Abstract
The Catsper1 gene, whose expression is restricted to male germ cells, has great importance in reproductive biology because of its function in sperm motility and fertilization. We previously reported that the promoter of this gene has transcriptional activity in either direction in a heterologous system. In the present study, we found that the Catsper1 promoter has in vitro transcriptional activity in either orientation in GC-1 spg mouse spermatogonial cells. The results also showed that this promoter regulates the expression of a new divergent Catsper1 gene named Catsper1au (Catsper1 antisense upstream transcript). Catsper1au is expressed in adult male mouse testis and liver tissues but not in female mouse liver or ovary tissues. In the testis, Catsper1au is expressed in embryos at 11.5 days post-coitum and from newborns to adults. This gene is also expressed in 1- to 3-week postnatal hearts and in 1-week to adult stage livers. The analysis of the 1402 bp whole genome sequence revealed that Catsper1au is an intronless and polyadenylated lncRNA, located in the nuclei of Sertoli and spermatogenic cells from adult testis. These data indicate that Catsper1au is divergently expressed from the Catsper1 promoter and could regulate gene expression during spermatogenesis.
Introduction
The genes expressed during spermatogenesis encode proteins and non-coding RNAs, which enable the progress of specific processes necessary for germ cell development1,2. The activation of these genes is highly controlled and includes mechanisms of transcriptional regulation in cis or trans that involve promoters, alternative splicing, transcriptional factors, enhancers, epigenetic mechanisms, and the more recently studied noncoding RNAs3–5. Male germ cells produce transcripts homologous to genes expressed in somatic cells, such as Gapdhs and alternative transcripts (Hk1s) expressed from the same gene in somatic cells6. There are also unique transcripts that do not have similarity to any other somatic transcripts, such as Prm1 and Catsper 3,7, and transcripts that encode proteins and non-coding RNAs whose function has not yet been characterized. CATSPER is a cation channel exclusively detected in sperm cells and located in the plasma membrane of the principal piece of the flagellum. CATSPER comprises a 4-protein family forming a tetrameric channel (CATSPER 1–4)8–10. The 4-member family associates with accessory proteins (CATSPERβ, CATSPERγ, and CATSPERδ)11–13. This channel mediates Ca2+ entry necessary for the hyperactivation motility and fertility since it is required to penetrate the zona pellucida surrounding the oocyte7. The four subunits are expressed at different stages of spermatogenesis. While Catsper1, 3 and 4 are transcribed in spermatids, indicating common transcriptional regulation, Catsper 2 is found in pachytene spermatocytes7,14–16. The Catsper1 gene was the first discovered in this family and differs from the other familial genes in its histidine-rich cytoplasmic domain, which senses changes in intracellular pH and activates the CATSPER channel7,17. However, the disruption of any of the four Catsper genes results in a nonfunctional channel leading to deficient hyperactivation motility and male infertility16. However, mutations in CATSPER1 and 2 have been associated with human infertility18,19. These facts make the CATSPER channel a target for developing a male contraceptive and for the study of male infertility. Previously, we characterized the promoter of human and mouse Catsper1 genes to understand the molecular mechanisms underlying the transcriptional regulation of this gene. Although in a heterologous system, the mouse Catsper1 promoter showed unusual bidirectional transcriptional activity even higher in antisense than in sense (Catsper1) orientation20. These data suggest that the mouse Catsper1 promoter could be the bidirectional promoter responsible for the transcription of a new gene in the antisense strand. However, there are no annotated genes in the antisense orientation in the 10.4-kb region upstream of Catsper1 transcriptional start site (TSS) and downstream of the Gm7074 pseudogene. Therefore, in the present study, the bidirectional transcriptional activity of the Catsper1 promoter was analysed in a homologous system and whether a new divergent Catsper1 gene is expressed. The Catsper1 promoter gene showed bidirectional transcriptional activity in spermatogonial GC-1 spg cells. Two TSSs were determined using 5′-RACE for the new gene named Catsper1au (Catsper1 antisense upstream transcript) in adult testis. Unlike Catsper1, the antisense transcript was not exclusive of the testis. The in silico analysis of the whole sequence revealed that Catsper1au is polyadenylated, intronless and does not contain translatable open reading frames (ORF). No protein was produced in an in vitro-coupled transcription and translation assay. Catsper1au was restricted to the nucleus of spermatogenic and Sertoli cells.
Results
Catsper1 promoter has antisense transcriptional activity in mouse spermatogonial GC-1 spg cells. Previous results have suggested that the Catsper1 promoter may act as a bidirectional promoter driving the expression of a divergent gene to Catsper1 20. To determine whether this promoter presents bidirectional transcriptional activity in a homologous system, spermatogonial (GC-1 spg) cells were transiently transfected with pGL3-800 and pGL3-1200 constructs. These constructs contain the Catsper1 promoter regions in either direction (coordinates −75 to +23 and −1192 to +23, relative to Catsper1 TSS + 1) upstream of the Photinus luciferase reporter gene (Fig. 1a). Both the 800- and 1200-bp promoter regions showed bidirectional activity; however, their antisense (Catsper1au) activities were up to 19.3- and 36.6-fold higher, respectively, while sense (Catsper1) activities showed only 7- and 4.4-fold higher activity compared with pGL3-Basic vector (Fig. 1b). These results indicate that the Catsper1 promoter has bidirectional transcriptional activity in spermatogonial cells.
Figure 1.
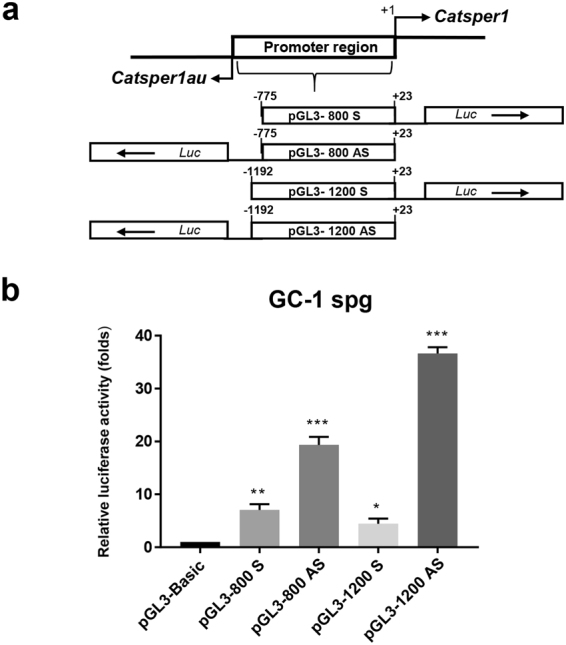
The Catsper1 promoter has bidirectional transcriptional activity in GC-1 spg spermatogonia cells. (a) Structure of the Catsper1 promoter constructs. Each construct was named according to the length of the promoter region inserted in either direction upstream of the Photinus luciferase gene (Luc). (b) Transcriptional activity in GC-1 spg spermatogonia cells. Transcriptional activity is expressed as a fold-increase of luciferase activity over the pGL3-Basic (empty vector), to which a value of 1 was assigned. The results represent the average activity of Photinus firefly luciferase normalized to the activity of Renilla reniformis luciferase as an internal transfection control. Bar graphs represent the means ± SD, n = 3. Statistical significance was evaluated with one-way ANOVA: *P < 0.05, **P < 0.01, ***P ≤ 0.001.
In silico analysis of the 10.4-kb region upstream of the mouse Catsper1 gene: Identification of the TSS of the new Catsper1au gene
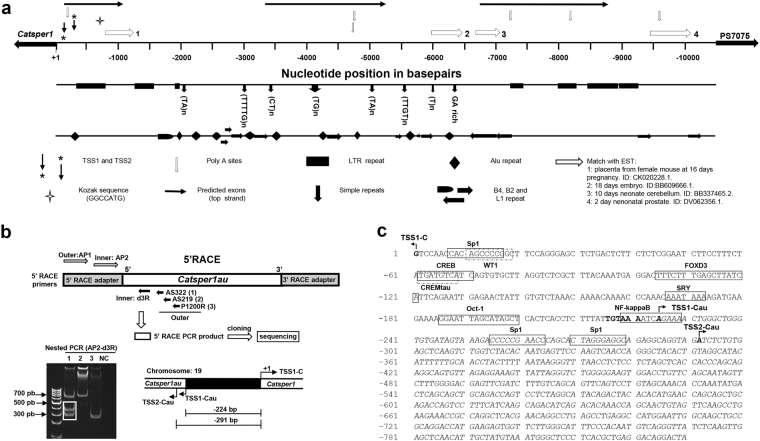
According to the mouse genome NCBI database, there are no annotated genes in the region upstream of Catsper1 in the complementary DNA chain. However, the antisense transcriptional activity of the Catsper1 promoter in vitro suggests the presence of a divergent gene in this region. Therefore, an in silico analysis of the 10.4-kb region downstream of the Gm7074 pseudogene (ID: 631868) and upstream of Catsper1 gene (ID: 225865) was performed (Fig. 2a). The sequence showed a Kozak sequence, poly (A) sites, and two TSSs, at 121 and 289 bp upstream of Catsper1 TSS, but no CpG islands were detected. Several matches with EST databases were also found. The analysis of this region also indicated a GC content of 45.10%. The presence of splicing donor/acceptor sites in this region predicted a putative Catsper1 divergent gene encoding a 4.4-kb transcript with three exons. Repetitive sequences are primarily located in the non-transcribed genomic regions or within intronic sequences21,22. The in silico analysis revealed that 58.02% of the 10.4 kb region contains repeats, such as: long terminal repeats (LTR); short interspersed nuclear elements (SINE), which include Alu (B1), B2 and B4; and long interspersed nuclear elements (LINE), such as L1 and simple repeats, whose content was 19.21, 28.23, 5.91 and 4.18% (Fig. 2a).
Figure 2.
In silico analysis of the 10.4 kb region upstream of the mouse Catsper1 gene and determination of the divergent gene TSSs. (a) The new putative Catsper1 divergent gene and its structure were predicted using different web servers (see Material and Methods). Catsper1 TSS is indicated. The new gene putative TSSs predicted at −121 and −289 bp are indicated. Predicted poly(A) and Kozak sequences, as well as exons and matches to EST 1–4, are indicated. LTR, simple repeats, Alu, B4, B2, and L1 repeat sequences are also indicated. (b) TSSs of the new gene Catsper1au. TSS were determined by 5′-RACE using a nested PCR as outlined in Materials and Methods. Two Catsper1au TSSs were confirmed as indicated by the sequencing of the 300 and 350 bp bands obtained in the second reaction of the nested PCR with primers AP1-AS322 (boxed in lane 1). The distances between Catsper1au TSS1 and TSS2 (labelled as TSS1-Cau and TSS2-Cau) from Catsper1 TSS-C were 224 and 291 bp, respectively. (c) Schematic representation of the Catsper1 bidirectional promoter in testis. Bent arrows indicate TSS-C (+1 bp) and TSS1-Cau (−225) and TSS2-Cau (−292). The arrows represent the direction of gene transcription. The predicted cis-acting elements in the 5′ upstream region of Catsper1au (transcriptional factors) are boxed; the TATA box (−216 bp) is bolded. The 5′ Catsper1au sequence, determined using 5′-RACE, is shown in italics.
Bidirectional promoters regulate the transcription of genes expressed in the same tissue and frequently have related functions23,24. Since Catsper1 is expressed only in spermatogenic cells7, we performed a 5′-RACE from adult mouse testis RNA to analyse whether the transcript predicted in silico was indeed expressed and to determine the distance between the new gene and Catsper1 TSS. To this end, primers specific for sequences within the first predicted exon were tested in nested PCR (Table 1 and see Materials and Methods). The amplified fragments from three different reactions were cloned into pJET and subjected to sequence analysis to confirm their identity (Fig. 2b). Two TSSs were identified for the new gene Catsper1au at −224 and −291 bp from the Catsper1 TSS (labelled as TSS1-C). The other products were nonspecific amplifications. Thus, the shortest intergenic distance was less than 1 Kb. These experimental TSSs were near the TSSs predicted in silico using the Network promoter programme. The intergenic sequence between Catsper1 and Catsper1au showed a TATA box located at −216 bp relative to TSS-C, and the presence of putative testis-specific transcription factor-binding sites, such as CREMτ and SRY25,26 (Fig. 2c).
Table 1.
Primers used.
| Name | Sequence (5′→3′) | Size | Experiment |
|---|---|---|---|
| d3R (R) | AGACCCTAATAACTTTCCTCTAACACTGCCTCTGC | 35 | 5′-RACE |
| AS322 (R) | TAGCTCCCTCTTAGTCCTGTC | 21 | 5′-RACE |
| AS219 (R) | CCAGTTGAGTGATGACGGTC | 20 | 5′-RACE |
| P1200 (R) | AGGGGTAACTTGGAGGAT | 18 | 5′-RACE |
| Adaptor Primer 1(AP1) (F/R) | CCATCCTAATACGACTCACTATAGGGC | 27 | 5′/3′-RACE |
| Nested Adaptor Primer (AP2) (F/R) | ACTCACTATAGGGCTCGAGCGGC | 23 | 5′/3′-RACE |
| Act (F) | TGACGGGGTCACCCACACTGTGCCCATCTA | 30 | RT-PCR |
| Act (R) | CTAGAAGCATTTGCGGTGGACGATGGAGGG | 30 | RT-PCR |
| Cat1 (F) | CTGAGCTAGAGATCCGAGGTG | 21 | RT-PCR |
| Cat1 (R) | CAATTAGCTTGAGGACTGCTTCT | 23 | RT-PCR |
| NGFORW225 (F) | AGAAAACTGGGCTGGGTGTGATAGTAAAG | 29 | RT-PCR, Catsper1au Cloning |
| NGFORW292 (F) | ATCTCTGTGAGCTCAAGTCTGGTCTACAC | 29 | RT-PCR |
| AS306REV (R) | GGCAGCTTGCCAATTCCATGGCCTCAGG | 28 | RT-PCR |
| NGFORW225RT(F) | AGAAAACTGGGCTGGGTGTGATAG | 24 | qPCR |
| REVD3(R) | TGGTGAGCTAGAGGAGAGGTTAAAC | 25 | qPCR |
| ActBForw (F) | AAGATCAAGATCATTGCTCCTCC | 23 | qPCR |
| ActBRev (R) | TAACAGTCCGCCTAGAAGCA | 20 | qPCR |
| 3′RACE-Forw1 (F) | CAGAGGCAGTGTTAGAGGAAAGTTATTAG | 31 | 3′RACE |
| 3′RACE-Forw2 (F) | GCTGAGGACAGGACTAAGAGGGAGC | 25 | 3′-RACE |
| NG-Reverse (F) | CGGACTTGATTTGGCATTACCCTAATGGG | 29 | RT-PCR, Catsper1au Cloning |
| ENGFORW225 (F) | GAATTCAGAAAACTGGGCTGGGTGTGATAGTAAAG | 35 | Catsper1au Cloning |
| BRACE3REV (R) | GGATCCTCGGACTTGATTTGGCATTACCCTAATGGG | 36 | Catsper1au Cloning |
| Rnu1a1 (F) | ATACTTACCTGGCAGGGGAGA | 21 | RT-PCR |
| Rnu1a1 (R) | CAGGGGAGAGCGCGAACGCA | 20 | RT-PCR |
| HNGF225 (F) | AAGCTTAGAAAACTGGGCTGGGTGTGATAGTAAAG | 35 | RNA-FISH/NB |
| ECOAS306 (R) | GAATTCGGCAGCTTGCCAATTCCATGGCCTCAGG | 34 | RNA-FISH/NB |
| ActH3 (F) | AAGCTTTGACGGGGTCACCCACACTGTGCCCATCTA | 36 | RNA-FISH |
| ActECO (R) | GAATTCCTAGAAGCATTTGCGGTGGACGATGGAGGG | 36 | RNA-FISH |
Abbreviations: R, reverse; F, forward; NB, Northern blot.
Expression profile of Catsper1au in different tissues
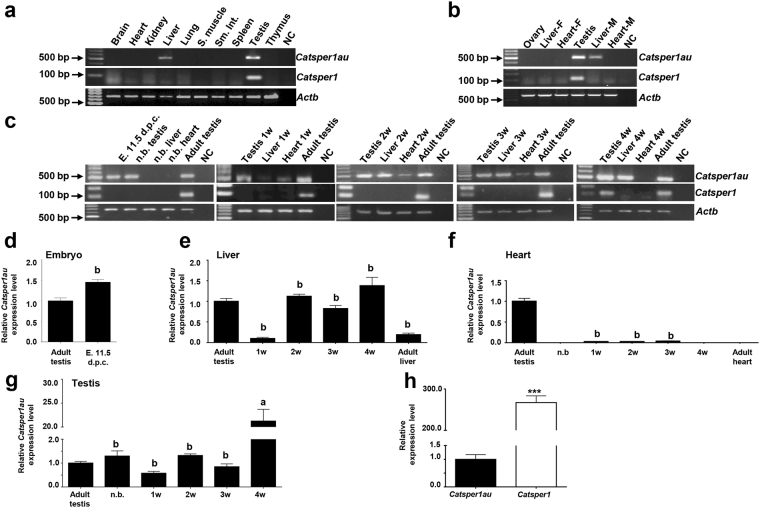
To ascertain whether similar to Catsper1, Catsper1au is only expressed in adult mouse testis, RT-PCR from various adult tissues was performed using oligonucleotides based on the 5′-mRNA sequence obtained by 5′-RACE. As a control, Actb expression was analysed. Surprisingly, Catsper1au was also expressed in the liver (Fig. 3a) but not in the adult brain, heart, kidney, lung, skeletal muscle, small intestine, spleen, or thymus. Additionally, Catsper1au was only detected in adult male mice since no signal was observed in adult female ovary, liver, or heart tissues (Fig. 3b). The expression profile of Catsper1au was also analysed at different stages during male mouse development. Catsper1au was expressed at the embryonic stage (11.5 d.p.c.), newborn testis and continued until the adult stage. The expression was detected from the first week until the adult stage in the liver. Interestingly, this gene was also expressed in heart tissue from 1-to 3-week-old mice (Fig. 3c). To verify these results and assess the relative expression levels of Catsper1au RNA, qRT-PCR was performed in tissues where Catsper1au was detected using RT-PCR and tissues where Catsper1au was not amplified, including adult, new born and 4-week heart (Fig. 3d–g). Interestingly, Catsper1au RNA expression was significantly higher in 4-week testis compared with adult testis (Fig. 3d). This finding may be relevant because this lncRNA could play an active role in adolescent mice, where Catsper1 expression is initiated in CD-1 mice (Fig. 3c). However, Catsper1 mRNA expression in adult mouse testis was higher compared with Catsper1au RNA expression (Fig. 3e).
Figure 3.
Expression profile of Catsper1au in male and female mouse organs. A cDNA panel was prepared from different organs and cells to amplify the Catsper1au 5′ region through PCR based on the sequences obtained using 5′-RACE. A product of 496 bp was produced using oligonucleotides AS-306REV and NG-FORW292. (a) Catsper1au expression from different adult mouse organs. S: smooth; Sm. Int.: small intestine. (b) Catsper1au expression in adult male and female gonads and organs. F: Female. M: Male. (c) Catsper1au expression profile at different stages of male mouse development. E: embryo; d.p.c: days post-coitum; n.b: newborn. W: week. A product of 93 bp was amplified for Catsper1 in 4-week and adult testis. Actb was analysed as an internal expression control and to verify the absence of genomic DNA. The negative control (NC) is shown in the last lane. (d–g) Catsper1au RNA expression levels analysed using qRT-PCR in different tissues. Expression levels were normalized with respect to Actb mRNA. Values are expressed as fold-variations of each tissue type relative to adult testis (to which a value of 1 was assigned). Statistical significance was evaluated using one-way ANOVA: a > b. (h) Catsper1 expression relative to Catsper1au. Catsper1 was expressed as fold-changes in the expression of Catsper1au (***P < 0.001; paired t test). The results are expressed as the means ± S.E.M., n = 3 for each tissue.
Catsper1au is a lncRNA
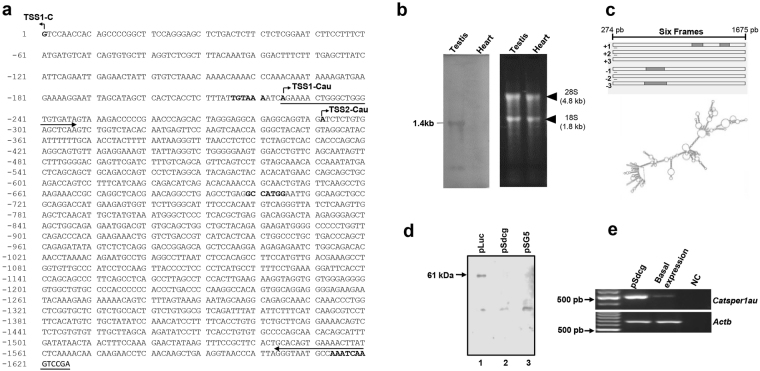
Catsper1au was characterized in mouse adult testis since at this stage Catsper1 and Catsper1au are expressed from the Catsper1 bidirectional promoter. To determine the full-length Catsper1au sequence, first the 3′-mRNA sequence was determined by 3′-RACE using the oligonucleotides indicated in Table 1. Catsper1au was subsequently amplified by RT-PCR using primers flanking the 5′ and 3′ ends. The resulting cDNA was cloned and sequenced to complete the whole nucleotide sequence. An unspliced 1402-bp transcript was obtained (Fig. 4a). This expression was confirmed using a Northern blot assay, where a ~1.4-kb band was detected (Fig. 4b). In addition, only a single band was apparently observed, although the two TSSs were determined by 5′-RACE in the same tissue. Either a single transcript is only produced or the difference of 67 bp between the two transcripts is difficult to distinguish from the Northern blot analysis. However, the size did not correspond with the in silico-predicted transcript length. A putative Kozak sequence starts at −699 bp and the poly (A) signal sequence with a single nucleotide change at −1,614 bp relative to the TSS-C. Surprisingly, no significant open reading frames (ORFs) containing a Kozak consensus sequence and longer than 100 base pairs were observed throughout the 1402-bp transcript (Fig. 4c). This analysis might indicate that this transcript is a lncRNA. In fact, the RNAfold server predicted a complex stable secondary structure for this transcript, a major feature of lncRNAs to fold into thermodynamically stable secondary and higher-order structures (Fig. 4c). To ascertain whether this RNA is translated, an in vitro-coupled transcription and translation assay was performed. To this aim, the pSdC1 construct, derived by cloning the 1402-bp Catsper1au gene under the T7 promoter in the pSG5 vector, was used to prime the reaction using a rabbit reticulocyte system in the presence of biotinylated lysine. The luciferase gene, encoding a 61-kDa protein, was used as a positive control (Fig. 4d). No protein products were detected, suggesting that Catsper1au is a lncRNA transcript. To verify whether Catsper1au is indeed transcribed, we transfected pSdC1 into adult primary germ cells, and Catsper1au was analysed using RT-PCR. As expected, compared with basal expression, Catsper1au was overexpressed (Fig. 4e).
Figure 4.
Sequence and characteristics of Catsper1au. The full length of the Catsper1au transcript (1402 bp) was determined by 5′/3′-RACE, cDNA cloning into pCG5 vector and sequencing. (a) A Kozak sequence (−699 bp) and the poly (A) signal sequence (−1,614 bp) are bolded. Bent arrows indicate TSS-C (+1 bp), TSS1-Cau (−225) and TSS2-Cau (−292). The 1402 bp transcribed region is indicated by the two bolded arrows from −225 to −1626 bp. (b) Expression of the 1.4 kb Catsper1au RNA in mouse adult testis. Total RNA from adult testis and heart was examined by Northern blot analysis using a digoxigenin-UTP-labelled riboprobe for Catsper1au (left panel). Ethidium bromide staining of ribosomal RNA (right panel). (c) The in silico analysis in all of the six reading frames did not reveal long open reading frame (ORF). (c) Predicted secondary structure of Catsper1au transcript. (d) Catsper1au transcript is not translated. A rabbit reticulocyte lysate system was used to express Catsper1au from pSG5 in vitro. Positive control, a construct coding 61-kDa luciferase (lane 1); pSdC1, a pSG5 derivative containing the 1.4 Kb RNA gene downstream of T7 promoter (lane 2); negative control, empty vector (pSG5) (lane 3). (e) Overexpression of Catsper1au RNA by RT-PCR in mouse germ cells. Actb was analysed as an internal expression control to verify the absence of genomic DNA.
Catsper1au is a nuclear lncRNA located in testis germ and somatic cells but not in sperm cells
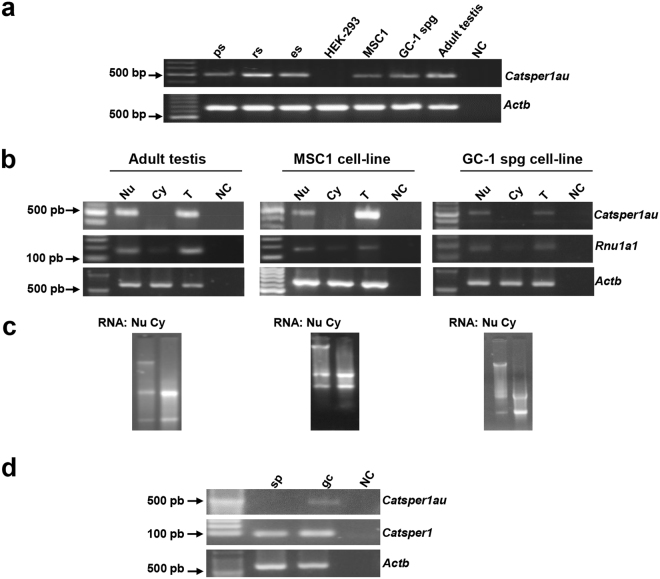
The subcellular localization of Catsper1au was analysed in pachytene spermatocytes, round and elongated spermatids, mouse Sertoli cells (MSC1) and GC-1 spg cells since Catsper1au was amplified from these cells using RT-PCR (Fig. 5a). Mouse adult testis tissue, MSC1, and GC-1 spg cells were fractionated into the nucleus and cytoplasm by differential centrifugation, and RNA was subsequently isolated (Fig. 5c). RT-PCR from Catsper1au was performed using specific primers. Catsper1au was only amplified from both total and nuclear RNA but not from cytoplasmic RNA (Fig. 5b). Rnu1a1, a small nuclear RNA (snRNA) component of the spliceosome located in the nucleus27, was used to assess the purity of the nuclear and cytoplasmic fractions. Amplification from the nuclear fraction was higher than that from the cytoplasmic fraction. Thus, Catsper1au RNA was predominantly detected in the nucleus, which is not surprising since numerous lncRNAs preferentially show nuclear localization28. To analyse Catsper1au expression in different subpopulations of germ cells, these cells were fractioned into pachytene spermatocytes, round and elongated spermatids using a BSA gradient. Catsper1au was specifically amplified by RT-PCR from these fractions (Fig. 5a); however, Catsper1au was not detected in differentiated sperm cells obtained from epididymis (Fig. 5d).
Figure 5.
The lncRNA Catsper1au displays a nuclear localization and is expressed in germ and somatic cells from testis. (a) Catsper1au expression in cell lines and testicular germ cells at different stages of spermatogenesis. (b) Nuclear and cytoplasmic expression of Catsper1au RNA in testis, MSC1, and GC-1 spg cells. Rnu1a1 was used as a control to verify the quality of fractions. (c) Electrophoretic analysis of RNA fractions in a 1.2% agarose gel. (d) Catsper1au is not expressed in sperm cells. Actb was analysed as an internal expression control and to verify the absence of genomic DNA. ps: pachytene spermatocyte; rs: round spermatid; es: elongated spermatid; sp: sperm cell; gc: germ cells.
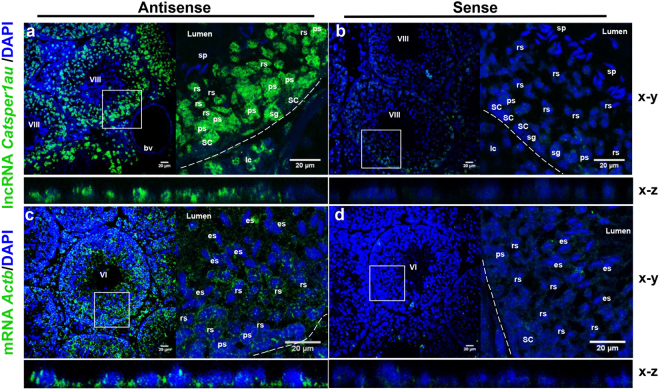
The subcellular localization and expression of Catsper1au in different germ cell subpopulations were further examined in vivo using RNA fluorescence in situ hybridization (FISH) on adult mouse testis, using an antisense probe derived from Catsper1au. Figure 6a shows that Catsper1au RNA localizes at the seminiferous tubules in the nucleus of somatic Leydig and Sertoli cells, spermatogonia cells, pachytene spermatocytes and round spermatids but is absent in the nucleus of sperm cells. A Catsper1au sense probe yielded a faint signal (Fig. 6b). In addition, RNA-FISH using an Actb antisense probe as a positive control, produced a specific signal both in the nuclei and cytoplasm of all cells present in the seminiferous tubule (Fig. 6c), while the Actb sense probe yielded a weak signal (Fig. 6d). Compared with Actb, higher concentrations of the riboprobe and anti-digoxigenin antibody (see Materials and Methods) were used to detect lncRNA Catsper1au based on its reduced expression observed using qRT-PCR (Fig. 3). The RNA-FISH analysis confirmed the nuclear localization of Catsper1au. These findings also corroborated the lack of Catsper1au signal in spermatozoa and its expression in pachytene spermatocytes, round and elongated spermatids (Fig. 5). Taken together, the RT-PCR and RNA-FISH results indicated that Catsper1au is a lncRNA present in the nuclei of germ and somatic cells of the testis, but not in sperm cells.
Figure 6.
Catsper1au is located in vivo in the nucleus of spermatogenic and somatic cells from the seminiferous tubule. (a) Catsper1au RNA was analysed in the mouse testis by FISH using antisense RNA probe derived from Catsper1au. (b) Sense Catsper1au RNA was included as a negative control. Antisense (c) and sense (d) Actb RNA probes were also included as controls. Dashed line, basal border of seminiferous tubule; VI and VIII, stages in the cycle of the seminiferous epithelium; SC: Sertoli cell; sg: spermatogonia cell; ps: pachytene spermatocyte; rs: round spermatid; es: elongated spermatid; sp: sperm cell. Blood vessel: bv; lc: Leydig cell.
Discussion
In the present study, we demonstrated that the Catsper1 promoter is bidirectional in GC-1 spg spermatogonial cells. A novel lncRNA (Catsper1au) is expressed divergently from the Catsper1 promoter. Catsper1au and Catsper1 TSSs are separated by 224 bp. Similar to Catsper, Catsper1au is expressed in adult testis and spermatogenic cells but also in Sertoli cells and liver tissues. No signal was observed in the ovary. In addition, Catsper1au is expressed from embryonic to adult stage in the testis, in the 1- to 3-week postnatal heart and in one week to adult stage liver, and unlike Catsper1, it is not detected in mature sperm cells. In addition, Catsper1au, which encodes a highly structured 1402-bp lncRNA, is located in the nucleus of Sertoli and germ cells from adult testis.
Because of the importance of Catsper1 in the biology of reproduction, the characterization of its promoter is an interesting study model since it is expressed with high specificity and site-temporality during spermatogenesis. The Catsper1 promoter bidirectional activity previously reported20 was confirmed in the present study in homologous GC-1 spg cells (Fig. 1b). However, a lower activity was detected in the sense direction, consistent with a lack of Catsper1 expression in spermatogonial cells in vivo. In addition, GC-1 spg cells do not express the CREMτ transcription factor, which is involved in gene expression in haploid spermatids during spermiogenesis25,29, a stage where Catsper1 is expressed15. Nevertheless, it has been reported that the activity of a bidirectional promoter may be regulated by orientation-dependent cis-control elements or trans-acting factors. These mechanisms could positively regulate the expression of Catsper1au in GC-1 spg cells24.
Another feature of bidirectional promoters is that the TSSs of divergent genes are separated by less than 1 kb30. The intergenic region between Catsper1 and Catsper1au divergent genes is consistent with the characteristics of bidirectional promoters30 since the two TSSs identified for Catsper1au are −224 and −291 bp upstream of the Catsper1 TSS (Fig. 2b). The majority of divergent gene pairs are co-regulated, have related functions, function in a common pathway and show tissue-specific expression24 similar to Col4a1 and Col4a2, HAND2 and HAND2-AS1 31,32. Although Catsper1au and Catsper1 are expressed in adult testis, suggesting that both genes are co-regulated through their shared proximal promoter, Catsper1au transcripts is unidirectionally expressed in adult liver. Therefore, these genes do not exhibit the same tissue-specific expression pattern (Fig. 3a). The genes regulated by a bidirectional promoter are not always co-expressed or co-regulated and may even be functionally unrelated, such as SERPINI1 and PDCD10 24. Interestingly, similar to Catsper1, Catsper1au is only expressed in male gonads but not in female gonads, potentially suggesting a specific role in the development of the male reproductive system (Fig. 3b), associated with the expression of Catsper1au in the embryonic stage (11.5 d.p.c.). Mammalian sex differentiation relies on the expression of the Sry gene, which in mouse begins at 10.5 d.p.c and peaks at 11.5 d.p.c33. At this stage, the participation of Sry and a set of male-specific genes in testis development might involve Catsper1au (Fig. 3c). However, Catsper1au was not expressed in the adult female liver (Fig. 3b). In this respect, male and female liver display sex-dependent gene-expression primarily regulated by pituitary GH, whose regulation is driven by oestrogen and testosterone sex hormones34. In this sense, SULT2A, a gene that participates in hydroxysteroid sulphate conjugation, is repressed in the adult male liver, while the transcript is only expressed in the adult female liver35. Thus, the Catsper1au expression in the male liver could be hormonally regulated. Additionally, Catsper1au is expressed in newborn to four-week testis and continues until the adult stage, while Catsper1 expression is restricted to adult testis. Accordingly, the expression throughout mouse life also suggests that Catsper1au might have a function in testis development and male germ cell differentiation. The first cycle of spermatogenesis in the mouse starts after birth, and the time points at which spermatogenic cell types appear are well defined36. Indeed, Catsper1au expression is higher in adolescent 4-week mice, where the spermatids reach the elongation phase, suggesting an important role at this stage.
Surprisingly, Catsper1au is expressed in 1-to 3-week-old but not in adult mouse hearts. The transition of cardiomyocytes to differentiated cells that are unable to proliferate occurs in the first two weeks post birth in the mouse heart37. At this time, the expression of genes, such as FRNK, has been implicated in this process, peaking at days 5 to 7 post birth and decreasing in the adult stage38, prompting the question of whether Catsper1au plays a role at this stage. Catsper1au expression in the liver was initiated from the first week and continued to the adult stage. Liver gene expression at one week after birth matches with the activation of genes involved in lipid and fatty acid metabolism39 and the genes that promote liver growth, such as Mest, Peg3, and Igf2 34. Catsper1 expression is initiated in the third week in Balb/C mice, unlike in CD-1 mice, where this expression is detected in four-week-old mice40. Diverse genes, with relevant functions in adult testis, show a differential expression pattern throughout mouse life, such as Dmrt1, Mtl5, NYD-SP5 41–43. Some of these genes are also expressed in other organs. For example, Mtl5n is also expressed in the foetal heart and ovary and in the adult heart42. Although Catsper1au has an expression profile different from that reported for Catsper1 7, these genes may be functionally related during their co-expression in adult testis.
The in silico analysis and the lack of translation products indicate that Catsper1au gene encode a lncRNA. These novel RNA molecules are more than 200 nt in length, primarily unspliced and polyadenylated, do not encode proteins and the primary sequence shows low conservation across evolution, unlike protein-coding genes2,44,45. Indeed, Catsper1au is unspliced and polyadenylated with a unique variation in a nucleotide, as reported for Mrhl lncRNA46, and this sequence was not detected in the human genome database (data not shown). Recently, novel TSSs in human lncRNAs genes, upstream of the previously annotated sites, have been identified using RACE-Seq. However, the expected transcripts have not been experimentally identified47. Thus, two Catsper1au isoforms could be expressed from the two TSSs identified using 5′-RACE. However, only one transcript was detected using Northern blotting. However, lncRNAs are expressed at much lower levels than protein-coding genes and exhibit a tissue/cell or developmental stage specific expression profile2,45,48. Indeed, the expression level of Catsper1au is less abundant than Catsper1 (Fig. 1e), which contrasts with the antisense in vitro bidirectional promoter activity (Fig. 1b). In the testis, it is likely that lncRNAs play an essential function in testis development and spermatogenesis2. For example, Tsx (Testes-specific X-linked), a lncRNA expressed in meiotic germ cells and brain tissue, is relevant for the development of germ cells because Tsx-null mice show smaller testis as a result of pachytene germ cell apoptosis49. Depending on the function, most of the genes expressed during spermatogenesis display exclusive expression profiles at the different stages of spermatogenic cells44. In this regard, Catsper1au is expressed both in spermatogonia cells, GC-1 spg cells, pachytene spermatocytes, round and elongated spermatids and in somatic Leydig and Sertoli cells (Figs 5 and 6), which may suggest a role for lncRNA-Catsper1au during spermatogenesis.
Bidirectional promoters in the testis may or may not co-regulate a protein-coding gene and a lncRNA. For example, a bidirectional promoter regulates the protein-coding gene Piwil1, which is important in meiosis and is exclusively expressed in spermatocytes and spermatid cells, and the lncRNA (AK016105). Both molecules show similar expression profiles in adult testis. An opposite expression profile was observed for Zfp148, which participates in the development of germ cells, and the lncRNA (AK160141). The expression of this lncRNA is lower than that of Zfp148 in newborn testis2,50,51. A bidirectional promoter that regulates the expression of a new lncRNA of unknown function named lncRNA-Tcam1 and Smarcd2 in testis has recently been reported. While lncRNA-Tcam1 is specifically expressed only in the testis in the nucleus of germ cells, Smarcd2 is expressed in all tissues. Therefore, this promoter is bidirectional in testicular germ cells and unidirectional in other tissues52, as is the case of the Catsper1 bidirectional promoter reported here.
Although most of the lncRNAs are located in the nucleus, some lncRNAs are located in the cytoplasm or both compartments53. The subcellular location of lncRNAs may be related to its function; for example, in the cytoplasm, lncRNA generally functions at the post-transcriptional level, such as BACE1-AS. In the nucleus, these molecules can act as guides for chromatin-modifying complexes at the epigenetic level. For example, XIST participates in the transcriptional regulation of proteins, such as Air and HOTAIR and regulates the alternative splicing of pre-mRNA, such as lncRNA-p21 54–56. Catsper1au plays an important role in gene regulation considering that this gene is expressed in the nucleus of germ and somatic cells in the testis, and this lncRNA was not detected in transcriptionally inactive mature sperm cells57. lncRNAs derived from bidirectional promoters can activate or repress the transcription of their neighbouring protein-coding genes in cis, such as Six3OS, or distant genes in trans, such as Vax2OS1, through epigenetic mechanisms54. However, additional studies are needed to characterize the Catsper1 bidirectional promoter and examine its role in the expression of both genes. It is also necessary to knock out Catsper1au in vitro and in vivo to assign a biological function to this novel lncRNA. However, these data strongly suggest that Catsper1au participates in the regulation of gene expression during spermatogenesis.
Materials and Methods
Source of tissues
ICR (CD-1) mice were obtained from the Institutional Animal Care and Use Committee (IACUC) of the Center for Research and Advanced Studies (IACUC-CINVESTAV). Animal handling and all experimental protocols were fully accredited and performed in accordance with the Ethical Guidelines and Procedures from the IACUC-CINVESTAV, protocol number 0113-14. IACUC-CINVESTAV is the regulatory office for the approval of research protocols involving the use of laboratory animals and fulfils the Mexican Official Norm (NOM-062-ZOO-1999) “Technical specifications for the Care and Use of Laboratory Animals”.
Transient transfection and luciferase assays
Mouse spermatogonial cells GC-1 spg (ATCC® CRL2053™) were cultured in DMEM (Sigma) supplemented with 10% (v/v) foetal bovine serum at 37 °C under a humidified atmosphere and 5% CO2. Cells were seeded in 24-well culture (2. 5 × 105 cells/well) at a density such that the cells reached 70–80% confluency at the time of transfection. GC-1 spg cells were transiently transfected using TurboFect™ in vitro (Fermentas) reagent. The transfection mixture was prepared with 1 µg of each pGL3-Basic derived constructs containing Catsper1 promoter in either direction upstream the luciferase reporter gene. pRL-CMV (0.2 ng) was used as a control to evaluate the transfection efficiency and normalize the data. Luciferase assay was performed using a Dual Luciferase system (Dual- Luciferase Reporter Assay; Promega) after 48 h of incubation.
In silico analysis of the 10.4-kb Catsper1 divergent region
The genomic sequence of the 10.4-kb region was analysed in silico to predict splicing sites and putative exons using GeneMark.hmm, NetGene2 and FGENESH 1.1 bioinformatics Web Servers. The TSS and poly(A) sites were analysed using Neural Network Promoter Prediction and POLYAH SOFTBERRY, respectively. The presence of CpG islands was analysed using CpGFinder. EST search was performed with NCBI BLAST service. RepeatMasker programme was used to identify DNA repeat sequences. RNA secondary structure was predicted with RNAfold. Transcription factor binding sites (TFB) were predicted with TRANSFAC database.
5′/3′-RACE
Catsper1au 5′ and 3′ ends were determined using the primers shown in Table 1 and the Marathon cDNA Amplification kit (Clontech) according to the manufacturer’s instructions. Reverse primers for 5′-RACE were designed downstream of the two putative TSSs predicted in silico for Catsper1au. For the first reaction of a nested PCR; AS322, AS219, and P1200R primers were used, each in combination with the forward primer adapter AP1, to generate the products AS322-AP1, AS219-AP1 and P1200R-AP1. In the second reaction, a 1:50 dilution of the cDNA products of the first reaction was used as template together with the reverse d3R primer and the forward internal adapter AP2. For the 3′-RACE, forward primers were designed based on the sequence obtained by 5′-RACE. In the first PCR, we used the 3′RACE-Forw1 primer, and the reverse adapter AP1. In the second reaction, 3′RACE-Forw2 and the reverse internal adapter AP2 were used. The PCR fragments generated in the 5′/3′-RACE were analysed using 5% acrylamide gel electrophoresis, cloned into pJET 1.2/blunt (ThermoFisher Scientific) and 20 clones were sequenced using the BigDye terminator sequencing kit Version 3.1 (Applied Biosystems). Catsper1au was amplified by RT-PCR using primers NGF225 and NG-Reverse that flanked the 5′ and 3′ sequences to generate and determine the full-length sequence; cloned into pJET 1.2/blunt and sequenced. The whole sequence of Catsper1au was annotated in the NCBI Database, accession number KX825862 and Catsper1au (Catsper1 antisense upstream transcript) was named according to The Jackson Laboratory, MGI Nomenclature Committee.
RT-PCR
Germ cells from adult (12-week-old) mouse testis were fractioned on a 2–4% BSA linear gradient into pachytene spermatocyte, round and elongated spermatids according to a previous report58. Sperm cells were obtained from epididymis as previously described59. Total RNA was isolated from mouse male and female tissues, germ cells, sperm cells; and from MSC1 and GC-1 spg cell lines by the phenol extraction method with TRIzol (Invitrogen™) and treated with RQ1 RNAse-free DNAse (Promega). RNA from mouse thymus and 11.5-day embryo was purchased from Clontech. The sex of the embryo was verified as shown in Supplementary Fig. 1. The cDNA was obtained from 2 µg of total RNA using a transcription High capacity RNA reverse transcription kit (Applied Biosystems). Actb was used as an internal expression control and to verify the absence of genomic DNA in the cDNA samples. Primers Act(F) and Act(R) generate a 660-bp Actb product and a band of 880-bp in the case of genomic DNA contamination. A 93-bp Catsper1 product was PCR amplified from testis cDNA (30 cycles and using 1 µl from the cDNA stock solution diluted 1:20) with primers Cat1 (F) and Cat1 (R). These primers were designed between exons 2 and 3 of Catsper1. Additionally, Catsper1 was amplified using 35 cycles and 1 µl of cDNA to verify the absence of genomic DNA in all the samples. A 1.2-kb product indicates genomic contamination. Primers NGF225 (F)-AS306 (R), NGF292 (F)-AS306 (R) and NGF225 (F)-NG-Reverse (R) were used to obtain the 496 bp, 429 bp and 1402 bp Catsper1au products, respectively. Catsper1au was PCR amplified from testis cDNA (40 cycles and using 1 µl from the cDNA stock solution). PCR products from Catsper1 and Catsper1au were sequenced to confirm the identity of the fragments.
Quantitative real-time PCR
The relative expression of Catsper1au RNA and Catsper1 was measured by real-time quantitative PCR (LightCycler® 480 System, Roche) using SYBR® Premix assay (Roche) with the primers NGFORW225RT (F)-REVD3 (R) and Cat1(F)-Cat1(R). The primers ActBForw (F) and ActBRev (R) were used for normalization and were designed from the Actb coding region (Table 1). The relative expression of mRNA was calculated using the ΔΔCt method60. Products were initially confirmed by sequencing. A final melting curve was also generated to ensure a single PCR product was produced in the reaction. Three independent experiments with three technical replicates were performed.
Northern blot analysis
Total RNA (10 µg) from adult testis or heart was loaded onto agarose gels containing guanidine thiocyanate (5 mM), run for 3 h and subsequently alkaline capillary transferred to a positively charged nylon membrane for 2.5 h. The blots were hybridized for 16 h at 62 °C with digoxigenin-UTP-labelled riboprobe for Catsper1au synthetized according to the manufacturer’s instructions in the DIG RNA labeling Kit (Sp6/T7) (Roche). A non-radioactive chromogenic method was used for probe detection. Briefly, after washing and blocking, the membrane was incubated with Anti-Digoxigenin-AP 1:2500 (Roche, Cat. 11093274910) and incubated for 16 h with BCIP®/NBT Liquid Substrate (Sigma, Cat. B1911).
In vitro coupled transcription-translation
The in vitro coupled transcription-translation reaction was performed with the TNT® Coupled Reticulocyte Lysate System. Catsper1au was amplified from adult testis cDNA using primers ENGFORW225 and BRACE3REV and cloned downstream of the T7 promoter into the pSG5 vector. Briefly, 1 µg of circular plasmid was added to TNT® Lysate and incubated in a 50-µl reaction for 90 min at 30 °C. To label the protein, Biotinylated lysine (Transcend™ Biotinylated tRNA) was included. The products were resolved on a 15% SDS-PAGE. A non-radioactive chemiluminescent system was used for protein detection. A luciferase plasmid encoding a 61-kDa protein and the pSG5 empty vector were used as positive and negative controls, respectively.
Overexpression of Catsper1au in primary culture of germ cells
Spermatogenic cells were obtained from two-month postpartum mouse. Testes maintained in DMEM were minced into small pieces, digested with 4-mg/ml type IV collagenase (Sigma-Aldrich) at room temperature for 30 min and washed with DMEM to obtain seminiferous tubules. Germ cells were obtained by pipetting the seminiferous tubules. Cell clots were removed and cells selected by filtration in 80 μm mesh. Two million cells were seeded onto 6-well plates with DMEM/F12 supplemented with nonessential amino acids, 15 mM HEPES, 0.12% sodium bicarbonate, 100 IU/ml penicillin, 100 μg/ml streptomycin, 30 mg/ml pyruvic acid, 5 µg/ml insulin, 2 mM L-glutamine, 5 µM vitamin E, 550 ng/ml FSH, 0.1 µM testosterone, and 10% FBS. The cells were subsequently transiently transfected with pSG5 plasmid (which expresses Catsper1au transcript) and Lipofectamine 3000 (Invitrogen), harvested 48 h later and maintained in TRIzol for RT-PCR analysis.
Subcellular fractionation
Mouse adult testis tissue, MSC1, and GC-1 spg cells were fractionated into nucleus and cytoplasm by differential centrifugation, and total RNA was isolated. Briefly, cells were suspended in 1 × PBS and centrifuged at 300 × g. The pellet was resuspended in ice-cold cell lysis buffer (50 mM Tris-HCl at pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.5% (v/v) Nonidet P-40, 25 U/µl RNasin® (Promega), and centrifuged 10 min at 17,000 × g and 4 °C. The cytoplasmic RNA was isolated from the supernatant by phenol extraction. The nuclear pellet was resuspended in lysis buffer and incubated for 15 min on ice, and sucrose was subsequently added to 2.3 M and centrifuged at 20,000 × g 4 °C for 30 min. Nuclear RNA was isolated using TRIzol and treated with RQ1 RNAse-free DNAse.
RNA-FISH
Testis from adult mice was cut into 8 μm sections using a cryostat (Leica MC1510) and mounted at −4 °C on gelatin- coated slides. Slides were fixed with 4% p-formaldehyde and treated with 10-μg/ml proteinase K. The sections were acetylated and permeabilized with 0.4% Triton X-100 in PBS. Samples were prehybridized for 30 min in prehybridization buffer (50% formamide, 4x SSC, 20% dextran sulphate sodium) and incubated with digoxigenin-UTP-labelled riboprobe for 12 h at 60 °C (800 and 200 ng/ml for Catsper1au and Actb probes, respectively). The sections were first washed with 50% formamide, 4x SSC and 10% SDS and subsequently with 50% formamide and 2x SSC. After blocking with 0.1% Tween and 3% BSA in PBS, the sections were incubated for 1 h with anti-digoxigenin-fluorescein Fab fragments diluted 1:100 and 1:200 for Catsper1au and Actb probes, respectively (Roche, Cat. 11207471910). Slides were mounted with Vectashield with DAPI (Vector Burlingame, CA), and the images were captured using a confocal microscope (Leica Sp8). The sense and antisense riboprobes for Catsper1au and Actb were synthesized according to the manufacturer’s instructions in the DIG RNA Labeling Kit (Sp6/T7) (Roche). Briefly, Catsper1au and Actb were amplified from adult testis cDNA using primers HNGF225 (F)-ECOAS306 (R) and ActH3 (F)- ActECO (R) respectively and cloned downstream of the SP6 promoter and upstream of the T7 promoter into the pSPT 18 vector.
Statistics analysis
Statistical analysis was performed with a one-way ANOVA test using GraphPad Prism 7.01 following Dunnett’s multiple comparisons test for luciferase data analysis. The level of significance was *P < 0.05, **P < 0.01, ***P ≤ 0.001. Data were expressed as the mean ± standard deviation (SD), n = 3. qRT-PCR data were also analysed using one-way ANOVA, followed by Bonferroni post hoc test. Differences among groups are indicated as letters, where a > b (Fig. 3d). Statistical significance was ***P ≤ 0.001 using paired 2-tailed t-test (Fig. 3e). Data were expressed as the means ± standard error (S.E); n = 3.
Electronic supplementary material
Acknowledgements
The authors thank Raúl González Pantoja and María de los Angeles Romero Tlalolini for technical assistance and G. Aguilar-González for automatic sequencing. This work was supported by CONACyT México (Grant 27975 to J.H.S.) and Instituto Mexicano del Seguro Social (Grant FIS/IMSS/PROT/G13/1214 to NO).
Author Contributions
S.J.B. performed the experiments and data analysis and drafted the manuscript. C.H.G. and L.G.M. contributed to RNA-FISH. J.H.S. and N.O. designed the experiments and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13867-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eddy EM. Regulation of expression during spermatogenesis. Seminars in Cell & Developmental Biology. 1998;9:451–457. doi: 10.1006/scdb.1998.0201. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS One. 2013;8(10):e75750. doi: 10.1371/journal.pone.0075750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy EM. Male Germ Cell Gene Expression. Recent Progress in Hormone Research. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, et al. Novel development-related alternative splices in human testis identified by cDNA microarrays. J Androl. 2005;26:189–196. doi: 10.1002/j.1939-4640.2005.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 5.Luk AC, Chan WY, Rennert OM, Lee TL. Long noncoding RNAs in spermatogenesis: insights from recent high-throughput transcriptome studies. Reproduction. 2014;147:131–141. doi: 10.1530/REP-13-0594. [DOI] [PubMed] [Google Scholar]
- 6.Mori C, Welch JE, Fulcher KD, O’Brien DA, Eddy EM. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol Reprod. 1993;49:191–203. doi: 10.1095/biolreprod49.2.191. [DOI] [PubMed] [Google Scholar]
- 7.Ren D, et al. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413:603–609. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson AE, et al. Identical phenotypes of Catsper1 and CatSper2 null sperm. J Biol Chem. 2005;280:32238–32244. doi: 10.1074/jbc.M501430200. [DOI] [PubMed] [Google Scholar]
- 9.Jin JL, et al. Catsper3 and catsper4 encode two cation channel-like proteins exclusively expressed in the testis. Biol Reprod. 2005;73:1235–1242. doi: 10.1095/biolreprod.105.045468. [DOI] [PubMed] [Google Scholar]
- 10.Lobley A, Pierron V, Reynolds L, Allen L, Michalovich D. Identification of human and mouse CatSper3 and CatSper4 genes: characterization of a common interaction domain and evidence for expression in testis. Reprod Biol Endocrinol. 2003;1:1–15. doi: 10.1186/1477-7827-1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung JJ, Navarro B, Krapivinsky G, Krapivinsk L, Clapham DE. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nature Communications. 2011;2:153. doi: 10.1038/ncomms1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Xia J, Cho KH, Clapham DE, Ren D. CatSperbeta, a novel transmembrane protein in the CatSper channel complex. J Biol Chem. 2007;282:18945–18952. doi: 10.1074/jbc.M701083200. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Liu J, Cho KH, Ren D. A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol Reprod. 2009;81:539–544. doi: 10.1095/biolreprod.109.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci USA. 2003;100:12201–6. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li HG, Ding XF, Liao AH, Kong XB, Xiong CL. Expression of CatSper family transcripts in the mouse testis during post-natal development and human ejaculated spermatozoa: relationship to sperm motility. Mol Hum Reprod. 2007;13:299–306. doi: 10.1093/molehr/gam009. [DOI] [PubMed] [Google Scholar]
- 16.Qi H, et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci USA. 2007;104:1219–1223. doi: 10.1073/pnas.0610286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirichok Y, Lishko PV. Rediscovering sperm ion channels with the patch-clamp technique. Molecular Human Reproduction. 2011;17:478–499. doi: 10.1093/molehr/gar044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avidan N, et al. CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur J Hum Genet. 2003;11:497–502. doi: 10.1038/sj.ejhg.5200991. [DOI] [PubMed] [Google Scholar]
- 19.Avenarius., et al. Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet. 2009;84:505–10. doi: 10.1016/j.ajhg.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata-Rocha M, et al. Molecular cloning and analysis of the Catsper1 gene promoter. Mol Hum Reprod. 2013;19:336–47. doi: 10.1093/molehr/gat003. [DOI] [PubMed] [Google Scholar]
- 21.Wick N, et al. Induction of short interspersed nuclear repeat-containing transcripts in epithelial cells upon infection with a chicken adenovirus. J Mol Biol. 2003;328:779–90. doi: 10.1016/S0022-2836(03)00363-2. [DOI] [PubMed] [Google Scholar]
- 22.McVicker G, Green P. Genomic signatures of germline gene expression. Genome Res. 2010;20:1503–1511. doi: 10.1101/gr.106666.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang MQ, Koehly LM, Elnitski LL. Comprehensive annotation of bidirectional promoters identifies co-regulation among breast and ovarian cancer genes. PLoS Comput Biol. 2007;3(4):e72. doi: 10.1371/journal.pcbi.0030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PY, et al. Two non- homologous brain diseases-related genes, SERPINI1 and PDCD10, are tightly linked by an asymmetric bidirectional promoter in an evolutionarily conserved manner. BMC Molecular Biology. 2007;8:2. doi: 10.1186/1471-2199-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martianov I, et al. Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics. 2010;11:530. doi: 10.1186/1471-2164-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 27.Black DL, Chabot B, Steitz JA. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–50. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 28.Bergmann JH, et al. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 2015;5:1336–46. doi: 10.1101/gr.189027.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, San Agustin JT, Witman GB, Kilpatrick DL. Novel role for a sterol response element binding protein in directing spermatogenic cell-specific gene expression. Mol Cell Biol. 2004;24:10681–10688. doi: 10.1128/MCB.24.24.10681-10688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poschl E, Pollner R, Kuhn K. The genes for the alpha 1(IV) and alpha 2(IV) chains of human basement membrane collagen type IV are arranged head-to-head and separated by a bidirectional promoter of unique structure. Embo J. 1998;7:2687–2695. doi: 10.1002/j.1460-2075.1988.tb03122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voth H, et al. Co-regulated expression of HAND2 and DEIN by a bidirectional promoter with asymmetrical activity in neuroblastoma. BMC Mol Biol. 2009;10:28. doi: 10.1186/1471-2199-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–14. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- 34.Conforto TL, Waxman DJ. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol. Sex Differ. 2012;3:9. doi: 10.1186/2042-6410-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kocarek TA, Duanmu Z, Fang HL, Runge-Morris M. Age- and sex-dependent expression of multiple murine hepatic hydroxysteroid sulfotransferase (SULT2A) genes. Biochem Pharmacol. 2008;76:1036–1046. doi: 10.1016/j.bcp.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laiho A, et al. Transcriptome Profiling of the Murine Testis during the First Wave of Spermatogenesis. PLoS One. 2013;8(4):e61558. doi: 10.1371/journal.pone.0061558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paradis AN, Gay MS, Zhang L. Binucleation of cardiomyocytes: the transition from a proliferative to a terminally differentiated state. Drug Discov Today. 2014;19:602–9. doi: 10.1016/j.drudis.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill TJ, 4th, Mack CP, Taylor JM. Germline deletion of FAK-related non-kinase delays post-natal cardiomyocyte mitotic arrest. J Mol Cell Cardiol. 2012;53:156–164. doi: 10.1016/j.yjmcc.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, et al. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics. 2009;93:235–242. doi: 10.1016/j.ygeno.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikpoor P, Mowla SJ, Movahedin M, Ziaee SA, Tiraihi T. CatSper gene expression in postnatal development of mouse testis and in subfertile men with deficient sperm motility. Hum Reprod. 2004;19:124–8. doi: 10.1093/humrep/deh043. [DOI] [PubMed] [Google Scholar]
- 41.Matson CK, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olesen C, Møller M, Byskov AG. Tesmin transcription is regulated differently during male and female meiosis. Mol Reprod Dev. 2004;67:116–26. doi: 10.1002/mrd.20007. [DOI] [PubMed] [Google Scholar]
- 43.Yin LL, Li JM, Zhou ZM, Sha JH. Identification of a novel testis-specific gene and its potential roles in testis development/spermatogenesis. Asian J Androl. 2005;7:127–137. doi: 10.1111/j.1745-7262.2005.00041.x. [DOI] [PubMed] [Google Scholar]
- 44.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 45.Zhang B, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishant KT, Ravishankar H, Rao MRS. Characterization of a mouse recombination hot spot locus encoding a novel non-protein-coding RNA. Mol. Cell. Biol. 2004;24:5620–5634. doi: 10.1128/MCB.24.12.5620-5634.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagarde J, et al. Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq) Nat. Commun. 2016;7:12339. doi: 10.1038/ncomms12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward M, McEwan CD, Mills J, Janitz M. Conservation and tissue-specific transcription patterns of long noncoding RNAs. J Hum Transcr. 2015;1:2–9. doi: 10.3109/23324015.2015.1077591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anguera MC, et al. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7(9):e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi A, et al. Heterozygosity with respect to Zfp148 causes complete loss of fetal germ cells during mouse embryogenesis. Nat Genet. 2003;33:172–176. doi: 10.1038/ng1072. [DOI] [PubMed] [Google Scholar]
- 52.Kurihara M, Shiraishi A, Satake H, Kimura AP. A conserved noncoding sequence can function as a spermatocyte-specific enhancer and a bidirectional promoter for a ubiquitously expressed gene and a testis-specific long noncoding RNA. J Mol Biol. 2014;426:3069–93. doi: 10.1016/j.jmb.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 53.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–14. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Froberg JE, Yang L, Lee JT. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425:3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 57.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liévano A, et al. T-type Ca2+ channels and alpha1E expression in spermatogenic cells, and their possible relevance to the sperm acrosome reaction. FEBS Lett. 1996;388:150–154. doi: 10.1016/0014-5793(96)00515-7. [DOI] [PubMed] [Google Scholar]
- 59.Baltierrez-Hoyos R, Roa-Espitia AL, Hernandez-Gonzalez EO. The association between CDC42 and caveolin-1 is involved in the regulation of capacitation and acrosome reaction of guinea pig and mouse sperm. Reproduction. 2012;144:123–134. doi: 10.1530/REP-11-0433. [DOI] [PubMed] [Google Scholar]
- 60.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.