Abstract
Hepatocellular carcinoma (HCC) is one of the most common neoplasms, and metastasis is the most important feature for HCC-related deaths. Mounting evidence implies the dynamic regulatory role of SIRT2, a histone deacetylase, in cancer cells. Unfortunately, the role of SIRT2 and the antitumor activity of its inhibition are not known in HCC. The present study aims to evaluate the biological function of SIRT2 in HCC and identify the target of SIRT2 as well as evaluate its therapeutic efficacy. We found that SIRT2 was upregulated in HCC tissues compared to adjacent normal tissues, and this was correlated with reduced patient survival. Although CCK8 and colony-formation assays showed that SIRT2 inhibiton marginally promotes proliferation in HCC cell lines, SIRT2 knockdown decreased the invasion of HCC cells. We demonstrated that downregulation of SIRT2 could inhibit its downstream target phosphoenolpyruvate carboxykinase 1 and glutaminase, which is related to mitochondrial metabolism and the E-Cadherin pathway. These results demonstrate, for the first time that downregulation of SIRT2 decreases migration as well as invasion in human HCC cells, indicating that inhibiting SIRT2 may be an effective therapeutic strategy for treating HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common neoplasms and the third most frequent cause of cancer death worldwide [1]. Globally, there are approximately 750,000 new cases of HCC reported per year. Incidence rate continues to approximate the death rate, indicating that most patients who develop HCC die as a result of it [2]. HCC is highly aggressive and likely to metastasize. HCC cells migrate through the extracellular matrix and vascular wall into vascular circulation and can promote the formation of new blood vessels as well. The treatment of patients with HCC is particularly challenging. Not only is resistance to conventional anticancer drugs becoming increasingly commonplace, but there too remains an urgent need for the identification of relevant biomarkers. Unfortunately, oncogenic pathways identified by genetic studies have proven difficult to target therapeutically. New therapies targeting HCC are needed [1].
Posttranslational modifications in metabolism regulation have received close attention due to their ability to respond to changes in cellular metabolic status as well as regulation by upstream signaling pathways. Acetylation has emerged as one such key posttranslational modification and is identified in metabolic enzymes in cellular regulation [3]. Deacetylases are designated as belonging to four classes (I-IV), depending on their amino acid sequence structure. Sirtuins (also known as SIRTs) are NAD+-dependent class III histone deacetylases (HDACs). Seven SIRT homologues have been identified as SIRT1 to 7 in mammals [4]. Among the sirtuin family members, which have been conserved in evolution from bacteria to mammalian species, SIRT2 catalyzes a wide range of biological processes including gene expression, development, and metabolism. Its enzymatic reaction removes the acetyl group from lysine residues and is accompanied with hydrolysis of NAD to generate nicotinamide, lysine, and O-acetyl-ADP-ribose. Nicotinamide can inhibit this enzymatic reaction [4]. SIRT2 is primarily a cytoplasmic protein, and both tubulin and phosphoenolpyruvate carboxykinase 1 (PEPCK1) are known substrates of this deacetylase [3], [4]. Mounting evidence implies a dual role for SIRT2 in tumorigenesis: the expression of SIRT2 is significantly reduced in gliomas and melanomas, while high SIRT2 level in breast cancer is associated with increased time to recurrence [5], [6], [7]. Studies have shown that the SIRT2 inhibitor has antitumor, anti-inflammatory, and antidiabetic properties [8], [9], [10]. How SIRT2 impacts metabolism and molecular mechanisms in HCC cells has not yet been reported.
Phosphoenolpyruvate carboxykinase (PEPCK) is the rate-limiting enzyme of gluconeogenesis and catalyzes the conversion of oxaloacetate into phosphoenolpyruvate. PEPCK is found in two forms: cytosolic (PEPCK1) and mitochondrial (PEPCK2) [11], [12], [13], [14]. Due to the important role of PEPCK1, its regulation has been extensively studied. Both yeast PEPCK1 and human PEPCK1 have been found to have acetylation, with their catalytic activity inactivated following this acetylation [3], [15]. Acetylation of Lys70, Lys71, and Lys594 of human PEPCK1 leads to decreased protein stability, reduced protein levels, and decreased gluconeogenesis without affecting mRNA levels [3]. PEPCK1 is an important marker in the evaluation of type II diabetes and can promote cancer cell proliferation by increasing glucose and glutamine utilization toward anabolic metabolism [16]. For glutamine metabolism, glutaminase (GLS) is the key enzyme in the conversion of glutamine to glutamate and is expressed in many tissue cells and cancer cells [17]. However, little is known about the role of PEPCK1 and GLS in HCC tumorigenesis.
In the present study, we discovered that SIRT2 plays a critical role in promoting HCC metastasis and invasion by directing neoplasm metabolism. SIRT2-mediated deacetylation in protein posttranslational modification stabilizes the protein level of PEPCK1 and GLS, promoting glucose utilization and inhibiting the signal of E-Cadherin.
Materials and Methods
Ethics, Consent, and Permissions
All experiments utilizing patients and cells were approved by the Ethics Committee of Medical Research of Fudan University.
Differentially Expressed Genes (DEGs) of Paired HCC from The Cancer Genome Atlas (TCGA) Data
The HCC RNA-Seq data were downloaded from the TCGA database using The GDC Data Portal (https://gdc-portal.nci.nih.gov/). The number of mRNA expression values was 60,483. The mRNA expression data included a total of 100 samples consisting of 50 normal sample and 50 paired-HCC samples. The sequencing data were all publicly available, and no ethical issues were involved. The edgeR package in Bioconductor was used to screen the DEGs in HCC and normal liver tissue samples. The edgeR package is based on the negative binomial distribution, which can correct the overdispersion problem in RNA-seq data by using a Poisson model and a Bayes procedure. The data with expression values of zero were removed. The genes were deemed to be DEGs if |FoldChange| >2, respectively, both with P value < .01 and false discovery rate (FDR) < 0.05.
Functional Annotation
The Database for Annotation Visualization and Integrated Discovery online tool (https://david.ncifcrf.gov/) was used to conduct the functional and pathway enrichment analyses in our study. We performed Gene Ontology (GO) and KEGG pathway enrichment analyses to detect the potential biological functions and pathways of the high- and low-expression genes in HCC.
Immunohistochemistry
Tumor specimens were fixed in 4% formalin and embedded in paraffin. Sections were treated with immunoperoxidase using the DAB kit (Zsbio) and then scored. The tissue microarray slides were obtained from Guge BIOTECH (Wuhan, China). Staining intensity was graded as follows: absent staining = 0, weak = 1, moderate = 2, and strong = 3. The percentage of staining was graded as follows: 0 (no positive cells), 1 (<25% positive cells), 2 (25%-50% positive cells), 3 (50%-75% positive cells), and 4 (>75% positive cells). The score for each tissue was calculated by multiplication, and the range of this calculation was therefore 0 to 12 [18].
Cell Culture and Reagents
Human liver cell lines (HepG2, Huh-7, Hep3B, 7721, and Chang's) were donated by the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and the Zhao lab of Fudan University (Shanghai, China). Cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Invitrogen), penicillin (Invitrogen) (100 U/ml), and streptomycin (Invitrogen) (100 U/ml). Full-length PEPCK1 (wild type, 3K/R and 3K/Q) and SIRT2 (wild type and H187Y) plasmids were also donated from the Zhao lab of Fudan University (Shanghai, China). Antibodies against Flag (Sigma, St. Louis., MO), PEPCK1 (Santa Cruz), SIRT2 (Sigma), α-tubulin (CST, Danvers, MA), acetylated α-tubulin (Abcam, Cambridge, UK), and β-actin (Sigma) were all purchased, and the polyclonal antibody against acetyl-lysine was a donation from the Zhao lab. Selisistat (Selleck, Houston, TX), MG132 (Sigma), salermide (Sigma), and CHX (Sigma) were all purchased. Control and siSIRT2 adenovirus were purchased (Vector Biolabs, Malvern, PA).
Western Blot
Standard procedures were followed for Western blot, except for the detection of acetylation, which used 50 mM Tris (pH 7.5) with 10% (v/v) Tween 20 and 1% peptone (AMRESCO, Solon, OH) as a blocking buffer. Primary and secondary antibodies were diluted in 50 mM Tris (pH 7.5) with 0.1% peptone. Signals were probed using the chemiluminescence ECL plus reagent (Thermo, Grand Island, NY) and detected using a Typhoon FLA9500 scanner (GE, Fairfield, CT).
Apoptosis Assay
Cell apoptosis was measured by flow cytometry using the AnnexinV-FITC/PI Apoptosis Detection Kit (BD, Franklin Lakes, NJ) following the manufacturer's instructions.
Deacetylation Assay
Cells were lysed in NP-40 buffer containing 50 mM Tris-HCl (Sigma), 150 mM NaCl (Sangon), 0.5% Nonidet P-40 (Sigma), 1 μg/ml aprotinin (Sigma), 1 μg/ml leupeptin (Sigma), 1 μg/ml pepstatin (Sigma), 1 mM Na3VO4 (Sigma), and 1 mM PMSF (Sigma), pH = 7.5. For immunoprecipitation, 500 μl of cell lysate was incubated with Flag-beads (Sigma) for 12 hours at 4°C with rotation, and the beads were washed three times with lysis buffer before proteins were dissolved in loading buffer. The SIRT2 assay was done using bacterial expression and purification (Biovision, Milpitas, CA). Deacetylation assays were carried out in the presence of 5 μg enzyme and 0.3 μg peptide in 30-μl reaction buffer [30 mM HEPES (Sigma), 0.6 mM MgCl2 (Sangon), 1 mM DTT (Sigma), 1 mM NAD+ (Sigma), and 10 mM PMSF (Sigma)]. The deacetylation reaction was incubated for 3 to 5 hours at 37°C before the mixture was desalted by passing it through a C18 ZipTip (Millipore). The desalted samples were analyzed using a MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Grand Island, NY). The acetylated peptide used in the assay was GILRRLKAcKAcYDNCWL (Glssale, Shanghai, China).
Cell Viability Assay
Cells viability was determined using the CCK-8 colorimetric assay in 96-well plates (2 × 103 cells/well) (Dojindo, Minato-ku, Tokyo, Japan). The absorbance at 450 nm was recorded using a microplate reader.
HCC Metastasis Model
Nude mice were purchased from the Department of Laboratory Animal Science, Nanjing Drum Tower Hospital. HepG2 cells (5 × 106) in FBS-free RPMI-1640 were injected into the tail vein of mice. Mice were treated with salermide at a dose of 20 mg/kg bodyweight in 200 μl volume via intraperitoneal injection twice a week for 6 weeks. The Animal Welfare Committee of Nanjing Drum Tower Hospital approved all procedures involving animals.
Statistics
Data was expressed as mean ± standard error of the mean (SE). The data were analyzed through one-way ANOVAs followed by post hoc Duncan tests (SPSS 17.0). P < .05 was considered significant.
Results
Metabolic Abnormalities and Metastasis of Liver Cancer
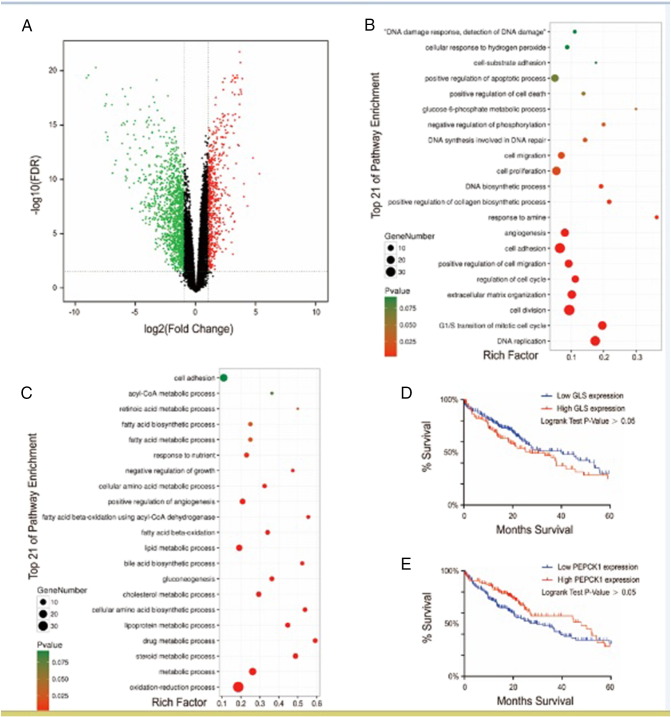
Tumors can change their metabolic status through differential gene expression, thereby promoting their malignant biological behavior [19]. Consequently, we analyzed the DEGs in liver HCC from TCGA data. The HCC RNA-Seq data were downloaded from the TCGA database using The GDC Data Portal (https://gdc-portal.nci.nih.gov/). There were a total of 60,483 mRNA expression values. The mRNA expression data further included 100 samples consisting of 50 normal samples and 50 paired-HCC samples. We identified the differentially expressed genes (Figure 1A), and we performed GO and KEGG pathway enrichment analyses to detect the potential biological functions and pathways of the high- and low-expression genes in HCC (Figure 1, B and C); the two most relevant include metabolic and metastatic pathways in HCC. PEPCK1 and GLS serve as important targets of energy and building blocks for many tumor cells. They promote the biological characteristics of cancer cells by increasing glucose and glutamine utilization for anabolic metabolism [16], [20]. Next, we queried the TCGA database, which contains clinically annotated genomic data from 286 HCC samples [21], [22], and found that both PEPCK1 and GLS reduced patient survival (P > .05, log-rank test) (Figure 1, D and E). These results suggest that metabolic abnormalities and metastasis are important features of liver cancer—moreover, these results underscore complex and unclear regulatory mechanisms.
Figure 1.
Metabolic abnormalities and metastasis are important features of liver cancer.
(A) Heat map of DEGs in liver HCC and paired normal liver samples. Heat map was drawn using the gplots package in Bioconductor. DEGs with fold change > 2 were shown in red; DEGs with fold change <− 2 were in blue (P < .01 and FDR < 0.05). (B) GO annotation pathways of high-expression genes in HCC. (C) GO annotation pathways of low-expression genes in HCC. (D) Kaplan-Meier curves for HCC patients' overall survival in patients with high and low GLS expression (n = 286, P = .0127). (E) Kaplan-Meier curves for HCC patients' overall survival in patients with high and low PEPCK1 expression (n = 286, P = .0127). GO annotation pathways were generated using the ggplot2 package in R language. The size of the dots represents the number of genes. Dot color represents the P value. Red: high degree of enrichment, green: low degree of enrichment.
SIRT2 Expression in HCC
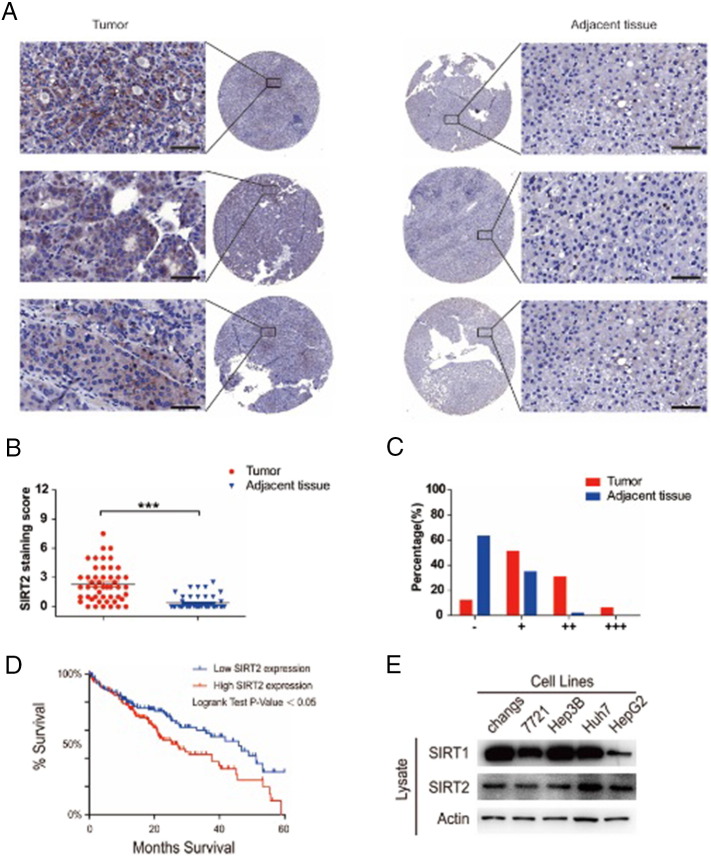
Acetylation has emerged as one such key posttranslational modification and is identified in metabolic enzymes in cellular regulation [3]. As the role of SIRT2 in HCC has not yet been elucidated, we queried tissue microarrays which contain clinically annotated data from 52 HCC samples and found that SIRT2 was significantly increased in tumor tissues as compared to adjacent tissues (Figure 2, A, B, and C). We queried the TCGA database, which contains clinically annotated genomic data from 286 HCC samples [21], [22], and found that a high SIRT2 level in HCC reduced patient survival (P = .0127, log-rank test) (Figure 2D). We further assessed the expression of SIRT1 and SIRT2 in HCC cell lines (changes, 7721, Hep3B, Huh7, and HepG2) and found that it was SIRT2, and not SIRT1, that was significantly expressed in these HCC cell lines (Figure 2E). These results suggest that SIRT2 expression was increased in HCC and was associated with reduced patient survival.
Figure 2.
SIRT2 expression was increased in HCC.
(A) SIRT2 protein levels in tumor and adjacent normal tissues from 52 HCC patients were detected and (B) quantified. The magnification is ×200. Scale bars, 100 μm. (C) After scoring, the different proportions of tumor and adjacent normal tissues were shown. (D) Kaplan-Meier curves for HCC patients' overall survival in patients with high SIRT2 expression and low SIRT2 expression (n = 286, P = .0127). (E) The expression of SIRT1 and SIRT2 in HCC cell lines (changes, 7721, Hep3B, Huh7, and HepG2) was determined by Western blot. Data represent the mean ± SEM, n ≥ 3. *P < .05; **P < .01. A score of “−”, no staining or membrane staining in less than 10% of the tumor cells; a score of “+”, a faint and partial membrane staining in greater than 10% of the tumor cells; scores of “++” and “+++”, a weak to moderate and a strong complete membrane staining in greater than 10% of the tumor cells, respectively.
The Effect of SIRT2 in HCC Cells
Studies have shown that the levels of SIRT2 expression and activity play a critical role in cell epigenetic alterations associated with cancer malignant biological behaviors [4]. Since most of the protein targets promote tumor proliferation, we evaluated the relationship between cell proliferation and SIRT2 expression in two human HCC cell lines (HepG2 and Hep3B). The effectiveness of the SIRT2 knockdown and overexpression was validated through Western blotting, and the protein levels of SIRT2 did not affect HCC cell proliferation (Figure 3, A and B). Previous research indicates that the SIRT2 inhibitor salermide has an antitumor effect [8], [9]. Three different cell lines including 7721, HepG2, and Hep3B were treated with different concentrations of salermide. After inhibiting SIRT2, cell growth was marginally affected (Figure 3C). Consistent with CCK8 assays, HCC cells displayed marginal difference in phenotypes after salermide treatment (Figure 3D). Similarly, after inhibition of SIRT2 by salermide, no significant difference in the amount of apoptotic cells was observed (Figure 3E). Collectively, these results reveal that SIRT2 did not affect the proliferation and apoptosis of HCC cells.
Figure 3.
SIRT2 did not affect proliferation and apoptosis in HCC cells.
(A) HepG2 and Hep3B cell proliferation was analyzed via CCK8 assay following siRNA transfection. (B) HepG2 and Hep3B cell proliferation was analyzed via CCK8 assay following transfection with SIRT2 overexpression vector. (C) 7721, HepG2, and Hep3B cells were treated with different concentrations of salermide and quantified via CCK-8 assay. (D) 7721, HepG2, and Hep3B cells were treated with salermide, and the morphological changes were observed. Scale bars, 200 μm. (E) 7721, HepG2, and Hep3B cells were treated with salermide; apoptotic cells were measured by flow cytometry (left) and quantified (right). Data represent the mean ± SEM, n ≥ 3. *P < .05, **P < .01.
SIRT2 Induced HCC Cell Migration
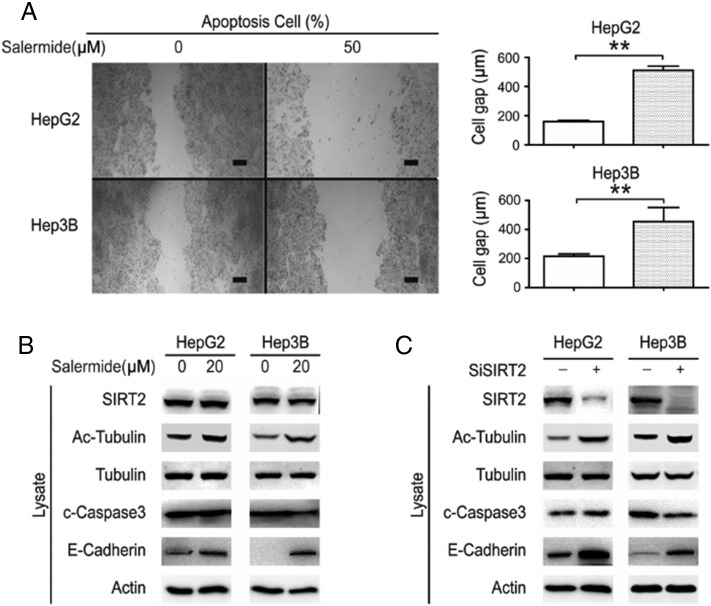
We next performed a wound-healing assay to observe the effect of inhibiting SIRT2 on cell migration—an important event in metastasis. The wound-healing assay was performed after treatment of HepG2 and Hep3B with salermide. A significant inhibitory effect on cell migration was observed when SIRT2 was inhibited (Figure 4A). To further explore the mechanism of SIRT2 induced cell migration, we investigated the effects of SIRT2 on their downstream targets (including Ac-tubulin) after inhibition of SIRT2 by either salermide treatment or siSIRT2 [4]. Western blotting demonstrated that acetylated α-tubulin was increased, and the molecular targets of apoptosis did not change after SIRT2 was inhibited (Figure 4, B and C). Interestingly, E-cadherin expression was increased when SIRT2 was inhibited, suggesting that SIRT2 induced the migration of HCC cells via the E-cadherin pathway (Figure 4, B and C).
Figure 4.
SIRT2 induced HCC cell migration by E-cadherin.
(A) HepG2 and Hep3B cells were treated with salermide; cell migration was photographed (left) and quantified (right). (B) HepG2 and Hep3B cells were treated with salermide; the downstream targets of SIRT2 (including Ac-tubulin, tubulin, c-caspase3 and E-cadherin) were visualized by Western blot. (C) SIRT2 was knocked down in HepG2 and Hep3B cells; the downstream targets of SIRT2 were visualized by Western blot. Scale bars, 500 μm.
SIRT2 Promote Mitochondrial Metabolism
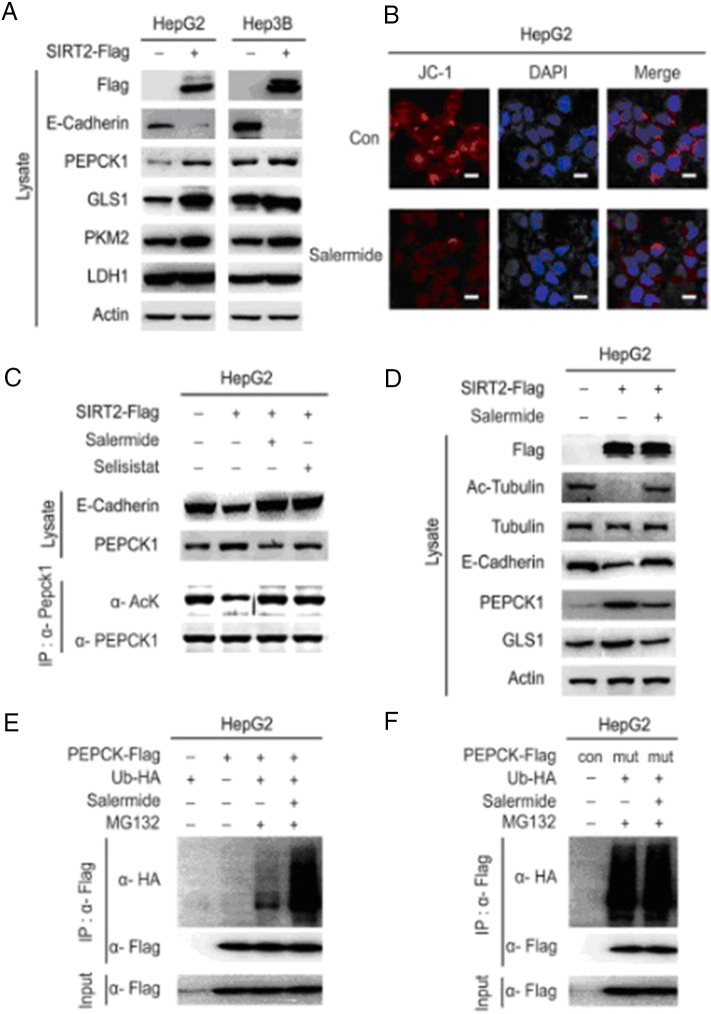
Sirtuins sense cellular energy requirements and availability, as well as direct metabolic pathways to ensure that energy production matches consumption. Accordingly, we aimed to identify SIRT2 downstream targets (PEPCK1, GLS1, PKM2, and LDH) involved in mitochondrial metabolism. We transfected Flag-SIRT2 into HCC cells and found that endogenous PEPCK1 and GLS1 were increased significantly (Figure 5A). Synergistically, we found that mitochondrial activity was significantly decreased after SIRT2 inhibition using mitochondrial activity staining with JC-1 (Figure 5B). This suggests that SIRT2 is a key target in promoting mitochondrial metabolism, as well as in the invasion of HCC cells. Wild-type SIRT2 overexpression reduced the acetylation of PEPCK1 and increased PEPCK1 protein levels. Conversely, SIRT2 inhibition by salermide decreased PEPCK1 by increasing the acetylation of PEPCK1 (Figure 5, C and D). It is worth noting that the protein marker of migration, E-cadherin, had a positive correlation with PEPCK1 (Figure 5, C and D). Consistent with these results, active PEPCK1 ubiquitinylation was detected, and the SIRT2 inhibitor significantly increased PEPCK1 ubiquitination (Figure 5E) after Flag-tagged PEPCK1 and HA-tagged ubiquitin were coexpressed in HCC cells. However, the role of SIRT2 inhibitor as a promoter of PEPCK1 ubiquitination was not observed after lysines (K) on three acetylation sites of PEPCK1 mutated to glutamine (Q) (Figure 5F). Overall, these results suggest that SIRT2 supported the migration of HCC by stabilizing downstream metabolic targets and promoting mitochondrial metabolism.
Figure 5.
SIRT2 affects the migration of HCC cells by promoting mitochondrial metabolism.
(A) SIRT2 overexpression vector was transfected into HepG2 and Hep3B cells, and the protein levels of E-cadherin, PEPCK1, GLS1, PKM2, and LDH were investigated by Western blot. (B) HepG2 cells were treated with salermide and stained with JC-1 and DAPI; the morphological changes were observed. Scale bars, 50 μm. (C) HepG2 cells were transfected with flag tagged SIRT2 plasmid, as well as treated with 20 μM salerminde for 24 hours. Cells were harvested and visualized by Western blotting. (D) HepG2 cells were transfected with flag tagged SIRT2 plasmid, as well as treated with 20 μM salerminde or selisistat for 24 hours. Cells were harvested and immunoprecipitated with an anti-PEPCK1 antibody, then visualized by Western blotting. (E) Flag-tagged PEPCK1 and HA-tagged ubiquitin were coexpressed in HepG2 cells and then treated with indicated salermide for 24 hours with or without 10 μM MG132 for 4 hours. Ubiquitination levels of affinity purified Flag-PEPCK1 proteins were detected and visualized by Western blotting. (F) Different phenotypic Flag-tagged PEPCK1 and HA-tagged ubiquitin were coexpressed in HepG2 cells and then treated with indicated salermide for 24 hours with 10 μM MG132 for 4 hours. Ubiquitination levels of affinity purified Flag-PEPCK1 proteins were detected and visualized by Western blotting.
Inhibitory Effect of SIRT2 in HCC Migration
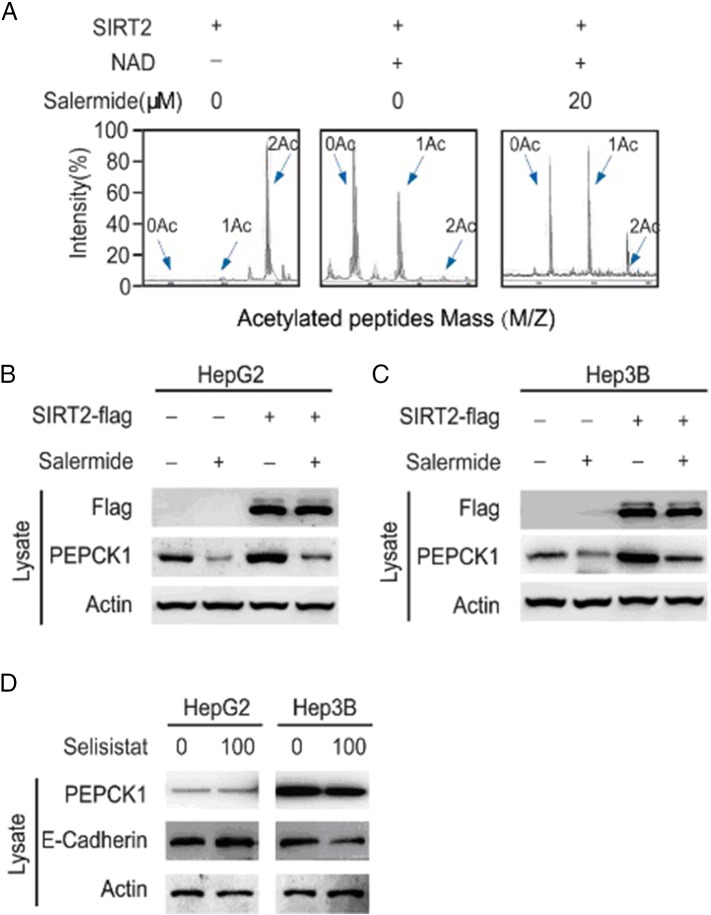
As a NAD+-dependent class III deacetylase, SIRT2 deacetylates and stabilizes PEPCK1 [4]. We therefore investigated whether SIRT2 is primarily responsible for decreasing protein levels of PEPCK1 with salermide. Since Lys70, Lys71, and Lys594 were identified as key acetylated sites of human PEPCK1 and these are deacetylated by SIRT2 [3], [4], we synthesized a peptide containing these three acetylated sites of PEPCK1 and incubated it with purified SIRT2 protein. We then assessed the peptides by mass spectrometry (Figure 6A). We found that SIRT2 overexpression reduced specific acetylation while salermide reversed this effect. As such, HepG2 and Hep3B cells were transiently transfected with Flag-SIRT2 or a control vector. The levels of PEPCK1 expression in both cells were significantly lower when cells were treated with salermide (Figure 6, B and C). Due to the high level of homology between the SIRT1 and SIRT2, salermide possibly had a weak inhibitory effect on SIRT1. In order to demonstrate that SIRT2 is the target of HCC, we employed the SIRT1-specific inhibitor selisistat to treat HCC cells and found that selisistat had a marginal effect on PEPCK1 and its downstream target E-cadherin (Figure 6C). Altogether, these data demonstrated that SIRT2 was a key molecule in the metastasis of HCC, and SIRT2 inhibition attenuated the migration of HCC.
Figure 6.
Inhibitory effect of SIRT2 reduces HCC migration.
(A) Purified SIRT2 protein was incubated with acetylated peptides with or without salermide, and the rate of deacetylation was determined using mass spectrometry. (B) HepG2 and Hep3B cells were transiently transfected with Flag-SIRT2 and then treated with or without indicated salermide for 4 hours. Cells were harvested and visualized by Western blot. (C) HepG2 and Hep3B cells were treated with selisistat, and cells were harvested as well as visualized by Western blot.
Effects of SIRT2 on Tumor Metastasis
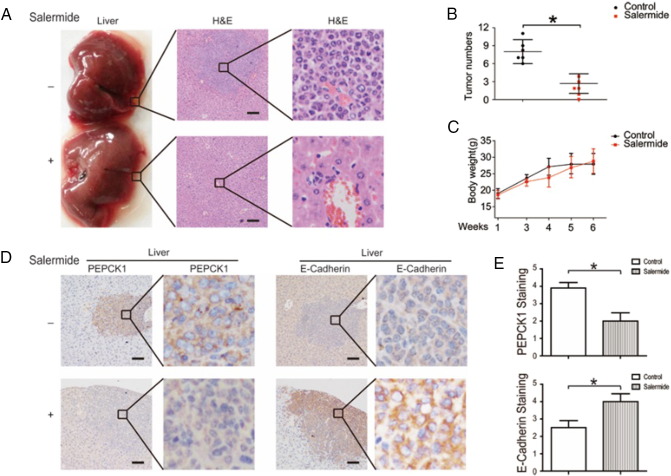
We employed the tail vein metastases model by injecting HCC cells to evaluate the in vivo antimetastasis effects of SIRT2 inhibition, and found that salermide administration significantly inhibited the formation of metastatic tumors (Figure 7, A and B). The body weights of treated mice were used as indicators of health [23]. Salermide treatment did not affect mouse body weight, suggesting that the mice did not experience evident toxicity in vivo (Figure 7C). Consistent with in vitro results, SIRT2 inhibition blocked the related downstream metabolic targets (Figure 7, D and E). Altogether, these data demonstrate that SIRT2 is a key molecule in the metastasis of HCC, and SIRT2 inhibition can attenuate the tumor metastases of HCC in vivo.
Figure 7.
Effects of SIRT2 inhibition on tumor metastasis.
(A) Systemic delivery of SirReal2 suppresses HCC cell liver metastasis in nude mice. Levers were photographed after all animals were sacrificed. Xenograft samples were stained with hematoxylin and eosin. Scale bars, 200 μm. (B) The number of liver metastases. (C) The body weights of tumor-burdened mice. (D-E) The immunohistochemistry staining of liver metastasis in nude mice. Xenograft samples were stained as indicated. Scale bars, 100 μm. Data represent the mean ± SEM, n = 5. *P < .05, **P < .01.
Discussion
HCC is one of the most common malignant cancers worldwide and the third most frequent cause of cancer-related mortality as a result of its highly aggressive nature and propensity for metastasis [24]. Chemical antimetastasis strategies have been widely developed in the past several decades. Nonetheless, new chemical anti-HCC drugs remain few and far between such that developing new strategies for treating HCC in the clinic, as well as identifying new tumor markers, remains an urgent necessity. Growing evidence suggests that acetylation is a conservative protein modification and that regulation of metabolic enzymes may affect cellular regulation. This provides a new strategy to develop anti-HCC drugs by regulating protein acetylation [3], [25]. In this study, we uncovered that downregulation of SIRT2 inhibits the invasion of hepatocellular carcinoma by inhibiting glycolysis.
Class III HDACs, also known as sirtuins, are NAD+-dependent protein deacetylases [4]. Seven sirtuin homologues have been identified in mammals as SIRT1 to 7. Cytosolic functions of SIRT2 include the regulation of microtubule acetylation, control of myelination in the central and peripheral nervous system, and gluconeogenesis. Both tubulin [26] and PEPCK [3] are known substrates of this deacetylase. Though studies have demonstrated that SIRT2 inhibitors may have antitumor and anti-inflammatory effects [8], [9], [10], the impact of SIRT2 on metabolism and possible molecular mechanisms in HCC cells have not yet been reported. Our results demonstrate that SIRT2 significantly promotes HCC cell migration and invasion, suggesting that SIRT2 may have an oncogenic role in HCC. This is consistent with previous studies, which suggested that SIRT2 is a key promoter of cell invasion in pancreatic cancer [27], [28], [29]. However, SIRT2 has also been reported as a tumor suppressor gene in a knockout mouse model and other cancer tissues [6], [21], [22], [30]. SIRT1 has likewise been noted to have tumor promoter and suppressor actions in a context-dependent manner. Therefore, it is possible that SIRT2 may promote tumor progression under certain circumstances, such as in human pancreatic cancer and HCC, as well as suppress tumor progression in other circumstances. Notably, we found that SIRT2 marginally promoted cell proliferation. This may be due to the various downstream targets of SIRT2. As in pancreatic cancer, LDH-A is the downstream target of SIRT2, and high levels of SIRT2 were able to block LDH-A degradation by reducing its acetylation. LDH-A overexpression in pancreatic cells led to proliferation [27], [31]. However, we found no difference between LDH-A levels in SIRT2-overexpressed HCC cells, indicating that different substrates of SIRT2 contributed to different malignant biological behaviors of the tumor.
PEPCK1 plays an important role in metabolism, and its regulation has been extensively studied. Both yeast and human PEPCK1 demonstrate acetylation, and catalytic activity is inactivated following this acetylation [3], [15], [28], [29]. Acetylation of Lys70, Lys71, and Lys594 of human PEPCK1 leads to decreased protein stability, reduced protein levels, and decreased gluconeogenesis without affecting mRNA levels [3]. PEPCK1 can promote glucose and glutamine utilization for anabolic metabolism [16]. Glutamine metabolism is also emerging as one aspect of dysregulated metabolism of tumors. GLS is the key enzyme in the conversion of glutamine to glutamate and is expressed in many tissue cells and cancer cells [17]. However, little is known about the role of PEPCK1 and GLS in HCC tumorigenesis. Consistent with previous studies, our data demonstrate that PEPCK1 and GLS stability is controlled via the balance between acetylation and deacetylation in response to SIRT2.
It is reported that some HDACs share 82% of their identity with each other [32]. Due to the high level of homology among HDACs, the inhibitory effects of the drug on other HDACs (especially SIRT1) besides SIRT2 could not be ignored. SIRT1, one of the most widely reported members of the SIRT family, is known to regulate cell proliferation, apoptosis, and migration [33], [34]. Inhibition of SIRT1 causes hyperacetylation of p53, Ku70, and FOXO3a, as well as phosphorylation of MAPK, leading to significant cytotoxic effects on breast and colon cancer cells [35], [36]. However, our cell viability test demonstrated that salermide was well tolerated in the absence of other noxious stimuli. This may be due to fewer malignant cells and the absence of stress stimuli (eg, cell starvation, salermide overdose, and chemotherapeutic agents) in our study. Therefore, selective SIRT2 knockdown is critical in order to determine whether SIRT2 is an anticancer target. Salermide, a new SIRT2 inhibitor, can be considered as an isotype-selective drug-like inhibitor with optimized potency and physicochemical properties compared with selisistat and other SIRT2 inhibitors. We evaluated this SIRT2 inhibitory effect by employing mass spectrometry. At the molecular level, downregulation of SIRT2 not only partially reversed cell migration and invasion but also reversed the PEPCK1 and GLS protein levels and the downstream E-cadherin pathway. This suggests that SIRT2 blocking inhibited HCC cell migration and invasion.
Previous studies have shown that mitochondrial metabolism [37] and E-cadherin may be critical in cancer cell invasion and metastasis [38]. The fueled TCA cycle leads to enhanced mitochondrial metabolism and is associated with tumor metastasis [37]. We found that SIRT2-related PEPCK1 and GLS degradation negatively correlates with the E-cadherin pathway in HCC, suggesting that this pathway is important in SIRT2-mediated HCC.
In the present study, we provide insight into the regulation of SIRT2 on HCC cellular migration and invasion. Based on our results, PEPCK1 and GLS are the downstream targets of SIRT2, and the inhibition of SIRT2 activity plays an important role in HCC cell migration as well as invasion. Thus, SIRT2 may be a novel molecular target for HCC therapy and may shed light on the underlying HCC treatment mechanism of salermide.
Author Contributions
Mingming Zhang, Robert G. Dorfman, and Tingsheng Ling designed the study; Dehua Tang, Qian Zhou, and Yida Pan did the cell experiments; Lixing Zhou, Yuyao Yin, Yuming Wang, and Wenjia Liu collected the tissue samples; Qian Zhou, Shan Huang, and Yang Li performed the protein analysis; Mingming Zhang, Shan Huang, and Zhenguo Zhao drafted the manuscript and performed the immunohistochemistry experiment; Robert G. Dorfman worked on manuscript preparation and editing; Shan Huang and Mingming Zhang supported the study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgements
We thank the Zhao lab for offering their help.
Footnotes
Grant support: This work was supported by grants from the National Natural Science Foundation of China (No. 81602076), the Jiangsu Clinical Medical Center of Digestive Disease (BL2012001), the Natural Science Foundation from the Department of Science & Technology of Jiangsu Province (BK20160113) and the Fundamental Research Funds for the Central Universities (No. 021414380244).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2017.09.006.
Contributor Information
Robert Gregory Dorfman, Email: rob.dorfman@yahoo.com.
Tingsheng Ling, Email: chinalts@126.com.
Mingming Zhang, Email: doczmm@126.com.
Appendix A. Supplementary data
The following are the supplementary data related to this article.
(A) Heat map of DEGs in liver HCC and paired normal liver samples. Heat map was drawn using the gplots package in Bioconductor. DEGs with fold change > 2 were shown in red; DEGs with fold change < −2 were in blue (P < .01) and FDR < 0.05. (B) KEGG pathways of high-expression genes in HCC. (C) KEGG pathways of low-expression genes in HCC. KEGG pathways were generated using the ggplot2 package in R language. The size of the dots represents the number of genes. Dot color represents the P value. Red: high degree of enrichment, green: low degree of enrichment.
References
- 1.Bruix J. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay JJ, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 5.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 6.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wölfel C, Huber C, Wölfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGlynn LM, Zino S, MacDonald AI, Curle J, Reilly JE, Mohammed ZM, McMillan DC, Mallon E, Payne AP, Edwards J. SIRT2: tumour suppressor or tumour promoter in operable breast cancer? Eur J Cancer. 2014;50:290–301. doi: 10.1016/j.ejca.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Heltweg B, Gatbonton T, Schuler AD, Posakony J, Li H, Goehle S, Kollipara R, Depinho RA, Gu Y, Simon JA. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Kim TH, Ahn MY, Lee J, Jung JH, Choi WS, Lee BM, Yoon KS, Yoon S, Kim HS. Sirtinol, a class III HDAC inhibitor, induces apoptotic and autophagic cell death in MCF-7 human breast cancer cells. Int J Oncol. 2012;41:1101–1109. doi: 10.3892/ijo.2012.1534. [DOI] [PubMed] [Google Scholar]
- 10.Liu FC, Day YJ, Liou JT, Lau YT, Yu HP. Sirtinol attenuates hepatic injury and pro-inflammatory cytokine production following trauma-hemorrhage in male Sprague-Dawley rats. Acta Anaesthesiol Scand. 2008;52:635–640. doi: 10.1111/j.1399-6576.2008.01592.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanson RW, Patel YM. Phosphoenolpyruvate carboxykinase (GTP): the gene and the enzyme. Adv Enzymol Relat Areas Mol Biol. 1994;69:203–281. doi: 10.1002/9780470123157.ch6. [DOI] [PubMed] [Google Scholar]
- 12.Nye CK, Hanson RW, Kalhan SC. Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J Biol Chem. 2008;283:27565–27574. doi: 10.1074/jbc.M804393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tannen RL. Ammonia metabolism. Am J Physiol. 1978;235:F265–277. doi: 10.1152/ajprenal.1978.235.4.F265. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarty K, Cassuto H, Reshef L, Hanson RW. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol. 2005;40:129–154. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 15.Lin YY, Lu JY, Zhang JM, Walter W, Dang WW, Wan J, Tao SC, Qian J, Zhao YM, Boeke JD. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montal ED, Dewi R, Bhalla K, Ou L, Hwang BJ, Ropell AE, Gordon C, Liu WJ, Deberardinis RJ, Sudderth J. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol Cell. 2015;60:1–13. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao Y, Wang J, Qin Y, Xuan Y, Jia Y, Hu W, Yu W, Dai M, Li Z, Yi C. Ku80 cooperates with CBP to promote COX-2 expression and tumor growth. Oncotarget. 2015;6:8046–8061. doi: 10.18632/oncotarget.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 20.Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 21.Temel M, Koc MN, Ulutas S, Gogebakan B. The expression levels of the sirtuins in patients with BCC. Tumour Biol. 2016;37:6429–6435. doi: 10.1007/s13277-015-4522-8. [DOI] [PubMed] [Google Scholar]
- 22.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 23.Wang LT, Liou JP, Li YH, Liu YM, Pan SL, Teng CM. A novel class I HDAC inhibitor, MPT0G030, induces cell apoptosis and differentiation in human colorectal cancer cells via HDAC1/PKCdelta and E-cadherin. Oncotarget. 2014;5:5651–5662. doi: 10.18632/oncotarget.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD(+)-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang P, Xu YH, Dong B, Xiong Y, Lei QY. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013;23:464–476. doi: 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Pan Y, Dorfman RG, Yin Y, Zhou Q, Huang S, Liu J, Zhao S. Sirtinol promotes PEPCK1 degradation and inhibits gluconeogenesis by inhibiting deacetylase SIRT2. Sci Rep. 2017;7:7. doi: 10.1038/s41598-017-00035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20:487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 33.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 34.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 35.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappa B-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu YF, Sheu JR, Lin CH, Yang DS, Hsiao G, Ou G, Chiu PT, Huang YH, Kuo WH, Hsu MJ. Trichostatin A and sirtinol suppressed survivin expression through AMPK and p38MAPK in HT29 colon cancer cells. Biochim Biophys Acta. 2012;1820:104–115. doi: 10.1016/j.bbagen.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 37.LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [1001–1015] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Heat map of DEGs in liver HCC and paired normal liver samples. Heat map was drawn using the gplots package in Bioconductor. DEGs with fold change > 2 were shown in red; DEGs with fold change < −2 were in blue (P < .01) and FDR < 0.05. (B) KEGG pathways of high-expression genes in HCC. (C) KEGG pathways of low-expression genes in HCC. KEGG pathways were generated using the ggplot2 package in R language. The size of the dots represents the number of genes. Dot color represents the P value. Red: high degree of enrichment, green: low degree of enrichment.