Abstract
Tripterygium wilfordii produces not only ent-kaurene, which is an intermediate of gibberellin (GA) biosynthesis in flowering plants, but also 16α-hydroxy-ent-kaurane, whose physiological role has not been characterized. The two compounds are biosynthesized from the universal diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) by diterpene synthases, which have been discovered and functionally characterized in T. wilfordii. Here, we described the functional characterization of four cytochrome P450 reductases (TwCPR) and one ent-kaurene oxidase (TwKO). Four TwCPRs were found to have relatively ubiquitous expression in T. wilfordii root, stem, leaf, and flower tissues. Co-expression of both a TwCPR and TwKO in yeast showed that TwCPR3 has a slightly better activity for providing the electrons required for these reactions, indicating that TwCPR3 is more suitable for use in the functional analysis of other cytochrome P450 monooxygenases. TwKO catalyzed the three-step oxidation of the C4α methyl of the tetracyclic diterpene intermediate ent-kaurene to form ent-kaurenoic acid as an early step in GA biosynthesis. Notably, TwKO could also convert 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid, indicating an important function of 16α-hydroxy-ent-kaurane in the anti-HIV principle tripterifordin biosynthetic pathway in planta. Homology modeling and molecular docking were used to investigate the unknown crucial active amino acid residue involved in the catalytic reaction of TwKO, and one key residue (Leu387) contributed to the formation of 16α-hydroxy-ent-kaurenoic acid, most likely by forming hydrogen bonds with the hydroxyl group (-OH) of 16α-hydroxy-ent-kaurane, which laid the basis for further investigation of the multifunctional nature of KO catalysis. Also, our findings paved the way for the complete biosynthesis of the anti-HIV principle tripterifordin.
Keywords: Tripterygium wilfordii, cytochrome P450 monooxygenase, cytochrome P450 reductase, molecular docking, site-directed mutagenesis, 16α-hydroxy-ent-kaurane, 16α-hydroxy-ent-kaurenoic acid
Introduction
Tripterygium wilfordii Hook. f. has a long history of use in traditional Chinese medicine, mainly to treat rheumatoid arthritis (Tu, 2009). Surprisingly, T. wilfordii also shows impressive and effective anti-inflammatory, immunosuppressive and antitumor activities (Titov et al., 2011; Chugh et al., 2012; Manzo et al., 2012; Wong et al., 2012; Zhou Z.L. et al., 2012). Much of the remarkable bioactivity can be attributed to bioactive diterpenoids, particularly the abietane-type diterpenoid triptolide. A major challenge to harnessing these natural products is that only small amounts are found in planta. Recently, many studies have been targeted toward the investigation of the triptolide biosynthetic pathway in order to directly obtain this bioactive compound using synthetic biology strategies (Andersen-Ranberg et al., 2016; Hansen et al., 2017; Inabuy et al., 2017). However, little attention has been paid to the biosynthesis of the kaurene-type diterpenoids, e.g., bioactive gibberellins (GAs) and anti-HIV principle tripterifordin (Chen et al., 1992).
GAs, which are biosynthesized from the key intermediate ent-kaurene through several steps, are a major class of diterpenoid hormones essential to a number of vascular plant growth and developmental processes (Hooley, 1994). To date, the upstream diterpene synthases, also known as ent-copalyl diphosphate synthase and ent-kaurene synthase, responsible for the formation of ent-kaurene has been well investigated in T. wilfordii (Andersen-Ranberg et al., 2016; Hansen et al., 2017; Inabuy et al., 2017). Interestingly, the identified T. wilfordii diterpene synthases can also catalyze the formation of 16α-hydroxy-ent-kaurane, this catalysis was reported in the moss Physcomitrella patens and in Populus trichocarpa (Hayashi et al., 2006; Irmisch et al., 2015). The downstream enzymes involved in the GA biosynthetic pathway are cytochrome P450 monooxygenases (Yamaguchi, 2008). One of them, ent-kaurene oxidase (KO) from the CYP701 family, has been reported to catalyze the three-step oxidation of the C4α methyl of the tetracyclic diterpene intermediate ent-kaurene to form ent-kaurenoic acid as an early step in GA biosynthesis (Morrone et al., 2010; Hedden and Thomas, 2012). Such catalytic activities strictly depend on their partner, NADPH-cytochrome P450 reductase (CPR), to provide the necessary electrons.
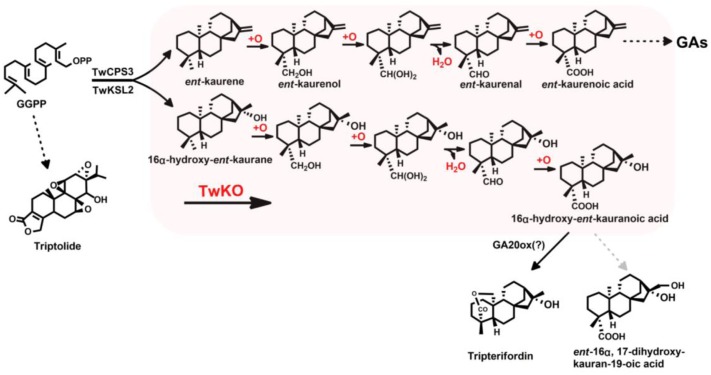
More recently, Mafu et al. (2016) reported the promiscuity of Oryza sativa KO (CYP701A6/OsKO2) and Arabidopsis thaliana KO (CYP701A3/AtKO). More than 20 labdane-related diterpenoid (LRD) olefins were used to thoroughly investigate the enzymatic promiscuity, and the results showed OsKO2 was much more specific (specific for ent-(iso)kaurene and the structurally closely related ent-trachylobane), whereas AtKO was found to exhibit much broader promiscuity, targeting the C4α methyl of ent-CPP-derived LRDs and the C2α and/or C3β of normal and syn-CPP-derived LRDs to produce hydroxylated or carboxylated derivatives (Mafu et al., 2016). These results prompted us to explore the promiscuity of TwKO and to investigate whether it reacted with ent-kaurene as well as 16α-hydroxy-ent-kaurane in T. wilfordii. Herein, we reported the identification of four CPRs and one KO from T. wilfordii, and TwKO was confirmed to catalyze ent-kaurene to form ent-kaurenoic acid, which is consisted with reports in Salvia miltiorrhiza, A. thaliana, O. sativa, Pisum sativum, and P. patens (Davidson et al., 2004; Sakamoto et al., 2004; Morrone et al., 2010; Miyazaki et al., 2011; Su et al., 2016). Notably, TwKO was discovered to have another new catalytic function, converting 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid, which most likely is an intermediate of the anti-HIV principle tripterifordin in T. wilfordii (Figure 1). In addition, the results of Mafu et al. (2016) have driven us to gain insight into the underlying structure–function relationship between this promiscuous enzyme TwKO and its substrates. However, except for GGPPS and terpene synthases (Chang et al., 2006; Mann et al., 2010; Zhou K. et al., 2012), no crystal structure of KO either in plant or in microorganisms has been reported. In the present study, homology modeling and molecular docking were used to construct the three-dimensional structure of TwKO, and one key active amino acid residue (Leu387) was identified to bind to the hydroxyl group (-OH) of 16α-hydroxy-ent-kaurane, which laid the basis for further investigation of the multifunctional nature of KO catalysis. Also, the forming of 16α-hydroxy-ent-kaurenoic acid catalyzed by TwKO, paved the way for the complete biosynthesis of the tripterifordin and provided the precursor for biosynthesis of other bioactive diterpenoids, e.g., ent-16α, 17-dihydroxy-kauran-19-oic acid (Chen et al., 1992; Sung et al., 2011).
FIGURE 1.
Role of KO in T. wilfordii gibberellin phytohormone biosynthesis.
Materials and Methods
Plant Materials
Tripterygium wilfordii suspension cells were cultured in Murashige and Skoog (MS) medium containing 30 g/L sucrose supplemented with 0.1 mg/L kinetin (KT), 0.5 mg/L indole-3-butyric acid (IBA), and 0.5 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D) and maintained at 25 ± 1°C while being shaken at 120 rpm in the dark (Su et al., 2014).
RNA Isolation, cDNA Synthesis, and Gene Cloning
The total RNA of the induced cells was extracted following the cetyltrimethylammonium bromide (CTAB) method (Del Sal et al., 1989). An aliquot (1 μg) of the total RNA was used to synthesize first-strand cDNA using the PrimeScript 1st Strand cDNA Synthesis Kit in accordance with the manufacturer’s protocol (Takara Bio, Dalian, China).
Mining of the T. wilfordii transcriptome (SRA accession number: SRR6001265) was performed, and gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (a manually annotated and reviewed protein sequence database), KEGG Ortholog database, and GO (Gene Ontology) databases. Then, one putative ent-kaurene oxidase and four putative CPRs with full-length coding sequences were identified. The full-length cDNAs for the open reading frames (ORFs) of the TwKO and TwCPR genes were cloned from the cDNA library using PrimeSTAR DNA polymerase (Takara Bio). The PCR products were purified and cloned into a pEASY-Blunt Simple Cloning Vector (TransGen Biotech, Beijing, China), then transformed into Escherichia coli Trans5α cells (TransGen Biotech), and cultured in Luria-Bertani (LB) medium at 37°C in the dark. The positive clones were sequenced. All PCR primers used in the experiments are listed in Supplementary Table S1.
Sequence Analysis
The sequences of the TwKO, TwCPR1, TwCPR2, TwCPR3, and TwCPR4 genes were analyzed using the NCBI database1. The ORFs and deduced amino acid sequences were analyzed using the online tool ORF Finder2 and the ExPASy online tool3, respectively. Multiple sequence alignment was implemented using DNAMAN software. Amino acid sequences for a variety of ent-kaurene oxidases were obtained from the NCBI database, and the phylogenetic tree was constructed with MEGA6 software based on the neighbor-joining method (Tamura et al., 2013). One thousand bootstrap replicates were performed during each analysis to define the level of confidence support.
qRT-PCR Analysis
Total RNA from the root, stem, leaf, and flower of T. wilfordii mature plant was extracted following the CTAB method. First-stand cDNA for real-time quantitative PCR was reverse transcribed from total RNA using the FastQuant RT Kit (Tiangen Biotech, Beijing, China). Quantitative reverse transcription PCR (qRT-PCR) were performed with gene specific primers (Supplementary Table S1) and a TransStart Top Green qPCR SuperMix (TransGen Biotech) on a Roche LightCycler 480 Real Time PCR System (Roche, Switzerland). Expression levels were evaluated using the 2-ΔΔCt method (Livak and Schmittgen, 2012) based on the eukaryotic translation elongation factor 1α (Ef1α) as reference gene and with triplicate measurements from five biological replicates.
Heterologous Expression and Affinity Purification of TwCPRs
To increase protein solubility, four TwCPR ORFs were subcloned into the N-terminal MBP fusion expression vector HIS-MBP-pET28a according to the protocol of the pEASY®-Uni Seamless Cloning and Assembly Kit (TransGen Biotech; HIS-MBP-pET28a was provided by Dr. Xiaohong Zhang. HIS, histidine; MBP, maltose-binding protein. The MalE gene encoding MBP was introduced into the NdeI and BamHI sites of pET28a). Heterologous expression of four TwCPRs in the E. coli strain Transetta (DE3) (TransGen Biotech) was performed as described previously (Su et al., 2017). Briefly, the selected transformants were incubated in 200 mL of LB medium that was supplemented with 50 mg/L kanamycin at 37°C to an OD600 of 0.6 ∼ 0.8, after which the temperature was lowered to 16°C for 0.5 h prior to induction with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside (Sigma, United States) for an additional 24 h at 200 rpm. The cell pellets were harvested by centrifugation (3000 g, 20 min, 4°C) and stored at -80°C.
The harvested cell pellets were resuspended in 5 mL of binding buffer (20 mM Na2HPO4, 0.5 M NaCl, 10 mM imidazole, 1 mM DTT, 1 mM PMSF, pH 7.4) and subjected to mild sonication for 20 min (lysed for 10 s, paused for 10 s). The supernatant of the centrifuged lysates (12,000 g, 30 min, 4°C) was mixed with Ni-NTA agarose (QIAGEN, WI, United States) at 4°C for 2 h, after which HIS-MBP-tagged purified proteins were acquired following gradient elution with elution buffer (20 mM Na2HPO4, 0.5 M NaCl, pH 7.4) containing different concentrations of imidazole (50, 100, 250, and 500 mM). The protein concentrations were determined using a Modified Bradford Protein Assay Kit (Sangon Biotech, Shanghai, China).
Enzyme Assays
Activities of TwCPRs were assayed as described (Yang et al., 2010). All assays and incubations were performed at 25°C in 200 μL of 50 mM Tris buffer (pH 7.4) containing TwCPR enzymes (∼20 μg/mL) and 100 μM cytochrome c. The reaction was started by addition of 100 μM NADPH, time-dependent absorbance change at 550 nm was monitored up to 3 min by a Varioskan Flash spectrophotometer (Thermo Fisher Scientific, CA, United States). A molar absorption coefficient of 21/mM/cm for equine heart cytochrome c was used for quantification (Guengerich et al., 2009). The reduction of dichlorophenolindophenol (DCPIP, 100 μM) was monitored at 600 nm (20.6/mM/cm); K3Fe(CN)6 (100 μM) at 424 nm (1.02/mM/cm).
Kinetic parameters for cytochrome c reduction were performed as described above with TwCPR enzymes (∼20 μg/mL), 100 μM NADPH and varying cytochrome c concentrations (0–200 μM). The kinetic parameters were determined by the non-linear Michaelis–Menten regression equation by using GraphPad Prism 5 (Yang et al., 2010).
Yeast Expression
Four TwCPRs genes were subcloned into the yeast epitope-tagging vector pESC-Leu under the control of the GAL1 inducible promoter using BamHI and ApaI sites (Agilent Technologies, United States). Then, the TwKO gene was introduced into the four recombinant plasmids above following the protocol of the pEASY®-Uni Seamless Cloning and Assembly Kit (TransGen Biotech). Each recombinant plasmid containing one of the TwCPR genes and the TwKO gene was co-expressed in the yeast BY-T20 strain (BY4742, ΔTrp1, Trp1::HIS3-PPGK1-BTS1/ERG20-TADH1-PTDH3-SaGGPS-TTPI1-PTEF1-tHMG1-TCY C1) (Dai et al. 2012, 2013; Shi et al., 2014). To provide the substrate (ent-kaurene and 16α-hydroxy-ent-kaurane) for TwKO, the substrate-producing module pESC-Trp::TwCPS3/TwKSL2 (whose major product is 16α-hydroxy-ent-kaurane) or pESC-Trp::TwCPS3/TwKSL2:A608M (whose major product is ent-kaurene) was also transformed into the BY-T20 strains.
The recombinant strains were selected on synthetic dropout medium-Trp-Leu (SD-Trp-Leu) supplemented with 20 g/L glucose and were grown at 30°C for 48–72 h. Single transformed yeast colonies were grown in SD-Trp-Leu liquid medium supplemented with 20 g/L glucose at 30°C during shaking at 230 rpm for 48 h. The yeast cells were pelleted and resuspended in 50 mL of SD-Trp-Leu liquid induction medium supplemented with 20 g/L D-galactose for an additional 48 h. Finally, the induced yeast cells were extracted three times with an equal volume of ethyl acetate. The organic fractions were dried and methylated with (trimethylsilyl)diazomethane (Aladdin Industrial Inc., Shanghai, China) as described previously (Su et al., 2016). The methylated samples were re-dried and then dissolved in 100 μL of ethyl acetate for gas chromatography–mass spectrometry (GC–MS) using a Thermo TRACE 1310/TSQ 8000 gas chromatograph equipped with a TG-5MS (30 m × 0.25 mm × 0.25 μm) capillary column. The GC conditions were as follows: the sample (1 μL) was injected in splitless mode at 250°C under a He flow rate of 1 mL/min in accordance with a temperature program of 1.5 min at 60°C, increased to 200°C at 25°C/min, then to 300°C at 5°C/min. The ion trap heating temperature was 250°C. The electron energy was 70 eV. Spectra were recorded in the range of 40–500 m/z.
Homology Modeling and Molecular Docking
Because the crystal structure of TwKO has not yet been resolved, homology modeling was used to construct the three-dimensional structure of TwKO. After search for templates in the protein database (PDB)4, several models were created based on alternative sequence alignments including secondary structure predictions. The crystal structure of cytochrome P450 monooxygenase CYP170A1 from Streptomyces coelicolor (PDB ID: 3DBG) was selected as suitable template in view of their high protein sequence similarity and closely related biological functions (Zhao et al., 2009). The academic version of MODELER 9v11 was used for homology modeling of TwKO (Eswar et al., 2003). The structural features in the template protein were used to derive spatial restraints to generate model protein structures using conjugate gradient and simulated annealing optimization procedures (Sali, 1995). After the addition of hydrogen atoms, the energy of the model structures was individually minimized using the staged minimization program of SYBYL X-1.2. First, the simplex method was used for 20 cycles prior to switching to the AMBER FF99 force field for 1000 iterations with the steepest descent calculation. Then, the conjugated gradient calculation was implemented until the convergence on the gradient reached 0.05 kcal/(Å mol). To identify the ligand binding site, the multichannel-surfaces searching method in SYBYL X-1.2 was used to investigate the cavities on the surface of TwKO. The cavity containing active amino acid residues in the catalytic A domain was selected as the active pocket surface to generate a protomol for molecular docking. The protomol was generated using the steric hydrophobic group (CH4), the hydrogen bond group (C = O), and the hydrogen acceptor (N–H) within 4.5 Å of the active pocket surface. Surflex-Dock, a well-recognized tool in the field of molecular docking (Wang et al., 2016; Wang X. et al., 2017), was used to calculate the ligand–receptor interaction. 16α-Hydroxy-ent-kaurane was prepared as described according to the following procedure: the structure was checked and the hydrogen atoms added, the atomic charges were then added according to the Gasteiger–Hückel method, and energy minimization was implemented using the Tripos force field with 1000 iterations. Then, the optimized compound was docked into the active site of TwKO including the heme in a heteroatom file using the default settings. After each Surflex-Dock run, the 10 best docked conformers or poses were sorted in a molecular spreadsheet; these conformers or poses represented binding affinities in -log 10 (Kd) based on Surflex-Dock scoring function [crash score (also pKd units), polar score, D-score, PMF-score, G-score, ChemSco and CScore] (Spitzer and Jain, 2012). The default parameters of the softwares were used if not stated.
Site-Directed Mutagenesis of TwKO
Site-directed mutants of TwKO, including the L387A, L387D, L387S, L387T, L387G, and L387R mutants (L, leucine; A, alanine; D, aspartic acid; S, serine; T, threonine; G, glycine; R, arginine), were constructed using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech). The mutagenic primer pairs are listed in Supplementary Table S1. The constructed mutants were verified by complete sequencing and then transformed into yeast strains containing the module pESC-Trp::TwCPS3/TwKSL2 and one of the TwCPR genes (TwCPR3) with higher catalytic activity. The products were extracted and analyzed by GC–MS as described above.
Statistical Analysis
All assays were performed as at least three independent experiments. The data were presented as the mean ± SD, and statistical comparisons between groups were tested using one-way ANOVA followed by the Dunnett’s test. Differences were considered significant when the p-value <0.05.
Results
Isolation and Identification of the TwCPR Genes
The appropriate CPR is typically required to support the activities of P450. The full-length cDNA sequences of four TwCPRs (TwCPR1, TwCPR2, TwCPR3, and TwCPR4) were then amplified by RT-PCR, and the total RNA from T. wilfordii suspension cells was used as a template.
The amino acid sequence comparison showed that these four CPRs are 84% identical to each other and that all share high sequence identities with known CPRs from other flowering plant species (62-76%) (Mizutani and Ohta, 1998; Guo et al., 2013). The alignment analysis revealed that the four TwCPRs shared many highly conserved amino acid residues and that all of the functional domains were involved in the binding of the P450 monooxygenase. The cofactors of FMN, FAD, and NADPH were also identified (Supplementary Figure S1).
Kinetics of Recombinant TwCPRs
The recombinant TwCPRs were heterologously expressed in E. coli, N-terminally fused to MBP for soluble expression, purified by affinity chromatography (Supplementary Figure S2), and then used for kinetic studies. The results showed that the requirement of the TwCPRs for an electron acceptor is non-specific: cytochrome c, K3Fe(CN)6, and DCPIP could all serve as acceptors (Table 1). The kinetic characteristics of the TwCPRs for reducing cytochrome c were determined. In general, the four TwCPRs showed similar levels of activity (Kcat value) (Supplementary Figure S3 and Table 2).
Table 1.
Specific activities of TwCPR1, TwCPR2, TwCPR3, and TwCPR4 in reducing cytochrome c, DCPIP, and K3Fe(CN)6, all at 100 μM, in the presence of 100 μM of NADPH.
| Specific activity (μmol/min/mg protein) | |||
|---|---|---|---|
| Cytochrome c | DCPIP | K3Fe(CN)6 | |
| TwCPR1 | 0.087 ± 0.007 | 0.056 ± 0.001 | 0.013 ± 0.006 |
| TwCPR2 | 0.074 ± 0.007 | 0.061 ± 0.006 | 0.038 ± 0.002 |
| TwCPR3 | 0.121 ± 0.012 | 0.040 ± 0.003 | 0.131 ± 0.015 |
| TwCPR4 | 0.120 ± 0.010 | 0.048 ± 0.002 | 0.102 ± 0.001 |
Values are presented as mean ± SD.
Table 2.
Kinetic constants of recombinant TwCPR1, TwCPR2, TwCPR3, and TwCPR4.
| Cytochrome c | ||||
|---|---|---|---|---|
| TwCPR1 | TwCPR2 | TwCPR3 | TwCPR4 | |
| Vmax (μmol/min/mg-1) | 0.246 ± 0.012 | 0.173 ± 0.005 | 0.236 ± 0.009 | 0.229 ± 0.010 |
| Km (μM) | 12.690 ± 3.848 | 12.201 ± 1.991 | 23.413 ± 3.187 | 23.609 ± 4.095 |
| Kcat (min-1) | 31.500 | 22.137 | 29.598 | 28.781 |
| Kcat/Km | 2.482 | 1.814 | 1.264 | 1.219 |
Kinetic parameters for cytochrome c was carried out with 100 μM NADPH and varying cytochrome c concentrations. Values are presented as means ± SD.
Expression Analysis of the Identified TwCPRs
To investigate the physiological roles of the identified TwCPRs in T. wilfordii, we performed qRT-PCR to evaluate mRNA transcript levels in various organs, including root, stem, leaf, and flower. Four functional TwCPRs were found to have relatively ubiquitous expression in all sampled organ tissues. TwCPR1 showed relative higher transcription levels in all tissues verse other three TwCPRs. On the contrary, TwCPR3 showed the lowest expression in all tissues (Figure 2).
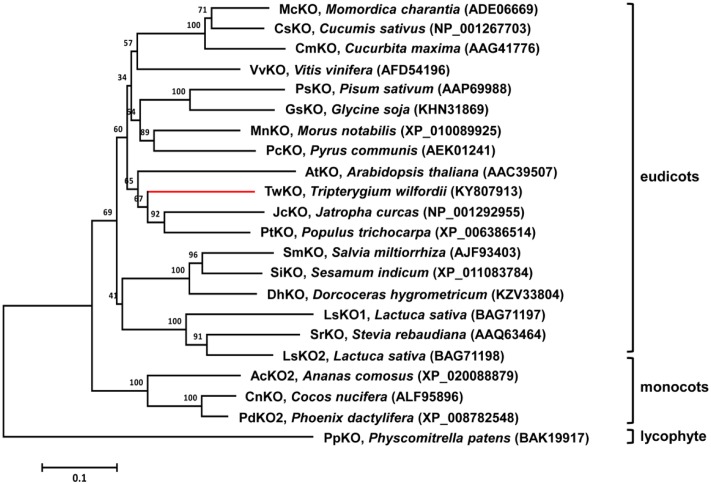
FIGURE 2.
Molecular phylogenetic analysis of selected KOs from different species. The neighbor-joining phylogenetic trees were constructed using the bootstrap method on MEGA6 and the number of bootstrap replications was1000.
TwCPRs Supported KO Monooxygenase Activity
Previous studies confirmed that TwCPS3 catalyzes the conversion of GGPP to ent-CPP. Subsequently, TwKSL2 converts ent-CPP to 16α-hydroxy-ent-kaurane as a major product via a complex bicyclization and ring rearrangement reaction. Interestingly, changing alanine (A) at the 608th aa of TwKSL2 to methionine (M) leads to the predominant production of ent-kaurene, with smaller amounts of 16α-hydroxy-ent-kaurane, from ent-CPP by the resulting mutant TwKSL2:A608M. Then, we constructed the recombinant plasmids pESC-Trp::TwCPS3/TwKSL2 and pESC-Trp::TwCPS3/TwKSL2:A608M, which we transformed into BY-T20 strains for producing 16α-hydroxy-ent-kaurane and ent-kaurene, respectively (Supplementary Figure S4).
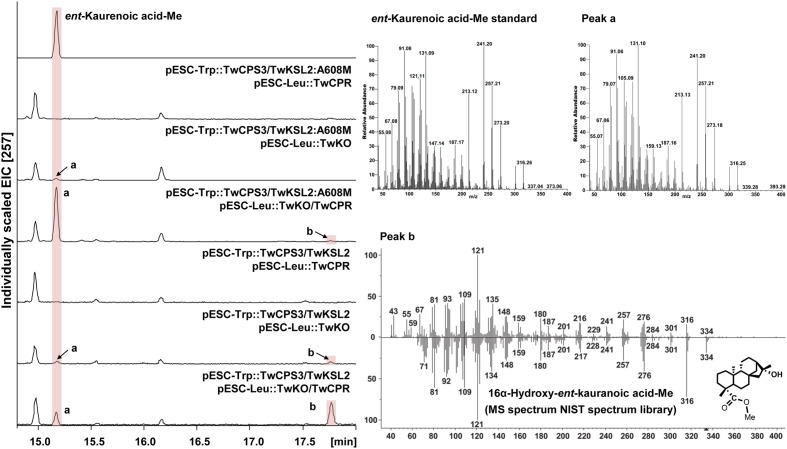
TwKO shares 67, 72, and 74% protein sequence identity with AtKO (A. thaliana), JcKO (Jatropha curcas) and PtKO (P. trichocarpa), respectively (Figure 3), indicating similar catalytic activity. To characterize the biochemical function of TwKO as well as the TwCPRs in vivo, the two complementary enzymes were co-expressed in the constructed ent-kaurene- and 16α-hydroxy-ent-kaurene-producing yeast strains mentioned above. After the extraction and methylation of the fermentation products, ent-kaurenoic acid methyl ester (ent-kaurenoic acid-Me) was detected by comparison with the authentic methylated standard (Sigma, United States). Another product with a major molecular ion peak at m/z 334 [M]+ was also identified as 16α-hydroxy-ent-kaurenoic acid methyl ester (16α-hydroxy-ent-kaurenoic acid-Me) by comparison with data from the NIST mass spectral database.
FIGURE 3.
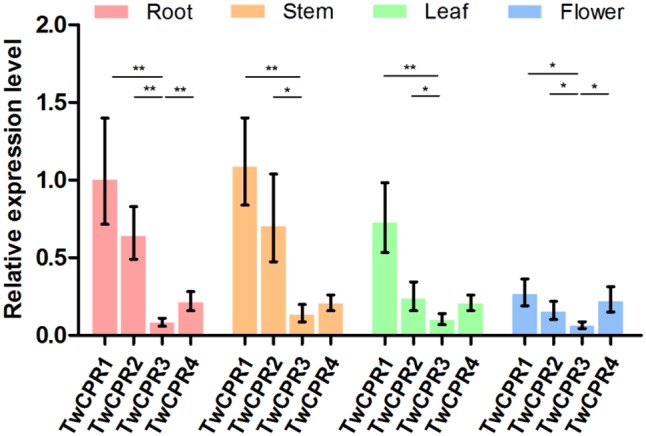
The mRNA expression levels of the TwCPRs in root, stem, leaf, and flower tissues from T. wilfordii. The expression level was normalized to that of actin. The error bars show the means ± SD value from three technical replicates of at least three biological replicates (∗P < 0.05, ∗∗P < 0.01).
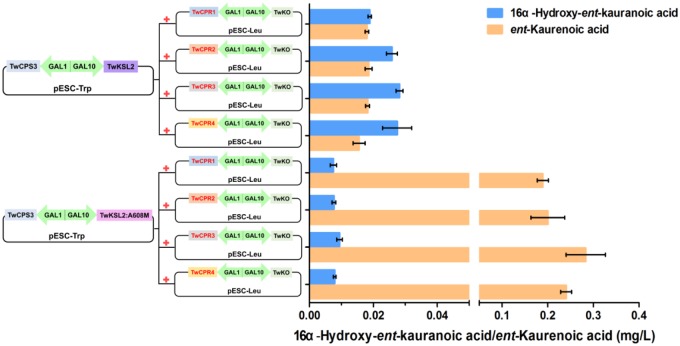
Co-expression of TwKO and the TwCPRs, in conjunction with the recombinant plasmid pESC-Trp::TwCPS3/TwKSL2:A608M, led to the predominant product ent-kaurenoic acid-Me, as well as smaller amounts of 16α-hydroxy-ent-kaurenoic acid-Me; while roughly equal amounts of 16α-hydroxy-ent-kaurenoic acid-Me and ent-kaurenoic acid-Me were produced, along with the recombinant plasmid pESC-Trp::TwCPS3/TwKSL2 (Figure 4). These results confirmed that TwKO encodes a functional ent-kaurene oxidase that is involved not only in the three-stage oxidation of ent-kaurene to ent-kaurenoic acid in the T. wilfordii GA biosynthetic pathway but also in the continuously oxidation of 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid in an unreported diterpenoid biosynthetic pathway. Different combinations of each TwCPR and TwKO produced distinct levels of 16α-hydroxy-ent-kaurenoic acid/ent-kaurenoic acid, and the combination of TwCPR3 and TwKO generated the highest content of 16α-hydroxy-ent-kaurenoic acid as well as ent-kaurenoic acid, indicating that TwCPR3 has slightly better activity for providing the electrons required for these reactions (Figure 5).
FIGURE 4.
GC–MS analysis of in vivo assay for TwKO activity. Peak a and b, methyl ester derivatives of the products from yeast strain BY-T20 cultures expressing TwKO combined with TwCPR, TwCPS3, and TwKSL2 (or TwKSL2:A608M), and their corresponding mass spectrums (Peak a, Rt = 15.17 min; Peak b, Rt = 17.77 min). Mass spectrum of peak a was identical to that of the authentic standard, and peak b was identified by comparison with the NIST spectrum library.
FIGURE 5.
16α-Hydroxy-ent-kaurenoic acid/ent-kaurenoic acid production by recombinant yeasts harboring modules overexpressing various enzymes. The strains were cultivated for 48 h in synthetic dropin media, and the products were extracted with equal volume of ethyl acetate and methylated with (trimethylsilyl)diazomethane. Values shown are means ± SD of at least three biological replicates.
Homology Modeling and Molecular Docking
TwKO has shown promiscuous enzymatic catalysis. This promiscuity was very recently reported for AtKO in A. thaliana (Mafu et al., 2016) and prompted us to screen the crucial active amino acid binding of a key group of substrates, providing insight into the catalytic mechanism. However, no crystal structure of KO in plants or in microorganisms has been reported. The crystal structure of cytochrome P450 CYP170A1 from S. coelicolor (Zhao et al., 2009) was selected as template structure used for homology modeling and molecular docking, and the values of max score, total score, query cover, E-value, and ident were 81.6, 81.6, 39%, 4e-16, and 29% for TwKO. We chose this structure as a template for establishing the 3D structures of TwKO. Modeller 4.0 software was used to generate the sequence alignment of the template protein with TwKO (Kuntal et al., 2010).
According to the multichannel-surfaces module, five cavities were produced from on the surface of TwKO. Previous studies suggested that the catalytic A domain of KO is associated with substrate binding and an oxygen pocket (Supplementary Figure S5) (Davidson et al., 2004). In this study, the corresponding location in TwKO, which covers most of the reported key amino acid residues, was selected as the active site to generate the protomol for molecule docking. 16α-Hydroxy-ent-kaurane was docked into the active site of TwKO using the Surflex-Dock program of the SYBYL X-1.2 software package. The binding mode of this compound with TwKO was predicted (Figure 6). So far, the docking can only detect the hydrogen-bond interaction. Only Leu387 interacted with the compound and showed a hydrogen-bond interaction with the compound, implying that Leu387 of TwKO is the key amino acid residue binding to this compound.
FIGURE 6.
Binding modes of TwKO with 16α-hydroxy-ent-kaurane (yellow) and heme (green carbon atoms). The models were generated using 3DBG (structure of human Streptomyces coelicolor cytochrome P450 monooxygenase CYP170A1) as a template. The hydrogen bonding interactions are displayed in dotted lines.
Site-Directed Mutagenesis of the TwKO Proteins
We predicted key catalytic sites by molecular docking. Structural comparisons suggested that a crucial amino acid, Leu387 of TwKO, may form hydrogen bonds with the hydroxyl group (-OH) of 16α-hydroxy-ent-kaurane, which is involved in continuous oxidation reactions. To preliminarily identify the biochemical impact of this key amino acid, we mutated Leu387 to six other amino acids that have different properties. Site-directed mutagenesis and enzyme assays demonstrated that the L387A, L387D, L387T, L387G, and L387R mutations led to an approximate 13.7–65.9% decrease; the L387S mutation led to an approximate 15.0% increase in TwKO activity toward ent-kaurenoic acid; and the L387A, L387S, L387T, and L387G mutations resulted in an approximate 62.7–69.0% decrease in TwKO activity toward 16α-hydroxy-ent-kaurenoic acid. Notably, both the L387D and L387R mutations caused the loss of the specific catalytic function from 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid (Figure 7).
FIGURE 7.
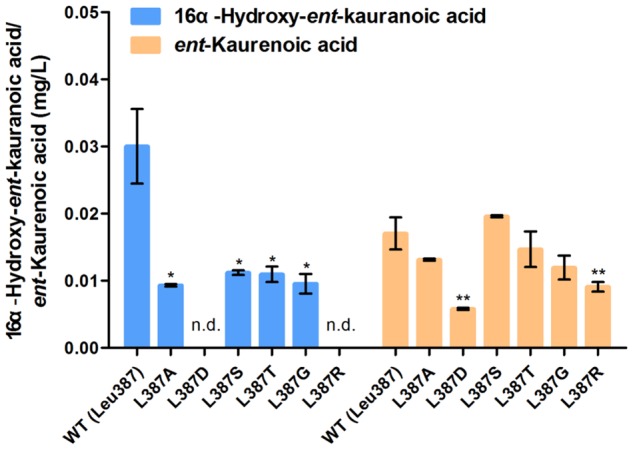
Relative catalytic activity of TwKO mutants when ent-kaurene and 16α-hydroxy-ent-kaurane were used as substrates. Means ± SD of three biological replicates are shown (∗P < 0.05, ∗∗P < 0.01).
Discussion
Since the discovery of the notable anti-inflammatory, immunosuppressive and antitumor activity of diterpenoids, particularly the abietane-type diterpenoid triptolide, multiple studies have been reported to target toward the investigation of the triptolide biosynthetic pathway in order to directly obtain this bioactive compound using synthetic biology strategies (Andersen-Ranberg et al., 2016; Hansen et al., 2017; Inabuy et al., 2017). However, little attention has been paid to the biosynthesis of the kaurene-type diterpenoids, e.g., bioactive GAs and anti-HIV principle tripterifordin. Here, we developed the T. wilfordii transcriptome library and searched in particular for enzymes involved in the kaurene-type diterpenoids biosynthesis. Then, we identified an ent-kaurene oxidase (TwKO), as well as four NADPH-CPRs (TwCPRs) in T. wilfordii.
Functional Characterization and Tissue-Specific Expression Analysis of the TwCPRs
In plants, thousands of metabolites are generated in response to environmental change, and P450s play vital roles in the biosynthesis of these metabolites. To support enzyme activities of different P450s, higher plants possess more than one CPR. These CPR genes include at least one constitutively expressed CPR to support their growth and development and other inducible expressed CPRs to cope with environmental challenges (Nicotra et al., 2010). In the work presented here, four TwCPR genes were isolated and characterized for their catalytic properties, and tissue-specific expression analysis.
To our knowledge, the amino acid sequences of CPRs in plants shared relatively high homology, but their potential as electron donors for P450 monooxygenases seem to be different. Guo et al. (2013) found that S. miltiorrhiza CPR1 (SmCPR1) was more competent than SmCPR2 to support the activity of the SmCYP76AH1 for producing ferruginol at a titer of 10.5 and 5.2 mg/L in the engineered yeasts. Although four TwCPRs showed similar levels of activity (Supplementary Figure S3 and Table 2), co-expression with TwKO in yeast showed TwCPR3 has slightly better activity for providing the electrons to support TwKO activity with D-galactose treatment (Figure 5), indicating that TwCPR3 was most likely to be the best choose for use in the functional analysis of other cytochrome P450 monooxygenases. TwCPRs were found to have relatively ubiquitous expression in all sampled organ tissues (root, stem, leaf, and flower). TwCPR1 showed relative higher transcription levels in all tissues verse other three TwCPRs, suggesting that TwCPR1 was predominant electron donors responsible for metabolites biosynthesis in planta (Figure 2). TwCPR3 showed the lowest expression in all tissues (root, stem, leaf, and flower), implying that TwCPR3 might be an inducible expressed CPR responded to exogenous environmental stimuli (e.g., D-galactose) similar to the reported Andrographis paniculata CPR2 gene (Lin et al., 2017).
Novel Catalytic Function of TwKO
Bioactive GAs are diterpene plant hormones that are biosynthesized through complex pathways and control diverse aspects of growth and development (Yamaguchi, 2008; Araújo and Fernie, 2012; Schwechheimer, 2012). T. wilfordii produces not only ent-kaurene as a key intermediate to GAs, but also 16α-hydroxy-ent-kaurane, whose physiological role is unknown. Functional characterization of TwKO revealed that it could catalyze three-step oxidation of the C4α methyl of the tetracyclic diterpene intermediate ent-kaurene to form ent-kaurenoic acid as an early step in GA biosynthesis. Notably, it could also convert 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid. In P. patens, 16α-hydroxy-ent-kaurane has no physiological role in growth and development and is directly released into the air (Von Schwartzenberg et al., 2004), while the role of this hydroxylated compound in P. trichocarpa could be the precursor for other yet unidentified compounds and function as an allelochemical or phytoanticipin (Irmisch et al., 2015).
In T. wilfordii, tripterifordin was reported to inhibit HIV replication in H9 lymphocyte cells with an EC50 of 1 μg/mL (6 μM), while its biosynthetic pathway is unknown. Here, we confirmed that 16α-hydroxy-ent-kaurane was transformed into 16α-hydroxy-ent-kaurenoic acid by TwKO, then could be converted to anti-HIV principle tripterifordin catalyzed by an unreported gibberellin 20-oxidase (GA20ox) (Phillips et al., 1995; Kawai et al., 2014). It is the first time to clarify the physiological role of 16α-hydroxy-ent-kaurane, which is the precursor for tripterifordin biosynthesis in T. wilfordii. Another bioactive compound ent-16α, 17-dihydroxy-kauran-19-oic acid in Siegesbeckia pubescens could regenerate epidermal tissue mainly through epithelial growth factor receptor phosphorylation, indicating this compound might be applied to wound-healing agents and to a basic materials used in cosmetics (Sung et al., 2011). 16α-Hydroxy-ent-kaurane can be also used as an intermediate to synthesize the bioactive ent-16α, 17-dihydroxy-kauran-19-oic acid, via one-step oxidation of the C17 methyl of 16α-hydroxy-ent-kaurenoic acid (Figure 1). In short, these findings paved the way for the complete biosynthesis of the tripterifordin and provided the precursor for biosynthesis of other bioactive diterpenoids.
Assumed Catalytic Mechanism of TwKO
In recent years, many reports have indicated that the substitution of a single amino acid in ent-kaurene synthase and cytochrome P450 oxygenases may change the reaction products (Xu et al., 2007; Jia and Peters, 2016; Scheler et al., 2016), driving us to find out the crucial residue related to the catalytic function of TwKO. The crystal structure of S. coelicolor CYP170A1 was used for homology modeling to construct the three-dimensional structure of TwKO (Zhao et al., 2009), and one key residue (Leu387) was identified to form hydrogen bonds with the hydroxy group (-OH) of 16α-hydroxy-ent-kaurane (Figure 6). All of the six mutants (L387A, L387D, L387S, L387T, L387G, and L387R) effect the TwKO activity toward ent-kaurenoic acid and 16α-hydroxy-ent-kaurenoic acid. In particular, L387D and L387R mutants led to an approximate 65.9 and 46.7% decrease in TwKO activity toward ent-kaurenoic acid, and caused the loss of the specific catalytic function from 16α-hydroxy-ent-kaurane to 16α-hydroxy-ent-kaurenoic acid (Figure 7).
These results suggested that Leu387 interacted with the hydroxy group (-OH) of 16α-hydroxy-ent-kaurane to form hydrogen bond, which contributed to form a catalytic conformation with high catalytic activity toward 16α-hydroxy-ent-kaurenoic acid. When the residue Leu387 was mutated to other amino acids, the space structure of protein residue would be affected and the hydrophilicity/hydrophobicity of the side chain might change, leading to the change of the distance of hydrogen bonding or hydrogen bonding sites. All these changes likely alter the catalytic conformation, which increases or reduces the entropy costs of substrate binding to the catalytic site and thus reduces or increases catalytic activity (Wang J. et al., 2017). Taking the L387D mutant as an example, the hydrophobic side chain of Leu was changed to the hydrophilic side chain of Asp, leading to the hydroxyl group of 16α-hydroxy-ent-kaurane interacted with the main chain of Arg388 to form H-bond (Supplementary Figure S6A). And this change maybe change the catalytic conformation and greatly increase the entropy costs of substrate binding to the catalytic site and thus reduce catalytic activity. As to the L387D and L387S mutants, the residue side chains were altered, resulting in hydrogen bonds could not be formed because of the increased steric hindrance in the bonding position (Supplementary Figures S6B,C). The entropy costs of substrate binding to the catalytic site were increased, thus reducing catalytic activity. Because ent-kaurene does not contain a group providing hydrogen-bonding interaction, the molecular docking results did not show hydrogen bonding interactions between TwKO with ent-kaurene, providing little useful information to explain the how these mutants affect the specific catalytic function from ent-kaurene to ent-kaurenoic acid. To confirm the assumed catalytic mechanism of TwKO calls for further study.
Author Contributions
PS, LH, and WG conceived and designed the research. PS, HG, YfZ, and LG performed the experiments. XW performed the homology modeling and molecular docking. YjZ, TH, JZ, BM, LT, and YT participated in the research and analyzed the data. PS, WG, and LH wrote the paper with contributions of all the authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Xueli Zhang for providing the yeast strain BY-T20 (Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, China), Dr. Xiaohong Zhang for providing HIS-MBP-pET28a plasmid (Karmanos Cancer Institute, United States).
Abbreviations
- CPR
cytochrome P450 reductase
- CTAB
cetyltrimethylammonium bromide
- 2,4-D
2,4-dichlorophenoxyacetic acid
- DCPIP
dichlorophenolindophenol
- ent-CPP
ent-copalyl diphosphate
- GAs
gibberellins
- GC–MS
gas chromatography–mass spectrometer
- GGPP
geranylgeranyl diphosphate
- GO
Gene Ontology database
- IBA
indole-3-butyric acid
- KO
ent-kaurene oxidase
- KOG/COG
Clusters of Orthologous Groups of proteins
- KT
kinetin
- LB
Luria-Bertani
- LRD
labdane-related diterpenoid
- MBP
maltose-binding protein
- MS
Murashige and Skoog
- Nr
NCBI non-redundant protein sequences
- Nt
NCBI non-redundant nucleotide sequences
- ORF
open reading frame
- Pfam
Protein family
- Swiss-Prot
a manually annotated and reviewed protein sequence database
Funding. This work was supported by the National Natural Science Foundation of China (81422053 and 81373906 to WG, and 81325023 to LH), Key project at central government level: The ability establishment of sustainable use for valuable Chinese Medicine Resources (2060302) to LH, National High Technology Research and Development Program of China (863 Program: 2015AA0200908) and Support Project of High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (CIT&TCD20170324) to WG.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01756/full#supplementary-material
References
- Andersen-Ranberg J., Kongstad K. T., Nielsen M. T., Jensen N. B., Pateraki I., Bach S. S., et al. (2016). Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew. Chem. Int. Ed. Engl. 55 2142–2146. 10.1002/anie.201510650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W., Fernie A. (2012). Systems biology of gibberellin induced plant cell growth. Front. Plant Sci. 3:173. 10.3389/fpls.2012.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. H., Guo R. T., Ko T. P., Wang A. H., Liang P. H. (2006). Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J. Biol. Chem. 281 14991–15000. 10.1074/jbc.M512886200 [DOI] [PubMed] [Google Scholar]
- Chen K., Shi Q., Fujioka T., Zhang D.-C., Hu C.-Q., Jin J.-Q., et al. (1992). Anti-AIDS agents, 4. tripterifordin, a novel anti-HIV principle from Tripterygium wilfordii: isolation and structural elucidation. J. Nat. Prod. 55 88–92. 10.1021/np50079a013 [DOI] [PubMed] [Google Scholar]
- Chugh R., Sangwan V., Patil S. P., Dudeja V., Dawra R. K., Banerjee S., et al. (2012). A preclinical evaluation of Minnelide as a therapeutic agent against pancreatic cancer. Sci. Transl. Med. 4 156ra139. 10.1126/scitranslmed.3004334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Liu Y., Huang L., Zhang X. (2012). Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 109 2845–2853. 10.1002/bit.24547 [DOI] [PubMed] [Google Scholar]
- Dai Z., Liu Y., Zhang X., Shi M., Wang B., Wang D., et al. (2013). Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab. Eng. 20 146–156. 10.1016/j.ymben.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Davidson S. E., Smith J. J., Helliwell C. A., Poole A. T., Reid J. B. (2004). The pea gene LH encodes ent-kaurene oxidase. Plant Physiol. 134 1123–1134. 10.1104/pp.103.032706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. (1989). The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7 514–520. [PubMed] [Google Scholar]
- Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., et al. (2003). Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P., Martin M. V., Sohl C. D., Cheng Q. (2009). Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 4 1245–1251. 10.1038/nprot.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Zhou Y. J., Hillwig M. L., Shen Y., Yang L., Wang Y., et al. (2013). CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U.S.A. 110 12108–12113. 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen N. L., Heskes A. M., Hamberger B., Olsen C. E., Hallstrom B. M., Andersen-Ranberg J. (2017). The terpene synthase gene family in Tripterygium wilfordii harbors a labdane-type diterpene synthase among the monoterpene synthase TPS-b subfamily. Plant J. 89 429–441. 10.1111/tpj.13410 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Kawaide H., Notomi M., Sakigi Y., Matsuo A., Nozaki H. (2006). Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580 6175–6181. 10.1016/j.febslet.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444 11–25. 10.1042/bj20120245 [DOI] [PubMed] [Google Scholar]
- Hooley R. (1994). Gibberellins: perception, transduction and responses. Plant Mol. Biol. 26 1529–1555. [DOI] [PubMed] [Google Scholar]
- Inabuy F., Fischedick J. T., Lange I., Hartmann M., Srividya N., Parrish A. N., et al. (2017). Biosynthesis of diterpenoids in Tripterygium adventitious root cultures. Plant Physiol. 175 92–103. 10.1104/pp.17.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S., Muller A. T., Schmidt L., Gunther J., Gershenzon J., Kollner T. G. (2015). One amino acid makes the difference: the formation of ent-kaurene and 16alpha-hydroxy-ent-kaurane by diterpene synthases in poplar. BMC Plant Biol. 15:262. 10.1186/s12870-015-0647-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia M., Peters R. J. (2016). Extending a single residue switch for abbreviating catalysis in plant ent-Kaurene synthases. Front. Plant Sci. 7:1765. 10.3389/fpls.2016.01765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Ono E., Mizutani M. (2014). Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. Cell Mol. Biol. 78 328–343. 10.1111/tpj.12479 [DOI] [PubMed] [Google Scholar]
- Kuntal B. K., Aparoy P., Reddanna P. (2010). EasyModeller: a graphical interface to MODELLER. BMC Res. Notes 3:226. 10.1186/1756-0500-3-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Wang J., Qi M., Guo J., Rong Q., Tang J., et al. (2017). Molecular cloning and functional characterization of multiple NADPH-cytochrome P450 reductases from Andrographis paniculata. Int. J. Biol. Macromol. 102 208–217. 10.1016/j.ijbiomac.2017.04.029 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2012). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Mafu S., Jia M., Zi J., Morrone D., Wu Y., Xu M., et al. (2016). Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 113 2526–2531. 10.1073/pnas.1512096113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann F. M., Prisic S., Davenport E. K., Determan M. K., Coates R. M., Peters R. J. (2010). A single residue switch for Mg2+-dependent inhibition characterizes plant class II diterpene cyclases from primary and secondary metabolism. J. Biol. Chem. 285 20558–20563. 10.1074/jbc.M110.123307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo S. G., Zhou Z. L., Wang Y. Q., Marinello J., He J. X., Li Y. C., et al. (2012). Natural product triptolide mediates cancer cell death by triggering CDK7-dependent degradation of RNA polymerase II. Cancer Res. 72 5363–5373. 10.1158/0008-5472.can-12-1006 [DOI] [PubMed] [Google Scholar]
- Miyazaki S., Katsumata T., Natsume M., Kawaide H. (2011). The CYP701B1 of Physcomitrella patens is an ent-kaurene oxidase that resists inhibition by uniconazole-P. FEBS Lett. 585 1879–1883. 10.1016/j.febslet.2011.04.057 [DOI] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. (1998). Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone D., Chen X., Coates R. M., Peters R. J. (2010). Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. 431 337–344. 10.1042/bj20100597 [DOI] [PubMed] [Google Scholar]
- Nicotra A. B., Atkin O. K., Bonser S. P., Davidson A. M., Finnegan E. J., Mathesius U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15 684–692. 10.1016/j.tplants.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Phillips A. L., Ward D. A., Uknes S., Appleford N., Lange T., Huttly A. K., et al. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Miura K., Itoh H., Tatsumi T., Ueguchi-Tanaka M., Ishiyama K., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. 10.1104/pp.103.033696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A. (1995). Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today 1 270–277. [DOI] [PubMed] [Google Scholar]
- Scheler U., Brandt W., Porzel A., Rothe K., Manzano D., Bozic D., et al. (2016). Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat. Commun. 7:12942. 10.1038/ncomms12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. (2012). Gibberellin signaling in plants – the extended version. Front. Plant Sci. 2:107. 10.3389/fpls.2011.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M. Y., Liu Y., Wang D., Lu F. P., Huang L. Q., Dai Z. B., et al. (2014). Construction of Saccharomyces cerevisiae cell factories for lycopene production. Zhongguo Zhong Yao Za Zhi 39 3978–3985. [PubMed] [Google Scholar]
- Spitzer R., Jain A. N. (2012). Surflex-dock: docking benchmarks and real-world application. J. Comput. Aided Mol. Des. 26 687–699. 10.1007/s10822-011-9533-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P., Cheng Q., Wang X., Cheng X., Zhang M., Tong Y., et al. (2014). Characterization of eight terpenoids from tissue cultures of the Chinese herbal plant, Tripterygium wilfordii, by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Biomed. Chromatogr. 28 1183–1192. 10.1002/bmc.3140 [DOI] [PubMed] [Google Scholar]
- Su P., Hu T., Liu Y., Tong Y., Guan H., Zhang Y., et al. (2017). Functional characterization of NES and GES responsible for the biosynthesis of (E)-nerolidol and (E,E)-geranyllinalool in Tripterygium wilfordii. Sci. Rep. 7:40851. 10.1038/srep40851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P., Tong Y., Cheng Q., Hu Y., Zhang M., Yang J., et al. (2016). Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 6:23057. 10.1038/srep23057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S. H., Park S. H., Song S. Y., Lee S. J., Lee H. W., Kim S. H., et al. (2011). Epidermal regeneration by ent-16alpha, 17-dihydroxy-kauran-19-oic acid isolated from Siegesbeckia pubescens. Cell Prolif. 44 527–536. 10.1111/j.1365-2184.2011.00786.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 k2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov D. V., Gilman B., He Q. L., Bhat S., Low W. K., Dang Y., et al. (2011). XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7 182–188. 10.1038/nchembio.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S. H. (2009). Difficulties and countermeasures in treatment of rheumatoid arthritis with Tripterygium. Chin J. Integr. Tradit. West Med. 29 104–105. 10.3321/j.issn:1003-5370.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Von Schwartzenberg K., Schultze W., Kassner H. (2004). The moss Physcomitrella patens releases a tetracyclic diterpene. Plant Cell Rep. 22 k780–786. 10.1007/s00299-004-0754-6 [DOI] [PubMed] [Google Scholar]
- Wang J., Kawasaki R., Uewaki J. I., Rashid A. U. R., Tochio N., Tate S. I. (2017). Dynamic allostery modulates catalytic activity by modifying the hydrogen bonding network in the catalytic site of human pin1. Molecules 22:E992. 10.3390/molecules22060992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Liu Q., Ai Z., Xiang Y., Qiao Y. (2016). Discovery of dual ETA/ETB receptor antagonists from traditional Chinese herbs through in silico and in vitro screening. Int. J. Mol. Sci. 17:389. 10.3390/ijms17030389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Yang Y., Wu X., Fan H., Qiao Y. (2017). Identification of berberine as a direct thrombin inhibitor from traditional Chinese medicine through structural, functional and binding studies. Sci. Rep. 7:44040. 10.1038/srep44040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. F., Yuan Y., Luk J. M. (2012). Tripterygium wilfordii bioactive compounds as anticancer and anti-inflammatory agents. Clin. Exp. Pharmacol. Physiol. 39 311–320. 10.1111/j.1440-1681.2011.05586.x [DOI] [PubMed] [Google Scholar]
- Xu M., Wilderman P. R., Peters R. J. (2007). Following evolution’s lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. U.S.A. 104 7397–7401. 10.1073/pnas.0611454104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59 225–251. 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yang C. Q., Lu S., Mao Y. B., Wang L. J., Chen X. Y. (2010). Characterization of two NADPH: cytochrome P450 reductases from cotton (Gossypium hirsutum). Phytochemistry 71 27–35. 10.1016/j.phytochem.2009.09.026 [DOI] [PubMed] [Google Scholar]
- Zhao B., Lei L., Vassylyev D. G., Lin X., Cane D. E., Kelly S. L., et al. (2009). Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J. Biol. Chem. 284 36711–36719. 10.1074/jbc.M109.064683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Gao Y., Hoy J. A., Mann F. M., Honzatko R. B., Peters R. J. (2012). Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J. Biol. Chem. 287 6840–6850. 10.1074/jbc.M111.337592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. L., Yang Y. X., Ding J., Li Y. C., Miao Z. H. (2012). Triptolide: structural modifications, structure-activity relationships, bioactivities, clinical development and mechanisms. Nat. Prod. Rep. 29 457–475. 10.1039/c2np00088a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.