Abstract
Earlier studies showed that human lens ALDH1A1 plays a critical role in protection against oxidative stress-induced cytotoxicity in human lens epithelial cells (HLEC), and opacification of rat and mouse lens. The complete coding sequence of ALDH1A1 was cloned from human lens cDNA library by using PCR methods and expressed it in Escherichia coli. The cloned human lens ALDH1A1 cDNA encodes a 501-amino-acid protein (molecular mass = 54.8 kD) that is 100% identical to human liver ALDH1A1 and shares significant identity with the same isozyme from other tissues and species. The purified recombinant human lens ALDH1A1 exhibited optimal catalytic activity at pH 8 and preferred NAD+ as cofactor and specifically catalyzed the oxidation of toxic lipid aldehydes such as 4-hydroxynonenal (HNE; Km = 4.8 µM) and malonaldehyde (Km MDA = 3.5 µM). Citral, disulfiram, and cyanamide were found to inhibit human lens ALDH1A1 at IC50 values of 55, 101, and 22610 µM, respectively, whereas diethylstilbestrol (DES) was found to be an activator (EC50, 1.3 µM). Further, modification of recombinant human lens ALDH1A1 with nitric oxide donors such as S-nitroso-N-acetylpenicillamine (SNAP) and S-nitrosoglutathione (GSNO) significantly inhibited the enzyme activity. It therefore appears that activation of ALDH1A1, which efficiently catalyzes the detoxification of lipid-derived toxic aldehydes, and/or prevention of its oxidative modification may be novel therapeutic interventions against oxidative stress-induced lens pathologies.
Cataracts are the leading cause of blindness globally (Resnikoff et al., 2002). Although cataract surgery with lens extraction and intraocular lens (IOL) implant is effective and successful, significant costs and unexpected complications associated with surgical procedures have encouraged investigators to seek more potential cost-effective therapeutic strategies (Steinberg et al., 1993). Etiology of cataracts remains obscure; however, oxidative stress due to aging, long-term ultraviolet (UV) exposure, and diabetes are established as important contributors to cataractogenesis (Lassen et al., 2008; Rafnsson et al., 2005; Spector, 1995; Srivastava et al., 1990). Under conditions of oxidative stress, peroxidation of polyunsaturated fatty acids forms cytotoxic lipid peroxidation products known as lipid-derived aldehydes (LDA). 4-Hydroxynonenal (HNE) is one of the most toxic and abundant LDA and known to interfere with normal cellular functions, including signal transduction, DNA synthesis, and enzyme activities, by attacking various amino acid groups with its reactive double bonds (C⩢C and C⩢O) to form Michael adducts or Schiff bases (Siems & Grune, 2003; Schaur, 2003). Our studies showed that micromolar concentrations of HNE induce significant apoptosis in human lens epithelial cells (HLEC) and cataract formation in rat lenses (Ansari et al., 1996; Choudhary et al., 2006; Xiao et al., 2004), suggesting that increased levels of toxic lipid aldehydes contribute to the pathophysiology of oxidative stress-induced cataractogenesis. Therefore, to prevent oxidative stress-induced toxicity and cataractogenesis, it is critical for the lens to rapidly metabolize HNE and other aldehydes generated from lipid peroxidation.
There are three known pathways associated with the detoxification of HNE: (1) conjugation with glutathione (GSH) to form GS-HNE catalyzed by glutathione S-transferase (GST) (He et al, 1996); (2) reduction of HNE to 1,4-dihydroxy-2-nonene (DHN) and GS-HNE to GS-DHN catalyzed by aldose reductase (AR) (Srivastava et al, 2000); and (3) oxidation to 4-hydroxy-2-nonenoic acid (HNA) catalyzed by aldehyde dehydrogenase (ALDH) isozymes (Choudhary et al., 2003; Pappa et al., 2003). Our earlier studies indicated that ALDH isozymes play a major role in the metabolism and detoxification of HNE in the lens (Choudhary et al., 2003; Pappa et al., 2003). In a previous study, Choudhary et al. (2006) found that transfecting HLEC with ALDH1A1-specific antisense or SiRNA reduced oxidation of 3H-HNE to 3H-HNA by approximately 40% without a concomitant increase in GS-HNE conjugate formation but with a parallel rise in apoptosis and decrease in cell viability. Rat lenses transfected with ALDH1A1-specific SiRNA showed approximately 40% fall in ALDH1A1 RNA and displayed a proportional decrease in the formation of HNA accompanied by an increase in the formation of protein-HNE adducts and accelerated opacification of the rat lens (Choudhary et al., 2003). Finally, Aldh1a1 transgenic knockout mice develop lens opacities later on in life that are associated with decreased proteasomal activity accompanied by increased protein oxidation and elevated levels of GSH and 4-hydroxy-2-nonenal– and malondialdehyde–protein adducts (Lassen et al., 2007). Therefore, regulation of lens ALDH1A1 might be a significant therapeutic option to prevent or delay cataractogenesis. Although ALDH1A1 proteins from various tissues and species such as human liver, chick retina, rat liver, and elephant shrew lens have been cloned and characterized (Hsu et al., 1985; Penzes et al., 1997), the structural and biochemical properties of human lens ALDH1A1 have not yet been reported. In order to understand the important role of human lens ALDH1A1 in oxidative cataractogenesis, studies were undertaken to clone human lens ALDH1A1 DNA, and this lens ALDH1A1 was expressed in E. coli. The biochemical properties of recombinant purified human lens ALDH1A1 were subsequently investigated after oxidative modifications.
MATERIALS AND METHODS
Synthesis and Cloning of Lens DNA
Lens cDNA constructed according to Bonaldo et al (1996) was obtained from the Human Eye cDNA Library in Genebank UI-E-DW (BU739505.). Single-stranded lens cDNA was primed with an oligo-dT primer containing a Not I site and a library tag sequence (CGATTAGCGA) located between the Not I site and the (dT) 18 tail. Synthesized double-stranded lens DNA was ligated to an EcorR I adaptor, digested with Not I, and cloned directly into pT7T3-Pac vector.
Subcloning of Human Lens ALDH1A1 DNA and Plasmid Construction
From the human eye lens cDNA library, ALDH1A1 was amplified by using ALDH1A1 specific primers by polymerase chain reaction (PCR). The primers used for PCR were (ALDH1A1–1NdeI-NH 5′-GGGAATTCCATATGTCATCCTCAGGCACG-3′ and ALDH1A1–501Xho-COH 5′-CCGCTCGAGTTATGAGTTCTTCTGAGA-3′). The amplified ALDH1A1 PCR product (1.5 kb of DNA) was isolated, purified, and inserted into an expression vector pET28b (Novagen, San Diego, CA). NdeI and XhoI (New England Biolabs, Ipswich, MA) were used as the cloning sites. The insertion of the human lens ALDH1A1 cDNA was verified by re-digestion with the restriction enzyme. pET28b is a kanamycin resistant plasmid and has both an N-terminal and a C-terminal his-tag. The N-terminal his-tag can be removed by thrombin.
Preparation of Recombinant Human Lens ALDH1A1 cDNA
Transformation of ALDH1A1 plasmid was made by using 100 µl of E. coli BL21DE3 competent cells (Navogen, San Diego, CA) with 50–100 ng of ALDH1A1 expression vector, seeded in agar petri dishes at 37°C overnight. Single clones were picked and grown on LB media with 50 µg/ml of kanamycin. Plasmid DNA was extracted and purified using Qiagen plasmid kit (Qiagen, Valencia, CA). Double-stranded ALDH1A1 plasmid (1 µg) with T7 forward and T7 reverse primers was used for DNA amplification (ALDH1A1-FORWARD 5′-CCTGCACTGAGCTGTGGAAACAC-3′ and ALDH1A1-REVERSE 5′-CATTAGAGAACACTGTGGGCTGG-3′). Sequence analysis was performed using ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Successfully transformed bacteria containing ALDH1A1 expression vector were stored in 30% glycerol at −80°C until used for the expression and purification of recombinant human lens ALDH1A1.
Expression, Purification, and Sequencing of Recombinant Human Lens ALDH1A1 Protein
ALDH1A1-transformed BL21DE3 cells were grown in LB media at 37°C with a shaking speed of 250 rpm for 12 h. Isopropyl-D-1-thiogalactopyranoside (IPTG, Sigma Aldrich, St. Louis, MO) at 1 mM was added to induce the production of ALDH1A1 protein until optical density (OD) reached 0.7–0.9. After the incubation, bacterial pellets were collected and sonicated by sonifier cell disruptor (Amphotech Ltd, St. Beverly, MA) in 20 ml homogenizing buffer (20 mM sodium phosphate, 150 mM sodium chloride, 2 mM EDTA, pH 7.8) with 1 ml protein inhibitor (Sigma Aldrich, St. Louis, MO) and 1 mM dTT. Centrifuged and filtered samples were loaded onto the Ni-His tag column (Novagen, San Diego, CA) and His-tag ALDH1A1 was processed with thrombin (Novagen, San Diego, CA), 1 mg ALDH1A1 protein per unit of thrombin, for 5 h at room temperature. ALDH1A1 protein was eluted by elution buffer (20 mM Tris-HCl, 150 mM NaCl, 2.5 mM EDTA, pH 8.4), and free thrombin was removed by streptavidin agarose (Novagen, San Diego, CA). Protein concentration and enzyme activity were determined with a Cary 100 ultraviolet–visible (UV-Vis) spectrophotometer (Palo Alto, CA). Fifty picomoles (2.5 µg) of purified ALDH1A1 protein was used for protein sequence analysis by the CLC Capillary 494 protein sequencing system (Applied Biosystems, Foster City, CA).
Substrate Specificity of Recombinant Human Lens ALDH1A1
ALDH1A1 activity was monitored for 5 min at 340 nm by using Cary 100-UV-Vis spectrophotometer in 100 mM sodium pyrophosphate buffer (pH 8) with 1 mM dTT, 2 mM NAD, 50 µg protein, and different substrates in various concentrations. Substrates such as acetaldehyde, benzaldehyde, propionaldehyde, trans-2-heptenal, all-trans-retinal, hexanal, MDA (all from Sigma Aldrich, St. Louis, MO), and HNE (Cayman Chemical, Ann Arbor, MI) were used. For IC50 determination, various inhibitors such as citral, disulfiram, and cyanamide (Sigma Aldrich, St. Louis, MO) were incubated with 50 µg of ALDH1A1 for 5 min; subsequently enzyme activity was determined using propionaldehyde as a substrate. For the determination of EC50, the activator DES (Sigma Aldrich, St. Louis, MO) was incubated with 50 µg ALDH1A1 for 5 min; subsequently enzyme activity was determined using propionaldehyde as substrate.
Regulation of Recombinant Human Lens ALDH1A1 Activity
Fifty micrograms of ALDH1A1 was incubated with: (1) 0.1, 0.2, 0.5, or 1 mM S-nitroso-N-acetylpenicillamine (SNAP, Sigma Aldrich, St. Louis, MO); (2) 50, 100, 150, or 200 µM S-nitrosoglutathione (GSNO, Sigma Aldrich, St. Louis, MO); and (3) 10, 20, 50, or 100 µM oxidized glutathione (GSSG, Sigma Aldrich, St. Louis, MO). After incubation for 0, 50, 100, 150, or 200 min, an aliquot of 100 µl was withdrawn to determine enzyme activity with 2 mM NAD+ and 1 mM propionaldehyde at 340 nm for 5 min. For NAD protection experiment, 50 µg ALDH1A1 was incubated with or without 0.1, 0.5, or 1 mM NAD for 5 min, and then 100 µM GSNO was added. After 0, 5, 10, 20, 50, 100, or 150 min, 100-µl aliquots were withdrawn to detect ALDH1A1 activity with 1 mM propionaldehyde.
RESULTS
Cloning and Expression of Recombinant Human Lens ALDH1A1
The complete cDNA sequence of human lens ALDH1A1 was generated from the human lens cDNA library by polymerase chain reaction with primers to both ends with a human liver cDNA template. The amplified cDNA was approximately 1.5 kb by gel electrophoresis and the DNA sequence analysis confirmed it to be a 1506-bp-long complete ALDH1A1 cDNA (data not shown). The cDNA sequence was submitted to DNA sequence database (GenBank AY390731). Human lens ALDH1A1 cDNA shows the closest nucleotide similarity with other ALDH1A1 previously cloned from human liver (100%; NM_000689), pongo kidney (98%; CR860845), rabbit cornea (91%; AY038801), cow liver (90%; BC105193), sheep liver (90%; NM_001009778), pig liver (90%; AK232310), rat liver (85%; U79118), and mouse liver (85%; M74570).
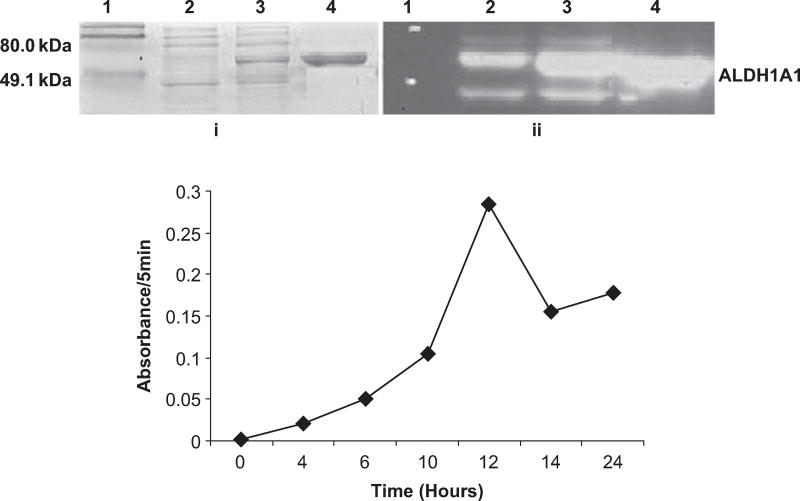
The cloned human lens ALDH1A1 DNA in E. coli. BL21DE3 cells was then expressed. Treatment with isopropyl-D-1-thiogalactopyranoside (IPTG) significantly increased the level of ALDH1A1 expression in E. coli (Figure 1A, i). To optimize IPTG induction time, proteins were harvested at time points 0, 4, 6, 10, 14, 16, and 24 h postinduction and the enzymatic activity of ALDH1A1 was measured. As shown in Figure 1B, the highest level of ALDH1A1 activity was achieved after 12 h of IPTG induction, and thus large-scale ALDH1A1 protein extraction was performed 12 h after IPTG treatment. The recombinant human lens ALDH1A1 was purified by using a Ni-His-tag column and His-tag was removed by processing with thrombin. The complete protein sequence analysis identified it as a 501-amino-acid protein (data not shown). The recombinant human lens ALDH1A1 was purified with an average yield of approximately 50 mg/L culture media with specific activity of 0.27 IU/mg and showed a molecular mass of approximately 55 kD. The recombinant human lens ALDH1A1 was cross-reacted with human liver ALDH1A1 antibodies, generously provided by Dr. Henry Weiner, Purdue University (Figure 1A, ii).
FIG. 1.
Characterization of recombinant human lens ALDH1A1: (A) 5-µg protein samples were run on a 12% SDS-PAGE gel, and (i) stained with Coomassie blue, (ii) immunoblotted with anti-ALDH1A1 antibodies. Lane 1: protein marker; lanes 2 and 3: protein before and after 12 -h IPTG induction; lane 4: purified ALDH1A1 through Ni-His-tag column. (B) Induction of human lens ALDH1A1 activity by IPTG. Protein extracts were obtained from aliquots of E. coli collected at time points 0, 4, 6, 10, 14, 16, and 24 h after IPTG treatment, and the enzymatic activity of ALDH1A1 was determined using propionaldehyde as the substrate and measured at 340 nm for 5 min by spectrophotometer.
Substrate Specificity of Recombinant Human Lens ALDH1A1
Subsequently, the catalytic efficiency of human lens ALDH1A1 was examined with various substrates and kinetic parameters calculated as shown in Table 1. Notably, 4-hydroxynonenal (HNE) and malonaldehyde (MDA) exhibited significantly lower Km values, 4.8 µM and 3.5 µM, respectively, compared to the other substrates examined. The catalytic efficiency of ALDH1A1 was highest for MDA (Vmax/Km = 109.03) due to its exceptionally high Vmax and low Km values. Despite having the lowest Vmax among the substrates tested, HNE was nevertheless efficiently catalyzed by ALDH1A1, as demonstrated by a comparably high Vmax/Km value of 28.1.
TABLE 1.
Kinetic Parameters of Human Lens ALDH1A1 Using Various Substrates
| Substrate |
Vmax (nmol/min/mg) |
Km (µM) | Vmax/Kmb |
|---|---|---|---|
| Acetaldehyde | 631.4 ± 10.9 | 238.2 ± 12.9 | 2.7 |
| Benzaldehyde | 750.3 ± 121.3 | 143.2 ± 6.4 | 5.2 |
| HNE | 135 ± 15.2a | 4.8 ± 0.12a | 28.1a |
| Propionaldehyde | 445.3 ± 34.4 | 121.4 ± 6.7 | 3.7 |
| Malonaldehyde | 381.6 ± 11.4 | 3.5 ± 0.6 | 109.0 |
| trans-2-Heptenal | 155.8 ± 7.3 | 177.1 ± 11.7 | 0.88 |
| NAD | 149.7 ± 4.9 | 59.4 ± 2.4 | 2.5 |
Note. Enzyme activities were measured in a mixture of 100 mM sodium pyrophosphate (pH 8.0) with 50 µg purified ALDH1A1, 1 mM dTT, 2 mM NAD+, and various concentration of substrates (10 µM to 3200 µM) by spectrophotometer at 340 nm for 5 min. For NAD+, 1 mM propionaldehyde was used as the substrate. Km and Vmax for various ALDH1A1 substrates and NAD+ were calculated using Sigma and Prizm software. Values are represented as mean ± SD.
Parameters were determined at pH 7.0.
Vmax/Km represents the catalytic efficiency of ALDH1A1.
Further, the enzymatic activity of human lens ALDH1A1 was determined after treatment with known ALDH inhibitors such as citral, disulfiram, or cyanamide and an activator such as diethylstilbestrol (DES). Human lens ALDH1A1 was incubated with various concentrations of inhibitors (10 to 25,600 µM) and activator (10 to 3200 µM) for different time intervals and enzyme activity was determined spectrophotometrically at 340 nm in the presence of 1 mM propionaldehyde as the substrate. The IC50 and EC50 values were calculated. Citral, disulfiram, and cyanamide demonstrated IC50 values of 55, 101, and 22,610 µM, respectively, for recombinant human lens ALDH1A1, whereas DES effectively activated human lens ALDH1A1 with EC50 as 11.3 µM (Table 2).
TABLE 2.
Inhibition and Activation of Human Lens ALDH1A1 Activity
| IC50 (µM) | EC50 (µM) | ||
|---|---|---|---|
| Citral | 55 | DES | 11.3 |
| Disulfiram | 101 | ||
| Cyanamide | 22,610 | ||
Note. Fifty micrograms ALDH1A1 was pre-incubated with either inhibitors citral, disulfiram, and cyanamide (10 µM to 25,600 µM) or the activator diethylstilbestrol (10 µM to 3200 µM) for 5 min, and then a reaction mixture containing 100 mM sodium pyrophosphate (pH 8.0), 1 mM dTT, 2 mM NAD+, and 1 mM propionaldehyde was added. The enzymatic activity of ALDH1A1 was monitored at 340 nm for 5 min by spectrophotometer. IC50 for citral, disulfiram, and cyanamide and EC50 for DES were calculated.
Regulation of Recombinant Human Lens ALDH1A1
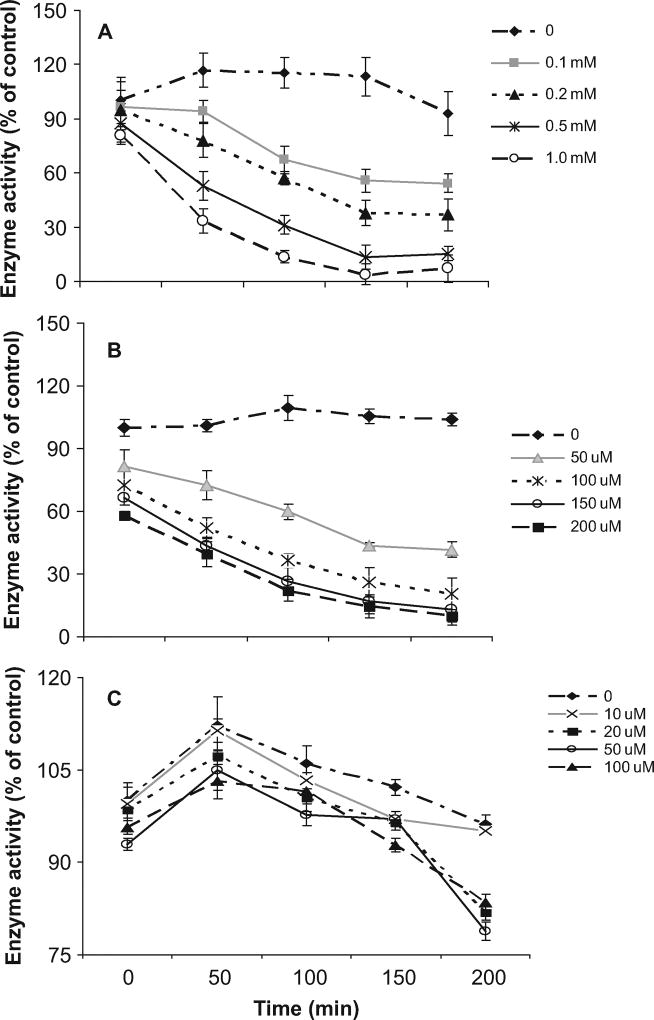
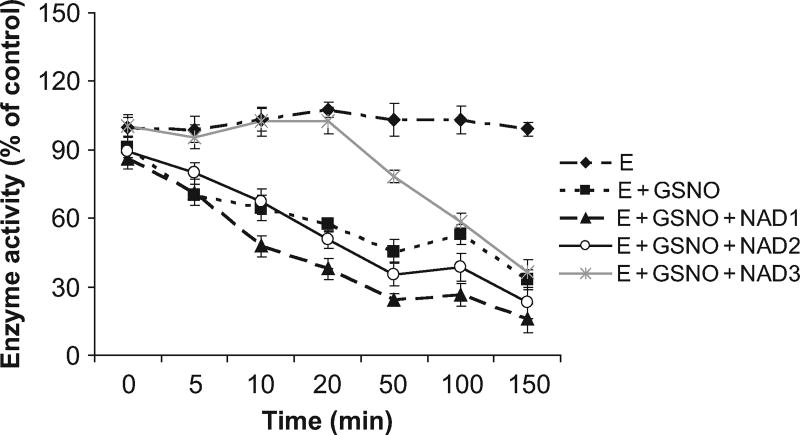
To further investigate the biochemical properties of recombinant human lens ALDH1A1, the enzyme with SNAP, GSNO, or GSSG was structurally modified. Incubation of ALDH1A1 with various concentrations of SNAP or GSNO resulted in a time- and concentration-dependent inhibition of ALDH1A1 activity (Figure 2, A and B). However, when treated with various concentrations of GSSG, ALDH1A1 activity was quantitatively increased until 50 min and reduced thereafter but not significantly (above 75% of control; Figure 2C). NAD imparted an effective protection to GSNO modified enzyme at high concentration (1 mM) within 30 min (Figure 3).
FIG. 2.
Inhibition of human lens ALDH1A1 activity by NO donors: 50 µg ALDH1A1 was incubated with (A) 0.1, 0.2, 0.5, and 1.0 mM SNAP or (B) 50, 100, 150, and 200 µM GSNO or (C) 10, 20, 50, and 100 µM GSSG. After 0, 50, 100, 150, and 200 min, an aliquot of 100 µl was withdrawn to determine enzyme activity with 2 mM NAD+ and 1 mM propionaldehyde at 340 nm for 5 min. The data are represented as percent of unmodified (control) ALDH1A1 activity. Values are mean ± SD.
FIG. 3.
NAD protects GSNO modified human lens ALDH1A1: 100 µM (NAD1), 500 µM (NAD2), and 1 mM (NAD3) of NAD were incubated with 50 µg of ALDH1A1 for 5 min, and then 100 µM GSNO was added. After 0, 5, 10, 20, 50, 100, and 150 min, a 100-µl aliquot was withdrawn to determine enzyme activity with 2 mM NAD+ and 1 mM propionaldehyde at 340 nm for 5 min. The data are represented as percent of unmodified (control) ALDH1A1 activity. Values are mean ± SD.
DISCUSSION
Previous studies by Choudhary et al. (2003) and King et al. (1998) showed that ALDH1A1 is the predominant ALDH isozyme in rat lenses and human lens epithelial cells (HLEC). In this study, ALDH1A1 was successfully amplified and cloned from human lens cDNA library. This is the first report on the cloning and expression of human lens ALDH1A1. The complete human lens ALDH1A1 cDNA encodes a 501-amino-acid protein, which shares a close sequence similarity with other ALDH1A1 isolated from human liver, pongo kidney, rabbit cornea, and rat and mouse liver. Identical sequence for human liver and lens ALDH1A1 implied a possible shared function, which may involve detoxification of reactive aldehydes via oxidation to their corresponding acids. The purified recombinant human lens ALDH1A1 has close structural and kinetic properties with ALDHA1s isolated from other species (Zheng et al., 1993; Bostian et al., 1978; Crabbe et al., 1991; Manzer et al., 2003).
To characterize the substrate specificity of recombinant human lens ALDH1A1, enzyme activity assay was conducted using aldehyde-based ALDH1A1 substrates such as acetaldehyde, propionaldehyde, MDA, and HNE. Among the various substrates tested, MDA and HNE demonstrated significant low Km values (Table 2), indicating their high reactivity with human lens ALDH1A1; probably both the substrates fit the active site well and rapidly induced the conformational change. Rabbit corneal ALDH1A1 exhibited similar properties with respect to MDA and HNE metabolism (Manzer et al., 2003). MDA and HNE are cytotoxic LDAs generated through lipid peroxidation, which is induced by oxidative stress resulting from aging, chronic UV exposure, or diabetes. Studies showed that MDA is the most common aldehyde and comprised 70% of the total aldehydes produced (Esterbauer et al, 1991; Townsend et al., 2001), and HNE is the most cytotoxic aldehyde, accounting for 5% of total aldehydes (Benedetti et al., 1980; Lindahl et al., 1991). In contrast to ALDH3A1, which prefers medium-chain lipid and aromatic aldehydes such as hexenal and benzaldehyde (Lindahl et al., 1991; Vasiliou et al., 1999; Pappa et al., 2003), ALDH1A1 prefers short-chain aldehydes such as malondialdehyde (Lindahl et al., 1984; Pappa et al., 2003). The accumulation of LDA in the lens contributes to the formation of cataracts, and their removal is therefore necessary to maintain lens transparency. Here data showed that human lens ALDH1A1 preferably oxidizes MDA and HNE, suggesting that ALDH1A1 may play a critical role in detoxifying MDA and HNE thus maintains lens clarity. This is further supported by the recent studies demonstrating that ALDH1A1-deficient mice later in life exhibit lens opacifications, which are associated with decreased proteasomal activity, increased protein oxidation, and elevated levels of GSH and of 4-hydroxy-2-nonenal– and malondialdehyde–protein adducts (Lassen et al., 2007). Based on all these data, it is apparent that inhibition or activation of this enzyme may change the lens ALDH1A1 function. Treating recombinant human lens ALDH1A1 with citral or disulfiram but not cyanamide effectively reduced its activity, as demonstrated by relatively low IC50 values of 55 and 101 µM for citral and disulfiram, respectively, compared to the significantly high IC50 of 22,610 µM with cyanamide (Table 2). Citral (3,7-dimethy l-2,6-octadienal) is a reactive and volatile α,β-unsaturated aldehyde that occurs naturally in herbs, plants, and citrus fruits. It was found that citral inhibits human liver ALDH via the formation of covalent bonds with sulfhydryl groups on the enzyme, and inhibition was reversed through the dilution of citral (Kikonyogo et al., 1999). In fact, citral may be regarded as not only an inhibitor but also a highly favored substrate of human liver ALDH, as demonstrated by a marked low Km value of 4.2 µM (Kikonyogo et al., 1999).
Disulfiram has been widely used in aversion therapy for alcoholics for over 50 yr. As reported by Lam et al. (1997), disulfiram is rapidly reduced in vivo to its corresponding metabolites, such as S-methyl-N,N-diethylthiocarbamoyl sulfoxide (MeDTC-SO) and S-methyl-N,N-diethylthiocarbamoyl sulfone (MeDTC-SO2), which are responsible for the inactivation of human cytosolic and mitochondrial ALDH, and disulfiram is a highly effective inhibitor of human liver ALDH1A1 with an IC50 as low as 0.15 µM. Cyanamide is commonly used as an alcohol consumption deterrent agent and must be bioactivated in vivo to become an active inhibitory species (Demaster et al., 1998). At low concentrations, cyanamide was shown to effectively inhibit rat liver mitochondrial ALDH after its enzymatic conversion to a reactive derivative (Svanas et al., 1985). Probably this is why our recombinant human lens ALDH1A1 in vitro exhibited a remarkably high IC50 to cyanamide; similar observations were made by Svanas et al. (1985), where affinity-purified ALDH was inactivated only by high concentrations of cyanamide (Choudhary et al., 2003).
Diethylstilbestrol, a steroid hormone, was found to activate sheep liver ALDH1A1 approximately 2.5-fold in the presence of 1 mM acetaldehyde and 10–50 µM NAD+ (Kitson et al., 1982). With a lower concentration of substrate and/or coenzyme the activation is less pronounced or is changed to mild inhibition (Kitson et al., 2001). Our data showed that DES treatment effectively increased the activity of human lens ALDH1A1 with an EC50 value of 11.3 µM. Further studies on ALDH1A1 activators will be helpful to look for and to design a specific activator to regulate ALDH1A1 activity in the lens and prevent lens opacity from oxidation stress.
Besides exogenous inhibitors such as citral and disulfiram, endogenous toxic metabolites such as oxidizers and nitric oxide (NO) may also inhibit ALDH1A1 enzyme activity. NO is well known to be highly reactive, diffusible, and unstable radical that plays an important role in the regulation of a wide range of physiological processes, including cellular immunity, angiogenesis, neurotransmission, and platelet aggregation. Previously, Chandra et al. (1997) and Ramana et al. (2003) showed that modification of recombinant aldose reductase (AR) by GSNO that forms a mixed disulfide with active site AR sulfhydryl produced time- and concentration-dependent inactivation of the enzyme. To test the effect of NO on the catalytic activity of human lens ALDH1A1, recombinant human lens ALDH1A1 was incubated with various concentrations of SNAP, GS-NO, or GSSG and enzyme activity was monitored over time. Our results showed that both SNAP and GS-NO treatment rapidly inactivated ALDH1A1 in a time- and concentration- dependent manner. Since SNAP might only nitrosate and GS-NO nitrosates as well as glutathiolates the sulfhydryl of ALDH1A1, it is likely that both glutathiolation and nitrosation might inactivate the enzyme. However, it is unlikely that glutathiolation inactivates ALDH1A1 because GSSG, which is an excellent glutathiolation reagent, did not inactivate the enzyme for up to 50 min and there after the inhibition was only milder. It therefore appears that inhibition of ALDH1A1 by SNAP and GSNO is via nitrosation. These studies further emphasize that under oxidative stress, nitrosation of ALDH1A1 may be a threat to the lens. Further studies showed that NAD is crucial for ALDH1A1 activity and may interfere with NO binding to catalytic site. In recent years there has been interest in determining the role of NO in cataractogenesis. Increased levels of NO were found in the aqueous humor of cataract patients (Kao et al., 2002), in patients with glaucoma (Chiou et al., 2001; Kotikoski et al, 2002), and in animal models of oxidative stress (Lam et al., 1997; Inomata et al., 2000; Inomata et al., 2001). It was reported that NO accelerates selenite cataract and inhibitors of NO synthase inhibit the selenite-induced lens opacification. It was postulated that increased NO in the aqueous humor, under oxidative stress, would be taken up by the lens epithelium as N2O3, which nitrosates GSH to GSNO, a potent glutathiolating agent. ALDH1A1, having a capacious entrance tunnel and an active Cys302, is a reliable candidate to be glutathiolated by GSNO (Moore et al., 199842). Glutathiolation of Cys302 would inactivate ALDH1A1 and thereby decrease the detoxification of LDA such as HNE and increase the HNE-associated toxicity. Furthermore, it appears that regulation of ALDH1A1 activity might be of clinical relevance for conditions in which NO levels increase, including millions of people who use nitroglycerine.
In summary, human lens ALDH1A1 was cloned, expressed, and purified and its kinetic properties were studied. Our data indicated that lens ALDH1A1 is an important defense enzyme against oxidative stress, since this enzyme detoxifies lens LDAs, including HNE and malonaldehyde, thereby protecting the lens from oxidative damage and maintaining its transparency. NO may contribute to the pathogenesis of oxidative cataracts through the inactivation of human lens ALDH1A1. Future studies will focus on investigating the role of NO in oxidation-induced cataractogenesis and devising ways to reduce its impact. In addition, studies on DES and structurally similar compounds are potentially important in designing specific activators that could function to maintain the activity levels of human lens ALDH1A1, thereby preventing or rescuing lens from oxidation-induced cataractogenesis.
Acknowledgments
We thank our colleagues for valuable discussions and critical reading of this article. This work was supported in parts by NIH grants EY13014 (NHA) and EY11490 (VV).
References
- Ansari NH, Wang L, Srivastava SK. Role of lipid aldehydes in cataractogenesis: Hydroxynonenal-induced cataract. Biochem. Mol. Med. 1996;58:25–30. doi: 10.1006/bmme.1996.0028. [DOI] [PubMed] [Google Scholar]
- Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochem. Biophys. Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Bostian KA, Betts GF. Kinetics and reaction mechanism of potassium-activated aldehyde dehydrogenase from Saccharomyces cerevisiae. Biochem. J. 1978;173:787–798. doi: 10.1042/bj1730787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Srivastava S, Petrash JM, Bhatnagar A, Srivastava SK. Modification of aldose reductase by S-nitrosogluthione. Biochemistry. 1997;36:15801–15809. doi: 10.1021/bi9714722. [DOI] [PubMed] [Google Scholar]
- Chiou SH, Chang CJ, Hsu WM, Kao CL, Liu JH, Chem WL, Tsai DC, Wu CC, Chou CK. Elevated nitric oxide level in aqueous humor of patients with acute angle-closure glaucoma. Ophthalmologica. 2001;215:113–116. doi: 10.1159/000050840. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Srivastava S, Xiao T, Andley UP, Srivastava SK, Ansari NH. Metabolism of lipid derived aldehyde, 4-hydroxynonenal in human lens epithelial cells and rat lens. Invest. Ophthalmol. Vis. Sci. 2003;44:2675–2682. doi: 10.1167/iovs.02-0965. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Xiao T, Vergara LA, Srivastava S, Nees D, Piatigorsky J, Ansari NH. Role of aldehyde dehydrogenase isozymes in the defense of rat lens and human lens epithelial cells against oxidative stress. Invest. Ophthalmol. Vis. Sci. 2005;46:259–267. doi: 10.1167/iovs.04-0120. [DOI] [PubMed] [Google Scholar]
- Crabbe MJC, Hoe ST. Aldehyde dehydrogenase, aldose reductase, and free radical scavengers in cataract. Enzyme. 1991;45:188–193. doi: 10.1159/000468888. [DOI] [PubMed] [Google Scholar]
- Demaster EG, Redfern B, Nagaswa HT. Mechanisms of inhibition of aldehyde dehydrogenase by nitroxy, the active metabolite of the alcohol deterrent agent cyanamide. Biochem. Pharmacol. 1998;55:2007–2015. doi: 10.1016/s0006-2952(98)00080-x. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radical Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- He NG, Singhal SS, Chaubey M, Awasthi S, Zimniak P, Partridge CA, Awasthi YC. Purification and characterization of a 4-hydroxynonenal metabolizing glutathione S-transferase isozyme from bovine pulmonary microvessel endothelial cells. Biochim. Biophys. Acta. 1996;1291:182–188. doi: 10.1016/s0304-4165(96)00064-5. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Tani K, Fujiyoshi T, Kurachi K, Yoshida A. Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2. Proc. Natl. Acad. Sci. USA. 1985;82:3771–3775. doi: 10.1073/pnas.82.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata M, Hayashi M, Shumiya S, Kawashima S, Ito Y. Aminoguanidine-treatment results in the inhibition of lens opacification and calpain-mediated proteolysis in Shumiya cataract rats (SCR) J. Biochem (Tokyo) 2000;128:771–776. doi: 10.1093/oxfordjournals.jbchem.a022814. [DOI] [PubMed] [Google Scholar]
- Inomata M, Hayashi M, Shumiya S, Kawashima S, Ito Y. Involvement of inducible nitric oxide synthase in cataract formation in Shumiya cataract rat (SCR) Curr. Eye Res. 2001;23:307–311. doi: 10.1076/ceyr.23.4.307.5455. [DOI] [PubMed] [Google Scholar]
- Kao CL, Chou CK, Tsai DC, Hsu WM, Liu JH, Wang CS, Lin JC, Wu CC, Peng CH, Chang CJ, Kao CL, Chiou SH. Nitric oxide levels in the aqueous humor in cataract patients. J. Cataract Refract. Surg. 2002;28:507–512. doi: 10.1016/s0886-3350(01)01102-6. [DOI] [PubMed] [Google Scholar]
- Kikonyogo A, Abriola DP, Dryjanski M, Pietruszko R. Mechanism of inhibition of aldehyde dehydrogenase by citral, a retinoid antagonist. Eur. J. Biochem. 1999;262:704–712. doi: 10.1046/j.1432-1327.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- King G, Holmes R. Human ocular aldehyde dehydrogenase isozymes: Distribution and properties as major soluble proteins in cornea and lens. J. Exp. Zool. 1998;282:12–17. [PubMed] [Google Scholar]
- Kitson TM, Grow KE. Enzymology of carbonyl metabolism: Aldehydrogenase and aldo/keto reductase. New York: Alan R. Liss; 1982. Activation of aldehyde dehydrogenase by diethylstilbestrol; pp. 37–52. [PubMed] [Google Scholar]
- Kitson TM, Kitson KE, Moor SA. Interaction of sheep liver cytosolic aldehyde dehydrogenase with quercertin, resveratrol and diethylstilbestrol. Chem. Biol. Interact. 2001;130–132:57–69. doi: 10.1016/s0009-2797(00)00222-2. [DOI] [PubMed] [Google Scholar]
- Lam JP, Mays DC, Lipsky JJ. Inhibition of recombinant human mitochondrial and cytosolic aldehyde dehydrogenases by two candidates for the active metabolites of disulfiram. Biochemistry. 1997;36:13748–13754. doi: 10.1021/bi970948e. [DOI] [PubMed] [Google Scholar]
- Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, Vasiliou V. Multiple and additive functions of ALDH3A1 and ALDH1A1: Cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J. Biol. Chem. 2007;282:25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Black WJ, Estey T, Vasiliou V. The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin. Cell Dev. Biol. 2008;19:100–112. doi: 10.1016/j.semcdb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Lindahl R, Evces S. Rat liver aldehyde dehydrogenase. I. Isolation and characterization of four high Km normal liver isozymes. J. Biol. Chem. 1984;259:11986–11990. [PubMed] [Google Scholar]
- Lindahl R, Peterson DR. Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharmacol. 1991;41:1583–1587. doi: 10.1016/0006-2952(91)90157-z. [DOI] [PubMed] [Google Scholar]
- Manzer R, Qamar L, Estey T, Pappa A, Petersen DR, Vasiliou V. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–238. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: The structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- Pappa A, Estey T, Manzer R, Brown D, Vasiliou V. Human aldehyde dehydrogenase 3A1 (ALDH3A1). Biochemical characterization and immunohistochemical localization in the cornea. Biochem. J. 2003;376:615–623. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Wang XS, Sperkova Z, Napoli JL. Cloning of a rat cDNA encoding retinal dehydrogenase isozyme type I and its expression in E. coli. Gene. 1997;191:167–172. doi: 10.1016/s0378-1119(97)00054-1. [DOI] [PubMed] [Google Scholar]
- Rafnsson V, Olafsdottir E, Hrfnkelsson J, Sasaki H, Arnarssp A, Jonasson F. Cosmic radiation increases the risk of nuclear cataract in airline pilots: a population-based case-control study. Arch. Ophthalmol. 2005;23:1102–1105. doi: 10.1001/archopht.123.8.1102. [DOI] [PubMed] [Google Scholar]
- Ramana KV, Chandra D, Srivastava S, Bhatnagar A, Srivastava SK. Nitric oxide regulates the polyol pathway of glucose metabolism in vascular smooth muscle cells. FASEB J. 2003;17:417–425. doi: 10.1096/fj.02-0722com. [DOI] [PubMed] [Google Scholar]
- Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull. WHO. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:149–159. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Siems W, Grune T. Intracellular metabolism of 4-hydroxynonenal. Mol. Aspects Med. 2003;24:167–175. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Spector A. Oxidative stress-induced cataract: Mechanism of action. FASEB J. 1995;9:1173–1182. [PubMed] [Google Scholar]
- Srivastava SK, Ansari NH, Bhatnagar A. Sugar induced cataractogenesis: a paradigm of oxidative tissue pathology? Lens Eye Toxicol. Res. 1990;7:161–171. [PubMed] [Google Scholar]
- Srivastava S, Dixit BL, Cai J, Sharma S, Hurst HE, Bhatnagar A, Srivastava SK. Metabolism of lipid peroxidation product, 4-hydroxynonenal (HNE) in rat erythrocytes: Role of aldose reductase. Free Radical Biol. Med. 2000;29:642–651. doi: 10.1016/s0891-5849(00)00351-8. [DOI] [PubMed] [Google Scholar]
- Steinberg EP, Javitt JC, Sharkey PD, Zuckerman A, Legro MW, Anderson GF, Bass EB, O’Day D. The content and cost of cataract surgery. Arch. Ophthalmol. 1993;111:1041–1049. doi: 10.1001/archopht.1993.01090080037016. [DOI] [PubMed] [Google Scholar]
- Svanas GW, Weiner H. Enzymatic requirement for cyanamide inactivation of rat liver aldehyde dehydrogenase. Biochem. Pharmacol. 1985;34:1197–1204. doi: 10.1016/0006-2952(85)90495-2. [DOI] [PubMed] [Google Scholar]
- Townsend AJ, Leone-Kabler S, Haynes RL, Wu YH, Szweda L, Bunting KD. Selective protection by stably transfected human ALDH3A1 (but not human ALDH1A1) against toxicity of aliphatic aldehydes in V79 cells. Chem. Biol. Interact. 2001;130–132:261–273. doi: 10.1016/s0009-2797(00)00270-2. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogena (ALDH) genes: Human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–434. [PubMed] [Google Scholar]
- Xiao TL, Choudhary S, Vergara LA, Van Kuijk FJGM, Ansari NH. Oxidative stress induced formation of protein-4-HNE adduct in lens epithelial cells: Implication in oxidative cataractoganasis. J. Tissue Res. 2004;4:181–188. [Google Scholar]
- Zheng CF, Wang TT, Weiner H. Cloning and expression of the full-length cDNA encoding human liver class 1 and class 2 aldehyde dehydrogenase. Alcohol Clin. Exp. Res. 1993;17:828–831. doi: 10.1111/j.1530-0277.1993.tb00849.x. [DOI] [PubMed] [Google Scholar]