas) of ηα(t). Right panel: Posterior mean estimates of coefficients and hazard ratios with corresponding 95% credibility intervals.
Association denotes the intercept of ηα(t), long the coefficients of the longitudinal submodel and survival the coefficients (i.e. the log hazard ratio) of the survival model.
Supplementary Figure 6. Individual and mean transformed titers of IAA autoantibodies after seroconversion per HLA genotype, stratified for progression to T1D.
Supplementary Figure 7. Posterior mean estimates (lines/dots) and 95% credibility intervals (shaded areas) of ηα(t, SEX) the time-varying log hazard ratio (HR) of the association between longitudinal autoantibody trajectories and T1D progression stratified for girls and boys (upper panel) and of the difference of the association between the groups over time, ηα(t, boys) – ηα(t, girls) (lower panel).