Baruch Blumberg discovered the Australia antigen when searching for immunological evidence of genetic polymorphisms. His hypothesis was that antibodies to blood proteins could be used to investigate human genetic diversity. The experiment was to look for antibodies in serum from patients who had been exposed to diverse blood sources via repeated transfusions. Blumberg and co-workers discovered a “protein with which they react [that] is very rare in populations of Western origin, but it is not uncommon in Micronesians, Vietnamese, Formosans and Australian aborigines” (1, 2). By 1967, Blumberg and others recognized that Australia antigen was correlated with hepatitis. In 1970 Australia antigen was identified as a structural component of the hepatitis B virus (HBV). That year Dane and co-workers published electron micrographs of 17–22 nm spherical HBV surface antigen (HBsAg) particles, 17–22 nm diameter filamentous HBsAg particles, and 45 nm diameter virions comprised of a HBsAg envelope and a 36 nm diameter core (3). HBV virions are known as Dane particles.
These early studies identified the salient features of the HBV virion and its epidemiology. The virus is outwardly simple but endemic and persistent. HBV is predominantly a virus of Southeast Asia, sub-Saharan Africa, and populations such as aboriginal Australians and Inuit. Worldwide, an estimated 240 million people have chronic HBV (4). This prevalence is consistent with predominantly vertical transmission of infection from mother to child. In young children HBV has a high probability of establishing chronic infection. HBV contributes to about 780,000 deaths each year, predominantly through hepatocellular carcinoma, cirrhosis, and liver failure (4). The HBV vaccine, now based on yeast-expressed HBsAg, the Australia antigen, is very effective and is changing the demographics of HBV disease (5), but cannot help those who are already infected.
Mature HBV is an enveloped, icosahedral virus. It packages a circular dsDNA genome with gaps in both strands and a reverse transcriptase (usually referred to as ‘P’) covalently bound to the 5′ end of the antisense strand; it also packages a number of host proteins. A description of the HBV lifecycle introduces the structural components of the virus and the basis for chronic infection.
To initiate infection, a Dane particle binds to the Na+-Taurocholate Co-transporter Polypeptide (NTCP) (6, 7). The virion is endocytically transferred to the cytoplasm; the mechanism of escape from the endocytic vesicle is not understood – release of duck HBV from endosomes appears to be insensitive to pH (8). The naked, DNA-filled core binds to importin α-importin β complexes via nuclear localization signals on the core protein (HBcAg) (9, 10). Core dissociation and genome release are associated with the nuclear pore complex (11). In the nucleus, P is removed from the DNA and the DNA “repaired” (12, 13). The resulting covalently closed circular DNA (cccDNA) becomes nucleosome-decorated and appears to function like host DNA. Two HBV proteins, HBcAg and the X protein, have been localized to cccDNA and may have regulatory roles (14–17).
In an infected cell, production of new virus starts with RNAs transcribed from cccDNA, presumably by Pol2. A terminally redundant, unspliced copy of the cccDNA (pregenomic RNA or pgRNA) is the messenger for HBcAg, an HBcAg variant known as the “e” antigen (HBeAg), and P protein. A set of sub-genomic RNAs encode the large, medium, and small variants of the surface antigens (L-, M-, and S-HBsAg). A final subgenomic RNA encodes the X protein; X is a non-structural protein that affects cccDNA persistence, cccDNA transcription, and mitochondrial Ca++ flux (15, 18–20). In the cytoplasm, a complex of P protein and pgRNA appears to nucleate assembly of the immature RNA-filled HBV core (21). A substantial number of empty capsids are also formed (22) (a capsid refers to the protein shell of the HBV core). Reverse transcription takes place within the core while it is resident in the cytoplasm, leading to synthesis of a relaxed circular dsDNA product from the linear pgRNA (23); large pores in the capsid surface allow nucleotides to diffuse in and out (24). The mature capsid, and also the empty capsid, carries signals that allow it to bind the L-HBsAg (possibly from late endoplasmic reticulum (ER) membranes) for envelopment and secretion or to bind importin proteins for nuclear transport to generate more cccDNA.
The goal of this review is to describe the structural biology of the HBV virion. This necessarily ignores much of the interplay between virus and host that is critical for infection. We also concentrate on human HBV, the type member of the hepadnavirus family. However, we should note that related hepadnaviruses, Woodchuck Hepatitis Virus (WHV) (25) and duck Hepatitis B Virus (dHBV) (26) have made great contributions to our understanding of hepadnavirus biology and pathology.
The Dane particle
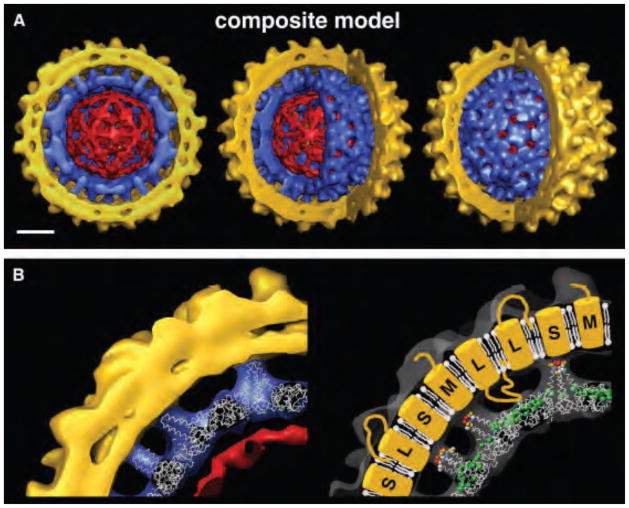
As little as a single virion, measured in terms of genomes, can lead to chronic HBV infection (27). Yet examination of electron micrographs reveals a startling heterogeneity. Particles typically show an enveloped core containing the three co-terminal HBsAg proteins – S, M and L – embedded in the membrane. However, the envelope may have a filamentous extension or the enclosed core may have a smaller diameter, 32 nm, or an aberrant shape (28). As many as 90% of secreted particles are actually empty (22). This suggests that cores containing correctly processed nucleic acid and empty cores both display a secretion signal (22). Efforts to apply cryo-electron microscopy (cryo-EM) image reconstruction to Dane particles revealed that the envelope and core do not share symmetry (29, 30). A composite map was built when the core and the envelope were independently reconstructed (by masking the other) and these reconstructions were then superposed (Figure 1). It was shown that HBsAg spikes, 40Å protrusions from the envelope, are arranged in a trigonal lattice such that positions of HBsAg and HBcAg did not necessarily coincide. Capsids were structurally indistinguishable from other cryo-EM reconstructions of capsid alone; there was also limited order in the encapsidated DNA due in part to icosahedral averaging during reconstruction. Packaged DNA was densest under fivefold and quasi-sixfold vertices, where HBcAg nucleic-acid binding domains are located (29) (Figure 1). Notably, in some Dane particles the envelope was only loosely associated with the core (30), this may be a function of the distribution of the different forms of the HBsAg. The capsid does not firmly constrain the distribution of HBsAg units in the envelope but provides a generalized template for packing (30).
Figure 1.
A composite cryo-EM reconstruction of a dane particle. (a) Cut away views of a composite model of the HBV virion comprised of an icosahedral capsid (blue) containing packaged DNA (red) and an outer envelope (gold) with protein projections spaced 60Å apart. Views are cross-sections (left), and two cut aways (b) X-ray crystal structure of recombinant capsid (36) docked into the cryo-EM density map of the virion capsid (left). The tips of the core spikes are in close apposition but do not penetrate the envelope. Additional details and cartoon of interpretation (right). The surface protein projections are ascribed to HBsAg and are designated as large (L), medium (M), and small (S) arbitrarily. These figures reproduced with permission from Dryden et al. (29).
HBcAg
Dimer structure
The basic soluble unit of HBcAg is a dimer (Figure 2a). A HBcAg monomer is 183 residues long (for in vitro studies we refer to it as core protein 183 or Cp183). The first 149 residues form the predominantly α-helical assembly domain (Cp149). The remaining 34 residues are the arginine-rich RNA-binding, C-Terminal Domain (CTD) (31, 32). This domain is located inside the capsid and has been identified to be intrinsically disordered based on sequence and Cryo-EM studies (33). The assembly domain alone is sufficient for assembly of morphologically regular empty capsids (32). The assembly domain has 5 α-helices connected by loops (34–36) (Figure 2a). Helices 1, 2 and 5 are part the chassis of the capsid. A tight proline-rich loop connects helix 5 to the CTD (37). Holding together a dimer, helices 3 and 4 from one half-dimer form a 4-helix motif with corresponding helices from the other half-dimer. A total of about 3200 Å2 of hydrophobic surface is buried by this interaction. This motif is flanked by salt bridges and hydrogen bonds that stabilize the hydrophobic interface. Helix 4 is kinked in the middle and is sometimes referred to as helices 4a and 4b. A disulfide bond can form over time between highly conserved C61 residues in helix 3, which contributes to the dimer interface (38–40).
Figure 2.
X-ray crystal structures of the assembly domain. (a) A HBcAg dimer in the context of a capsid. The helices in HBcAg are named 1–5 from N to C-terminus. The dimer interface comprises of a four-helix bundle created by two helices from each monomer. (b) a superposition of an HBcAg dimer in the context of a capsid (grey) on a free dimer (Y132A mutant) (blue). The free dimer is less compact than the dimer in the context of a capsid (c) An HBeAg dimer. The dimeric interface is drastically altered and stabilized by disulfide bonds (d) An HBcAg T=4 capsid with the asymmetric unit in color. The individual subunits are A (blue), B (red), C (green) and D (yellow) or AB and CD dimers.
Despite its conserved sequence and α-helical state, there is a degree of variability in the structure of the dimer. Dimers in the context of capsids are more compact than free dimers in solution (Figure 2b) (36, 41). Crystal structures of an assembly-incompetent mutant (Y132A) of HBcAg (41–43) show substantial variability, mainly at the spike tips and the C-termini. This variability in structure is thought to be indicative of the variety in functional roles for HBcAg in the viral life cycle. Regulating the structural state of HBcAg would induce/stabilize specific conformational states that match specific functional states. For example, an oxidized C61–C61 dimer results in weaker dimer-dimer interactions and slower capsid assembly rates in comparison to the reduced form (38); thus, the oxidized dimer favors a conformation (or conformations) that are unfavorable for capsid assembly. While the cytoplasm (where capsid assembly takes place during an infection) is a reducing environment, the proximity of the C61 residues is high may be sufficient to overcome the reducing potential. Data suggest that structural dynamics that allow HBcAg to transition between these states are an important key to its function (38, 41, 44, 45).
HBeAg
HBeAg is a secreted protein expressed by every member of the Hepadnaviridae family though its expression is not required to maintain infection. HBeAg is transcribed from the C gene using the first of the two start codons, HBcAg uses the second. The HBeAg pre-protein has 29 residues upstream of HBcAg (46). The first 19 residues, a signal peptide, trafficks nascent HBeAg to the ER where the signal and the CTD are proteolytically removed (47, 48). The remaining 10 residues upstream of HBcAg including a Cys at position -7. This C-7 forms an intramolecular disulfide with C61 to stabilize the structure (40). HBeAg is thought to have an immune-modulatory role that allows for humoral and cell-mediated immune evasion (49, 50). Paradoxically, loss of e antigen is correlated with clearance of acute HBV infection and establishment of chronic infection (51). In a similar vein, loss of HBeAg in culture correlates with increased HBV expression while in humans it correlates with decreased viremia (52).
HBeAg and HBcAg show important physical differences: the thermal melting temperature of HBeAg is 51°C while that of HBcAg dimer is 63°C (53). Reducing the C-7–C61 disulfide or mutating C-7 to Ala residue increased the stability of the HBeAg to near HBcAg levels (54). While oxidized HBeAg does not assemble into capsids, sedimentation analysis and electron microscopy showed that reduced HBeAg dimers could (53, 54). These results suggest that HBeAg could adopt a conformation similar to that of HBcAg (40, 53). Thus, it is possible that both HBeAg- and HBcAg-associated dimer conformations may be accessible to both proteins in solution. Assembly studies with reduced HBeAg dimers indicate that their capsid-like products are less regular than HBcAg-assembled capsids (53). This would suggest that co-assembly of HBcAg with HBeAg would have a negative impact on regular capsid assembly, both from a kinetic and thermodynamic point of view.
The 10-residue peptide upstream of HBcAg makes HBcAg and HBeAg serologically distinct, though there is also substantial cross reactivity (35, 55, 56). In the HBeAg crystal structure (Figure 2c) (54), the overall fold of an HBeAg monomer was the same as an HBcAg monomer, with some differences at the spike tips and C-termini. The dimer interface, however, was heavily altered; one half-dimer was rotated by 140°, so that the HBeAg four-helix bundle is almost antiparallel relative to the motif in HBcAg (Figure 2c). Thus, the tip of the spike must have evolved to support both parallel and antiparallel interaction. This gross change was stabilized by the disulfide from C-7 in the peptide to C61 of helix 3 (one per monomer), which replaces the HBcAg C61–C61 disulfide (36). The structure is also stabilized by hydrophobic contacts between the peptide and helix 3 and 4. The peptide covers some of the exposed hydrophobic surface at the base of helix 3, creating a new interface (54).
Capsid structure
HBcAg can self-assemble to form the icosahedral virus capsid. A 120-dimer T=4 capsid (Figure 2d) is the major assembly product (~95%) with a small amount T=3 capsid observed in infected human livers, recombinant E. coli capsids and in vitro assembled recombinant HBcAg subunits (24, 28, 57–59). Structures for T=4 and T=3 capsids have been determined by Cryo-EM (24, 34, 35, 37, 60, 61). Crystal structures have been determined for T=4 capsids and assembly-incompetent HBcAg dimers (36, 41, 42, 62).
Icosahedra are comprised of 60 asymmetric units. A T=4 asymmetric unit contains contains 4 HBcAg monomers, A through D, (or two dimers, AB and CD) that form the icosahedral asymmetric unit (Figure 2d). The subunits are in “quasi-equivalent” environments. The 4-helix motifs from the dimers are oriented perpendicular to the surface of the capsid, forming the spikes that punctuate the capsid. By convention, A monomers form the fivefold vertices and two sets of B, C and D monomers form the quasi-sixfold vertices (Figure 2d). A T=3 asymmetric unit has 3 quasi-equivalent monomers resulting in AB dimers and twofold symmetric CC dimers. Large holes fenestrate the capsid surface and provide a means for nucleotides and other small molecules to diffuse in and out of the particle.
Contacts between dimers are mediated by burial of about 1700 Å2 of hydrophobic surface. This is a tongue-and-groove contact where the groove is located near the junction where helix 5 emerges from the four-helix bundle at the “intradimer” contact. This groove is filled by the helix-turn-extended structure of an incoming subunit (36). Tyrosine 132 from the incoming subunit contributes about 10% of the buried surface are of this interaction (36); a Y132A mutant is assembly-incompetent (63) and has been the basis of HBcAg dimer structures (41–43). A peculiar pocket formed at this interface is the basis of assembly-directed antivirals, which will be discussed later in this review.
The HBcAg capsid is itself highly immunogenic and reported to induce both B- and T-cell responses (64), though these responses are not protective against HBV infection. Peptide mapping studies identified the spike tip, around residue 80, as the major epitope (56, 65) In a series of cryo-EM studies antibodies to capsids were shown to bind not only to a linear epitope on the spike tip but to conformational epitopes on the sides of the spike and its base (35, 66–68). Other epitopes have been observed at interdimer contacts (65, 68). Because of its immunogenicity, HBV capsids have been used as a carrier for epitopes (69, 70). To facilitate insertion of large immunogens into the spike tip while avoiding steric clashes, investigators have used proteolytically processed “split cores” or a tandem fusion of HBcAg that allows modification of one half dimer at a time (71, 72).
HBcAg C-terminal Domain, the CTD
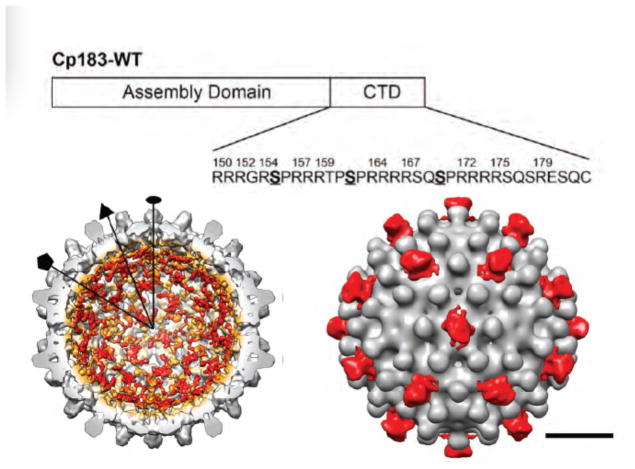
The 34-residue (or 36 residue, depending on genotype) arginine-rich HBcAg CTD (Figure 3a) is localized to the interior of the capsid (Figure 3b), though it can transiently be exposed on the capsid exterior (37, 73–75). CTDs regulate RNA packaging, reverse transcription, and binding to host proteins. The CTD can be considered a sequence of four arginine-rich repeats (Figure 3). The CTD has 7 conserved serines and a threonine which can be phosphorylated (168,169, 79, 148) (Figure 3a). Several host protein kinases have been implicated, including protein kinase C, serine/arginine protein kinase (SRPK) and cyclin dependent kinase 2 (Cdk2) (76–78). These kinases may act on HBcAg prior to assembly or in the context of capsid. One or more types of kinase also gets packaged in cores, with strong evidence supporting identification as Cdk2 (78). Phosphorylation of S155, S162, and S170 is critical for packaging RNA (Figure 3a) – substitution of these residues with alanine to mimic the unphosphorylated state suppresses pgRNA packaging (76, 79–81).
Figure 3.
The HBcAg CTD. (a) A schematic of the assembly domain and the CTD including the sequence of the CTD. S155, S162 and S170 are deemed to be critical for pgRNA packaging (173, 174) (b) cut-away view of a cryo-EM reconstruction of an empty Cp183 capsid. The density in color corresponds to the CTD based on the fitting of an X-ray crystal structure of a Cp149 capsid in the reconstruction. CTD density is prominent beneath the fivefold and quasi-sixfolds (c) A cryo-EM reconstruction of empty Cp183 capsids bound to SRPK molecules. SRPK (red) binds to transiently exposed CTDs at the quasi-sixfolds. These figures reproduced with permission from Selzer et al. (75) and Chen et al. (74).
Interaction of Cp183 with SRPK has been used to demonstrate flexibility of the CTD and a possible mechanism for regulating assembly. In vitro, SRPK binds Cp183 dimers and empty capsids (74). However, the affinity for dimer is 50-fold stronger suggesting a thermodynamic basis for stalling assembly; SRPK also is large enough that it would occlude assembly of bound dimers. However, assembly of SRPK-bound Cp183 can be reactivated by addition of ATP to initiate phosphorylation and release the bound SRPK. Thus, hypothetically SRPK can acts as a non-canonical chaperone that gates assembly. A cryo-EM reconstruction of pre-assembled empty Cp183 capsids bound to SRPK revealed that SRPK trapped CTDs that were transiently exposed to the capsid surface at twofold (quasi-sixfold) vertices (Figure 3c) (74). Transient exposure of CTDs in empty Cp183 capsids has also been documented by trypsin digestion studies (75). Mass spectrometry of trypsin-digested empty capsids showed ready accessibility as far as residue 157. Cryo-EM reconstructions of these products verified the cleavage and loss of CTD density underneath quasi-sixfold vertices (Figure 3b) (75).
There is a strong correlation between the positive charge of a viral capsid and its nucleic acid content (82, 83); this correlation extends to HBV (84–86). In vivo, a capsid with fully phosphorylated CTDs packages about 3400 nucleotides of pgRNA with its polyA tail, a ratio of 1.77 bases per arginine, and a fully dephosphorylated DNA-filled core has about 6300 bases, a ratio of 1.75 bases per arginine. The number of positively charged arginines in the CTD correlates with the amount of encapsidated RNA (84, 86). Decreasing the amount of positive charge in an HBV expression system, by truncating the CTD (85) or by replacing blocks of arginines with alanines (87), decreases the amount of pgRNA encapsidated in cell culture expression of HBV. Surprisingly, the decreased RNA content correlates with decreased representation of 3′ ends compared to 5′ ends, suggesting that RNA is packaged so that the 3′ end is particularly vulnerable to nucleases or that it is packaged in a stepwise process 3′ end last.
Phosphorylation appears to modulate structural interactions between CTDs that affect capsid stability and RNA organization within the capsid. In empty Cp183 capsids, external exposure of CTDs is decreased and capsid stability is strengthened by an S155E, S162E, S170E triple mutation, mimicking phosphorylation of the three critical serines (75). In a virus expression system, the triple mutant assembles without packaging RNA (85). Image reconstructions comparing this triple mutant to wild type capsids show that mutant CTDs, particularly around the fivefold vertices, cluster together, which was suggested to be a result of electrostatic interactions between “phosphorylations” on one CTD with arginines on an adjacent CTD (33).
The triple mutant phosphorylation mimic also modified the organization of packaged pgRNA in in vitro assembled particles (33). In vitro, Cp183 can be assembled on (single-stranded DNA) ssDNA and single-stranded RNA (ssRNA) but not on double-stranded (dsDNA) (88). The pgRNA in capsids with unphosphorylated CTDs formed an icosahedral cage of thick RNA ropes (Figure 4a)(33). In the phosphorylation-mimic mutant the RNA formed a more diffuse mesh-like network (Figure 4b) of thin and short stretches of electron density (33). Consistent with the ability of CTD modification to change the disposition of packaged RNA, the CTD has been shown to have RNA chaperone activity in the context of a capsid (89).
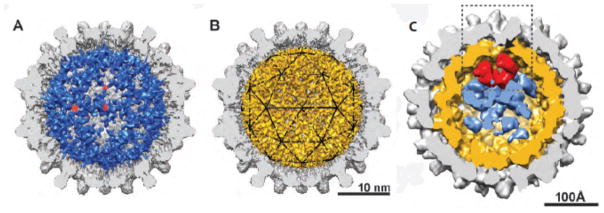
Figure 4.
RNA containing capsids. Icosahedral reconstructions of (a) wildtype Cp183 and (b) a phosphorylation mimic mutant with in vitro packaged pgRNA (blue and gold respectively). The pgRNA in the former forms an icosahedral cage while the same in the latter is more mesh-like (c) An asymmetric reconstruction of an RNA-containing virion shows density for pgRNA (gold), P protein (red) and other unassigned content. These figures reproduced with permission from Wang et al. (33) and Wang et al. (94).
Though CTDs are ostensibly on the capsid interior, in empty capsids and DNA-filled capsids they are accessible to the capsid exterior (74–76). While in RNA-filled capsids, the CTD is trapped on the capsid interior. External exposure of CTDs is biologically critical as the CTD has a role in nuclear translocation. Mature cores are imported into the nucleus through nuclear pores in a process that involves the binding of an Importins α-Importin β complex (11). Importin α binds to nuclear localization signals (NLSs), a sequence of three or four to six consecutive basic amino acids. When bound to an NLS, importin α exposes an importin β binding domain, a motif of 13 basic residues spread over 39 residues (90, 91). Emphasizing a fundamental difference between CTD exposure of DNA-filled versus empty capsids, in vitro empty capsids and free dimers bind Importin β without Importin α mediation (73). Furthermore, importin β appears to bind dimer and destabilize empty capsids. These observations suggest that intracellular transport of free dimer, empty capsids, and mature capsids use different mechanisms. Efforts to clarify the specific NLS sites have been complicated by the fact that they may overlap. Furthermore, the same sequences in the CTD have been shown to act as nuclear export signals, presumably for Cp183 dimer; this has been suggested as a mechanism for export of viral RNA from the nucleus (92).
Reverse Transcription
Reverse transcription (reviewed in (23, 93, 94)) is the target of most anti-HBV therapeutics now available. Capsid assembly is nucleated by a pgRNA-reverse transcriptase (P protein) complex, with P bound to the ε stem loop near the 5′ end of pgRNA. The P protein has four domains, a terminal priming domain, a linker, a polymerase domain, and an RNaseH. The polymerase, though it is monomeric, has been modeled based on the HIV reverse transcriptase (95). The RNAseH shares little sequence identity with other RNaseH proteins but enough to allow identification of active site residues and evaluate potential inhibitors (96). Within the capsid, using the loop in ε as its first template, a tyrosine in P’s N-terminal domain primes minus strand DNA synthesis, leaving the nascent chain covalently bound to the protein. After 3 bases, P switches template to a sequence (direct repeat 1, DR1) near the 3′ end of pgRNA and then completes minus strand DNA synthesis. The carboxy-terminal RNaseH domain of P digest the template pgRNA leaving only about 17 bases of the original RNA. ssDNA-filled capsids accumulate in cytoplasm, suggesting that the next step in DNA synthesis has a high kinetic barrier. The remaining RNA with the P-protein (covalently bound to minus strand DNA) now makes a second template switch to a region overlapping a copy of the DR1 sequence, DR2, near the 5′ end the minus strand. Synthesis of the plus strand begins there, using the RNA as a primer. After running out of template, the P complex with the growing plus strand switches template for the third time. Successful maturation of the HBV virion requires three template switches in which P and associated nucleic acids jump from one end of the template nucleic acid to the other. These template switches are facilitated by complementary sequences at the 5′ and 3′ ends of the nucleic acid (97, 98). For these to anneal, the encapsidated P protein and pgRNA adopt a specific quaternary arrangement.
There were two hypotheses to explain the process of reverse transcription: (i) The RNA lines the interior wall of the capsid allowing the P protein to travel on a nucleic acid track where 5′ and 3′ ends are topographically close to one another; (ii) The polymerase is directly bound to the inner wall of the capsid, requiring the nucleic acid to move through the enzyme like a conveyer belt. An asymmetric Cryo-EM reconstruction of RNA-filled cores with packaged P protein, determined to 16 Å resolution (Figure 4c), strongly supports the first hypothesis (94). Mutation of the priming tyrosine in P supported pgRNA packaging but arrested the core in the RNA-filled state (99). RNA density was irregular but lined the interior surface of the capsid, similar to in vitro-assembled RNA-filled capsids (Figure 4c). The RNA density was not resolved enough to fit individual strands. Internal to the RNA, a “donut-shaped” density was adsorbed to the RNA shell interior (under a threefold) consistent in size and shape to a polymerase (Figure 4c). Other unassigned internal density was observed and could be other packaged protein or could be an artifact arising from irregularity in the capsids and the difficulty of determining asymmetric orientation in an icosahedral particle.
The shell of the capsid as well as its interior surface impact reverse transcription. Mutation of conserved Val124 at the inter-dimer interface affects capsid assembly in a predictable way, changing capsid stability proportional to the change in buried surface (100). These mutations lead to defects in packaging of pgRNA, suggesting a competition between assembly of RNA-filled capsids with assembly of empty/defective ones. Reverse transcription was also profoundly affected (100). While first strand synthesis was proportional to the amount of pgRNA packaged, second strand synthesis was abolished (100, 101). This suggests that the capsid is not just an inert carrier of genomic material and its structural properties affect seemingly unrelated processes.
Interaction of HBV cores with Surface proteins
The HBV Surface antigen has distinct roles in the virus lifecycle. Intracellularly, HBsAg binds cores, attenuating core transport to the nucleus and effectively regulating cccDNA copy number (102). HBsAg is secreted as subviral particles that attenuate immune response to the virus (103); indeed, yeast-expressed S-HBsAg is the basis of the HBV vaccine. HBsAg is responsible for binding cores during assembly and receptor during infection. HBsAg is also a structural component of Hepatitis Delta Virus, a satellite of HBV that drastically increases morbidity and mortality (104).
The structure of HBsAg is known to low resolution only. At a basic level, HBsAg is a glycosylated integral membrane protein that has been localized to ER, pre-Golgi, and late endosomal membranes (105–107). HBsAg comes in three sizes, small, medium, and large, referred to as S-HBsAg, M-HBsAg, and L-HBsAg. The L-HBsAg has three domains, in order, pre-S1 (108 or 119 residues), pre-S2 (55 residues), and the S domain (226 residues). M-HBsAg, which is not required for infection, lacks pre-S1. S-HBsAg is comprised of only the S domain which is composed of four trans-membrane helices (108). The S domain forms disulfide crosslinked dimers, allowing formation of homo- and heterodimers.
Subviral particles are heterogeneous in chemical makeup and structure. Spherical subviral particles are enriched for S-HBsAg with only trace amounts of L-HBsAg whereas filaments have a 1:1:4 ratio of L-, M-, and S-HBsAg (109). Micrographs of subviral particles show serrations indicative of regular protrusions (Figure 5a, b). However, spherical particles appear to be heterogeneous in size and geometry – they are not icosahedral – frustrating efforts to generate image reconstructions (110). Filamentous particles also are heterogeneous but can be classified based on helical parameters (eg. one-, two-, and three-start helices) allowing successful reconstruction to 20Å resolution. Spikes in the reconstruction are similar in size to those in the micrograph. They project 40Å from the membrane and are separated by about 50Å. The volume of a given spike corresponds well with the expected volume for four S domains, suggesting the biologically relevant complex is a dimer of dimers (110). In a Dane particle reconstruction (29), HBsAg spikes have a similar spacing but are arranged with approximately trigonal geometry on the membrane surface; notably, the HBsAg lattice does not align with the core’s icosahedral lattice.
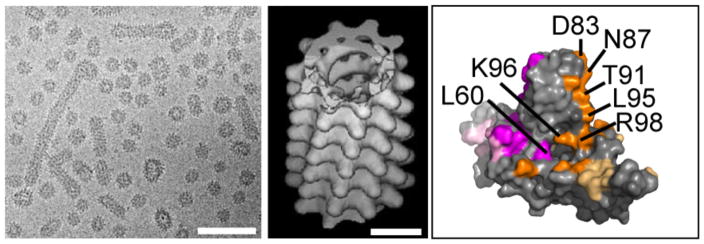
Figure 5.
HBsAg subviral particles. (a) A cryo-micrograph of a mixture of ~22nm spherical and filamentous particles. Note how some filaments vary in their diameter. The scale bars correspond to 100nm. (b) A helical real-space reconstruction of a self-consistent data set. This two-start helix has a subunit twist of 35° and rise of 9.8Å. (c) A HBcAg dimer highlighting residues that affect secretion of Dane particles but do not affect core assembly (117). Residues from different monomers are in magenta and orange, respectively. Residues that are partially obscured by interdimer interfaces are highlighted in muted colors. Panels (a) and (b) are reproduced with permission from Short et al. (110).
In most viruses, internal features of the envelope protein(s) interact with the virus core and external features mediate interaction between the virion and its receptor(s). In HBV, both of these activities are functions of the pre-S domains of L-HBsAg, a peptide that can change its localization over time (111) and possibly in response to external triggers. The N-terminus of S-HBsAg and the pre-S2 domain of newly translated M-HBsAg are in the ER lumen, presumably due to an internal signal sequence (112). In L-HBsAg pre-S1, which is myristoylated, and pre-S2 are largely cytoplasmic, based on proteolytic sensitivity and the absence of pre-S2 glycosylation. This places the pre-S domains where they can interact with newly matured HBV cores. Conversely, in Dane particles the pre-S1 and pre-S2 domains are accessible to external proteases.
Dane particles appear to first interact with host cells through a relatively weak electrostatic interaction to highly sulfated proteoglycan (HSPG); this interaction is a necessary precursor to infection (113, 114). In vitro other anionic glycans, e.g. heparin sulfate, can bind HBsAg. This interaction is modulated by a disulfide-stabilized motif on the S domain (115, 116). The virions then bind to NTCP (6, 7) in an interaction mediated specifically by a highly conserved N-terminal sequence of pre-S1, residues 2–48 (117). A myristoylated peptide based on this sequence, named MyrcludexB, is the basis for an HBV entry inhibitor that that displays an 80 picomolar IC50 (118, 119). Thus it is likely that binding to HSPG induces a conformational change exposing the N-terminal region of pre-S1 (116) – antibodies to this segment of pre-S1 do not precipitate Dane particles (120).
Interaction of cores with L-HBsAg is sensitive to the nucleic acid content of the core via a poorly characterized switch (22)(121). We have a rudimentary understanding of the structural basis of interaction of the pre-S domains with the HBV core that are associated with virion assembly and egress. Peptides based on the C-terminus of pre-S1 shows notable affinity for capsids based on binding of sequential peptides to capsids (122) and the ability of a series of L-HBsAg mutants with substitutions in the Pre-S1 and S2 sequences to support virion secretion (123). Additional determinants of interaction are associated with the S domain (122, 124). Ponsel and Bruss conducted a rigorous alanine mapping of HBcAg by evaluating the ability of the HBcAg mutants to support capsid formation and virion secretion (125). The amino acids identified by that study run down the solvent-exposed face of HBcAg helix 4 (Figure 5c). Using an alternative approach, phage display, a hydrophobic peptide with sequence similarity to the first 20 amino acids of pre-S1 was identified that bound at the core spike tips (126).
Helix 4 is particularly attractive as regulatory element for core-envelope interaction. Its conformation is responsive to assembly state and bound small molecules (Fig 5c) (41, 127). In a cryo-EM comparison of DNA-filled cores from HBV-expressing cells with RNA-filled capsids from E. coli, Roseman and Crowther observed small structural changes in the protruding HBcAg spike centering on a hydrophobic cavity bounded by residues K96 and F97 from one half-dimer and L60 from the other half (121) (Figure 5c). K96 and F97 are on the back face of helix 4.
Notably, mutation F97L (or I97L, depending on genotype) results in a core that is secreted in the immature ssDNA state while a L60V mutation inhibits secretion altogether (128–130). One cannot tell whether the pocket mutations affect the helix or vice versa. It is also possible that both the helix and pocket are involved in binding pre-S or that these mutations affect a dynamic state rather than a static structure. In Dane particle structures (29, 30), the spike tip is in contact with the envelope. Direct interaction with protein could not be discerned in these structures due to the relative disorder of the envelope. However, the spike tip has been suggested as a binding site for HBsAg (126, 131).
Assembly
HBcAg spontaneously assembles into a mixture of T=4 and T=3 particles (58). Self-association of the assembly domain, the first 149 residues of HBcAg (Cp149), has been investigated extensively. The assembly reaction can be summarized by two equations (132–134). The kinetics of assembly can be described as a rate-limiting nucleation step followed by rapid elongation by addition of one dimer at a time:
| 1) |
This simple equations can result in complex behavior (135, 136). While HBV is relatively simple, virus assembly reactions can be much more complex involving an array of metastable intermediates (137, 138). The importance of the nucleation step is that it prevents formation of kinetic traps, reactions where incomplete capsids accumulate but there are not enough subunits to complete them. To prevent kinetic traps, nuclei must be relatively rare either because they form slowly compared to elongation or because they are relatively unstable (132, 139). Assembly models predict a high energy barrier to disassembly (i.e. hysteresis to dissociation) that has been observed in experimental studies (140–142). Simulations suggest an array of intermediates during assembly (135, 136), biased towards smaller sizes, which have now been observed by ion-mobility mass spectroscopy (143). An implication of this kinetic model is that allostery between assembly incompetent and competent states can contribute to regulation of nucleation and elongation (144), an implication borne out by structural differences between free and capsid-bound HBV dimers (41, 44, 45). Hydrogen-deuterium exchange (45) and protease sensitivity (44, 75) indicate that HBV Cp149 is extremely dynamic, prone to conformational fluctuations and local melting of secondary structure.
At equilibrium, the assembly reaction reduces to a simple equilibrium expression:
| 2) |
The huge exponent in equation 2 leads to a pseudo-critical concentration (pcc), where the maximal concentration of free Cp149 dimer in solution is approximately equal to the pcc and all addition Cp149 is in the form of capsid and sometimes non-capsid polymer. Unlike a true critical concentration, equation 2 predicts assembly will occur at low Cp149 concentrations; this has now been observed using nanofluidic devices sensitive to very low protein concentrations (145). The term Kcapsid is a function of the pairwise interaction energy between two dimers, ΔGcontact (139). For HBV, and most other viruses, ΔGcontact is remarkably weak and assembly at physiological concentrations is only possible because subunits are multivalent. Weak association energy prevents entrapment of defects by favoring dissociation of misassembled components. Consistent with the hydrophobic inter-dimer contacts observed in capsids (36), assembly is entropy-driven (134). Kinetic traps can form when Cp149 association energy is strengthened by solution conditions; this was not evident until recent developments in charge detection mass spectrometry, a single particle technique (146). The same approach was used to show that Woodchuck Hepatitis Virus has a much greater tendency to form aberrant capsids (147, 148).
In vitro assembly on nucleic acid is much more complicated because of the combination of protein-protein and protein nucleic acid interactions (149, 150). In vitro assembly of Cp183 on ssRNA and ssDNA is fast and, based on the paucity of intermediates, highly cooperative (88). Assembly on dsDNA is problematic. The protein binds dsDNA with apparently high affinity. However, the assembly products are heterogeneous as if protein-protein interactions are insufficient bend and enclose the relatively rigid dsDNA substrate (88, 150) leading to a calculation that a dsDNA could destabilize capsids (88, 150). Consistent with this calculation, a growing body of evidence indicates that dsDNA-filled cores are not particularly stable – they are sensitive to proteases and nucleases and show alter biophysical properties (151, 152).
Capsids and capsid-directed antiviral strategies
HBcAg is an attractive antiviral target because dimer and capsid participate in several steps of the virus life cycle (153, 154). Misdirection of in vitro assembly by altering temperature and ionic strength can lead to formation of kinetically trapped intermediates or aberrant non-capsid structures. Forming these products under physiological conditions may be an effective antiviral strategy (155, 156).
In cell-based screens, ostensibly for non-nucleoside reverse transcriptase inhibitors, two enigmatic classes of molecules were discovered: the phenylpropenamides (PPAs) and the heteroaryldihydropyrimidines (HAPs) (157, 158). In both cases it was discovered the mode of action focused on the core protein, not the reverse transcriptase. PPAs led to loss of cytoplasmic RNA-filled cores and accumulation of empty capsids (159). HAPs led to loss of core protein (158, 160). Unsurprisingly, these molecules were insensitive to nucleoside analog-resistant mutants of HBV reverse transcriptase (161–163).
In vitro assembly experiments uncovered important clues to mechanism of action. HAPs and PPAs speed up assembly and stabilize protein-protein interaction (162, 164–166). HAPs, in concentrations that can saturate available HBcAg, lead to assembly of aberrant non-icosahedral complexes (165–167). On examination, HAPs favored formation of hexagonal repeats of core protein at the expense of fivefolds (165). Quantification of the effect of HAPs and PPAs on assembly shows that they strengthen dimer-dimer association energy, decreasing the pseudo-critical concentration of assembly from ca. 15μM down to ca. 30nM, and increasing the apparent rate of assembly by more than two orders of magnitude. Some of this effect may be due to simply filling a hydrophobic pocket – stabilizing intermediates will affect the observed rate of assembly as well as the yield of product (168). However, some the kinetic effect has been attributed to allosterically modulating HBcAg conformation; hence this molecules that drive assembly may be though of as Core protein Allosteric Modulators (CpAMs).
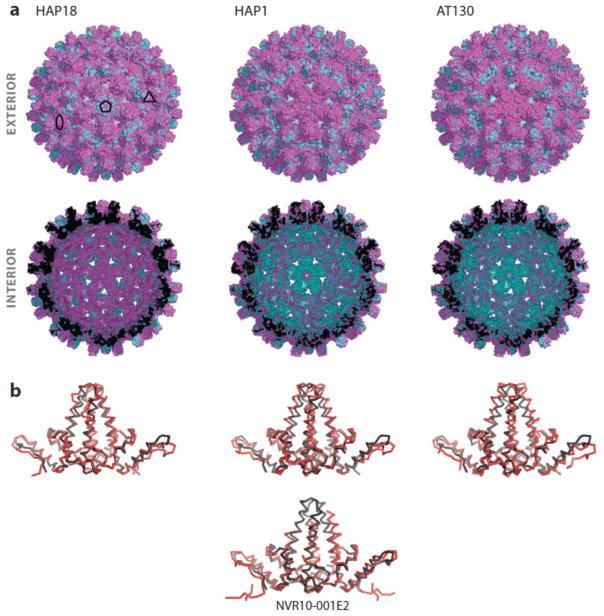
HAP1 led to substantial quaternary structural differences, an expansion at fivefold axes, when compared to an unliganded apo capsid structure (62). Any changes in tertiary structure were negligible at the observed 5Å resolution. CpAMs were found to wedge between dimers at the interfacial HAP pocket, which explained the strengthening of inter-dimer interactions. The site was made by a C-shaped four helix motif with helices 2, 4 and 5 forming a pocket and helix 5 from the interfacial monomer forming a cap over the pocket. Strong electron density was only observed in the pocket associated with C monomers that interfaced with a D monomer from a neighboring dimer; weak density was observed in the B pocket.
A crystal structure of capsid bound to AT130 showed similar quaternary structural changes but substantial tertiary structural changes in C and D monomers (127). Electron density for the CpAM was found in the HAP pocket at the B–C inter-dimer interface with weak density at the C–D interfacial pocket. The findings from this structure supported the hypothesis that CpAMs drive aggressive assembly towards misdirection by affecting the quaternary arrangement of dimers.
A third structure of capsid bound to HAP18, a derivative of HAP1, determined to 4Å resolution, surprisingly showed no quaternary structural changes and minimal changes in tertiary structure (169). HAP18 had been previously shown to affect assembly in vitro (leading to microns long 100 nm diameter tubes) and in vivo (162). Equally strong HAP18 density in both B–C and C–D sites though there were substantial differences between the different binding modes. Thus, HBV capsids have diverse structures and structural responses to CpAMs.
Efforts to obtain higher resolution structures have been based on the assembly-deficient Y132A mutant, with an apo-structure determined to 2.25Å resolution. The structure of a HAP1 derivative (NVR10-001E2) soaked into crystals of the mutant was determined to 1.95Å (43). The CpAM bound in HAP pocket in a similar orientation as CpAMs observed in capsids. However, the lack of a capsid-like environment and quasi-equivalence in the quaternary arrangement of the monomers in this structure limit its use in rational drug design, despite the resolution. The structure, however, does serve to validate the orientation of similar molecules in other low-resolution structures.
The range of quaternary structure changes in the presence and absence of CpAMs suggest that the altering the quaternary structure may not be a basis for CpAM mechanism. Changes in structure, rates of crystallization, effects of CpAM-emulating mutants (100, 101), the effect of antibodies damping global HD exchange (170) – the variety of structural responses – leads to the hypothesis there may be dynamic basis to CpAM activity. Damping capsid dynamics as an antiviral mechanism has been demonstrated previously in studies with picornaviruses (171, 172). This would have important implications in future CpAM design. For HBV this would also have larger implications for the mechanism of CpAM action in the viral life cycle because affecting dynamics would affect the role of the capsid in processes upstream and downstream of assembly.
Closing comments
There is no known cure for HBV infection. Eliminating or silencing the cccDNA in cells is essential to clearing infection permanently. Direct acting antivirals may be the key to cccDNA. The virus capsid is one such target. Reverse transcriptase is the primary target for available antiviral therapeutics, however even the best of these methods lead to only about 5% clearance of infection after five years of treatment (51). Clearly other targets are needed, perhaps in combination. The RNaseH domain of the reverse transcriptase is an independent and unutilized target with opportunity for synergism (96). Another direct target is the cellular entry process which has shown its value in protecting liver transplants (118, 119). The virus may also be blocked by interfering RNAs that block viral products (174). Recent advances indicate that HBV X protein has a direct role in preventing the host cell from silencing cccDNA, but do not suggest a mechanism for targeting it (175).
HBV is one of the smallest human pathogens, based on a 3200 bp genome encoding only four open reading frames. Yet structurally the virus shows a remarkable diversity of structural features, often with the same proteins. In part this is the parsimony of viruses, where a minimal number of proteins performs a wide variety of functions. However, a more important them is that weak interactions between components and irregular interactions between components lead to a highly dynamic system. In HBV this is manifested as a virion where the envelope proteins have multiple structures, the envelope capsid interaction is irregular, the capsid is a dynamic metabolic compartment that actively participates in structural changes to its encapsidated genome. Thus, direct acting antivirals have a limited repertoire of targets, but have the opportunity to be highly specific.
Figure 6.
CpAMs alter the structure and dynamics of the capsid. (a) Overlays of CpAM bound capsids (magenta) on apo-capsid (cyan) when viewed down the fivefold reveal systematic differences in capsid structure in the exterior (upper panels) and interior (lower panels). This figure reproduced with permission from Venkatakrishnan et al. (b) Superpositions of Cα traces of individual dimers from CpAM bound structures (red) on the apo structures (grey).
References
- 1.Blumberg BS. Polymorphisms of the serum proteins and the development of iso-precipitins in transfused patients. Bull N Y Acad Med. 1964;40:377–86. [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlich WH. Medical virology of Hepatitis B: how it began and where we are now. Virol J. 2013;10:239. doi: 10.1186/1743-422X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with australia-antigen-associated hepatitis. Lancet. 1970;1(7649):695–98. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 4.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of Hepatitis B Virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30(12):2212–19. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 5.Ni Y-H, Huang L-M, Chang M-H, Yen C-J, Lu C-Y, et al. Two decades of universal Hepatitis B vaccination in Taiwan: impact and implication for future strategies. Gastroenterology. 2007;132(4):1287–93. doi: 10.1053/j.gastro.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Zhong G, Xu G, He W, Jing Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human Hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146(4):1070–83. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Rigg RJ, Schaller H. Duck Hepatitis B Virus infection of hepatocytes is not dependent on low ph. J Virol. 1992;66(5):2829–36. doi: 10.1128/jvi.66.5.2829-2836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. Phosphorylation-dependent binding of Hepatitis B Virus core particles to the nuclear pore complex. J Cell Biol. 1999;145(1):45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kann M, Schmitz A, Rabe B. Intracellular transport of Hepatitis B Virus. World J Gastroenterol. 2007;13(1):39–47. doi: 10.3748/wjg.v13.i1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabe B, Vlachou A, Panté N, Helenius A, Kann M. Nuclear import of Hepatitis B Virus capsids and release of the viral genome. Proc Natl Acad Sci. 2003;100(17):9849–54. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J-T, Guo H. Metabolism and function of Hepatitis B Virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res. 2015;122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of Hepatitis B Viruses. Proc Natl Acad Sci. 2014;111(40):E4244–53. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, et al. Structural organization of the Hepatitis B Virus minichromosome. J Mol Biol. 2001;307(1):183–96. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 15.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in Hepatitis B Virus infection. J Hepatol. 2009;51(3):581–92. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y-H, Li Y-N, Zhao J-R, Zhang J, Yan Z. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics. 2011;6(6):720–26. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 17.Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci. 2015;112(42):E5715–24. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang B, Bouchard MJ. The Hepatitis B Virus X protein elevates cytosolic calcium signals by modulating mitochondrial calcium uptake. J Virol. 2012;86(1):313–27. doi: 10.1128/JVI.06442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benhenda S, Ducroux A, Rivière L, Sobhian B, Ward MD, et al. Methyltransferase PRMT1 is a binding partner of HBX and a negative regulator of Hepatitis B Virus transcription. J Virol. 2013;87(8):4360–71. doi: 10.1128/JVI.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, et al. Hepatitis B Virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55(5):996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11(9):3413–20. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, et al. Secretion of genome-free Hepatitis B Virus – single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 2011;7(9):e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassal M. Hepatitis B Viruses: reverse transcription a different way. Virus Res. 2008;134(1–2):235–49. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 24.Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, et al. Three-dimensional structure of Hepatitis B Virus core particles determined by electron cryomicroscopy. Cell. 1994;77(6):943–50. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 25.Menne S, Cote PJ. The woodchuck as an animal model for pathogenesis and therapy of chronic Hepatitis B Virus infection. World J Gastroenterol. 2007;13(1):104–24. doi: 10.3748/wjg.v13.i1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funk A, Mhamdi M, Will H, Sirma H. Avian Hepatitis B Viruses: molecular and cellular biology, phylogenesis, and host tropism. World J Gastroenterol. 2007;13(1):91–103. doi: 10.3748/wjg.v13.i1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, et al. The size of the viral inoculum contributes to the outcome of Hepatitis B Virus infection. J Virol. 2009;83(19):9652–62. doi: 10.1128/JVI.00867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stannard LM, Hodgkiss M. Morphological irregularities in Dane particle cores. J Gen Virol. 1979;45(2):509–14. doi: 10.1099/0022-1317-45-2-509. [DOI] [PubMed] [Google Scholar]
- 29.Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M. Native Hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell. 2006;22(6):843–50. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Seitz S, Urban S, Antoni C, Böttcher B. Cryo-electron microscopy of Hepatitis B virions reveals variability in envelope capsid interactions. EMBO J. 2007;26(18):4160–67. doi: 10.1038/sj.emboj.7601841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birnbaum F, Nassal M. Hepatitis B Virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64(7):3319–30. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassal M. The arginine-rich domain of the Hepatitis B Virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66(7):4107–16. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JC-Y, Dhason MS, Zlotnick A. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of Hepatitis B Virus. PLoS Pathog. 2012;8(9):e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the Hepatitis B Virus capsid by cryo-electron microscopy. Nature. 1997;386(6620):91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 35.Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, et al. Hepatitis B Virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279(5):1111–21. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 36.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human Hepatitis B Virus capsid. Mol Cell. 1999;3(6):771–80. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 37.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of Hepatitis B Virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci. 1997;94(18):9556–61. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selzer L, Katen SP, Zlotnick A. The Hepatitis B Virus core protein intradimer interface modulates capsid assembly and stability. Biochemistry. 2014;53(34):5496–5504. doi: 10.1021/bi500732b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nassal M, Rieger A, Steinau O. Topological analysis of the Hepatitis B Virus core particle by cysteine-cysteine cross-linking. J Mol Biol. 1992;225(4):1013–25. doi: 10.1016/0022-2836(92)90101-o. [DOI] [PubMed] [Google Scholar]
- 40.Nassal M, Rieger A. An intramolecular disulfide bridge between cys-7 and cys61 determines the structure of the secretory core gene product (e antigen) of Hepatitis B Virus. J Virol. 1993;67(7):4307–15. doi: 10.1128/jvi.67.7.4307-4315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Packianathan C, Katen SP, Dann CE, Zlotnick A. Conformational changes in the Hepatitis B Virus core protein are consistent with a role for allostery in virus assembly. J Virol. 2010;84(3):1607–15. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander CG, Jürgens MC, Shepherd DA, Freund SMV, Ashcroft AE, Ferguson N. Thermodynamic origins of protein folding, allostery, and capsid formation in the human Hepatitis B Virus core protein. Proc Natl Acad Sci. 2013;110(30):E2782–91. doi: 10.1073/pnas.1308846110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, et al. High-resolution crystal structure of a Hepatitis B Virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci. 2015;112(49):15196–201. doi: 10.1073/pnas.1513803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hilmer JK, Zlotnick A, Bothner B. Conformational equilibria and rates of localized motion within Hepatitis B Virus capsids. J Mol Biol. 2008;375(2):581–94. doi: 10.1016/j.jmb.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bereszczak JZ, Watts NR, Wingfield PT, Steven AC, Heck AJR. Assessment of differences in the conformational flexibility of Hepatitis B Virus core-antigen and e-antigen by hydrogen deuterium exchange-mass spectrometry. Protein Sci Publ Protein Soc. 2014;23(7):884–96. doi: 10.1002/pro.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Standring DN, Ou JH, Masiarz FR, Rutter WJ. A signal peptide encoded within the precore region of Hepatitis B Virus directs the secretion of a heterogeneous population of e antigens in xenopus oocytes. Proc Natl Acad Sci. 1988;85(22):8405–9. doi: 10.1073/pnas.85.22.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruss V, Gerlich WH. Formation of transmembraneous Hepatitis B e-antigen by cotranslational in vitro processing of the viral precore protein. Virology. 1988;163(2):268–75. doi: 10.1016/0042-6822(88)90266-8. [DOI] [PubMed] [Google Scholar]
- 48.Ou JH, Laub O, Rutter WJ. Hepatitis B Virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci. 1986;83(6):1578–82. doi: 10.1073/pnas.83.6.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen MT, Billaud J-N, Sällberg M, Guidotti LG, Chisari FV, et al. A function of the Hepatitis B Virus precore protein is to regulate the immune response to the core antigen. Proc Natl Acad Sci. 2004;101(41):14913–18. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milich DR, Jones JE, Hughes JL, Price J, Raney AK, McLachlan A. Is a function of the secreted Hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci. 1990;87(17):6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gish RG, Given BD, Lai C-L, Locarnini SA, Lau JYN, et al. Chronic Hepatitis B: virology, natural history, current management and a glimpse at future opportunities. Antiviral Res. 2015;121:47–58. doi: 10.1016/j.antiviral.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Samal J, Kandpal M, Vivekanandan P. Hepatitis B “e” antigen-mediated inhibition of HBV replication fitness and transcription efficiency in vitro. Virology. 2015;484:234–40. doi: 10.1016/j.virol.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 53.Watts NR, Conway JF, Cheng N, Stahl SJ, Steven AC, Wingfield PT. Role of the propeptide in controlling conformation and assembly state of Hepatitis B Virus e-antigen. J Mol Biol. 2011;409(2):202–13. doi: 10.1016/j.jmb.2011.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiMattia MA, Watts NR, Stahl SJ, Grimes JM, Steven AC, et al. Antigenic switching of Hepatitis B Virus by alternative dimerization of the capsid protein. Structure. 2013;21(1):133–42. doi: 10.1016/j.str.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imai M, Nomura M, Gotanda T, Sano T, Tachibana K, et al. Demonstration of two distinct antigenic determinants on Hepatitis B e antigen by monoclonal antibodies. J Immunol. 1982;128(1):69–72. [PubMed] [Google Scholar]
- 56.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from Hepatitis B Virus. J Virol. 1989;63(2):798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenney JM, von Bonsdorff CH, Nassal M, Fuller SD. Evolutionary conservation in the Hepatitis B Virus core structure: comparison of human and duck cores. Structure. 1995;3(10):1009–19. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 58.Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in escherichia coli: subunit composition, conformation analysis, and in vitro capsid assembly. Biochemistry. 1995;34(15):4919–32. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 59.Zlotnick A, Palmer I, Kaufman JD, Stahl SJ, Steven AC, Wingfield PT. Separation and crystallization of T = 3 and T = 4 icosahedral complexes of the Hepatitis B Virus core protein. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 3):717–20. doi: 10.1107/s090744499801350x. [DOI] [PubMed] [Google Scholar]
- 60.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of Hepatitis B Virus by electron cryomicroscopy. Nature. 1997;386(6620):88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 61.Yu X, Jin L, Jih J, Shih C, Hong Zhou Z. 3.5Å cryoEM structure of Hepatitis B Virus core assembled from full-length core protein. PLoS ONE. 2013;8(9):e69729. doi: 10.1371/journal.pone.0069729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourne CR, Finn MG, Zlotnick A. Global structural changes in Hepatitis B Virus capsids induced by the assembly effector HAP1. J Virol. 2006;80(22):11055–61. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bourne CR, Katen SP, Fulz MR, Packianathan C, Zlotnick A. A mutant Hepatitis B Virus core protein mimics inhibitors of icosahedral capsid self-assembly. Biochemistry. 2009;48(8):1736–42. doi: 10.1021/bi801814y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milich DR, McLachlan A, Stahl S, Wingfield P, Thornton GB, et al. Comparative immunogenicity of Hepatitis B Virus core and e antigens. J Immunol. 1988;141(10):3617–24. [PubMed] [Google Scholar]
- 65.Pushko P, Sallberg M, Borisova G, Ruden U, Bichko V, et al. Identification of Hepatitis B Virus core protein regions exposed or internalized at the surface of HBcAg particles by scanning with monoclonal antibodies. Virology. 1994;202(2):912–20. doi: 10.1006/viro.1994.1413. [DOI] [PubMed] [Google Scholar]
- 66.Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, et al. Diversity of core antigen epitopes of Hepatitis B Virus. Proc Natl Acad Sci. 2003;100(19):10884–89. doi: 10.1073/pnas.1834404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris A, Belnap DM, Watts NR, Conway JF, Cheng N, et al. Epitope diversity of Hepatitis B Virus capsids: quasi-equivalent variations in spike epitopes and binding of different antibodies to the same epitope. J Mol Biol. 2006;355(3):562–76. doi: 10.1016/j.jmb.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 68.Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, et al. Structure, assembly, and antigenicity of Hepatitis B Virus capsid proteins. Adv Virus Res. 2005;64:125–64. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 69.Peyret H, Stephen SL, Stonehouse NJ, Rowlands DJ. Viral Nanotechnology. CRC Press; 2016. History and potential of Hepatitis B Virus core as a VLP vaccine platform; pp. 177–86. [Google Scholar]
- 70.Pumpens P, Borisova GP, Crowther RA, Grens E. Hepatitis B Virus core particles as epitope carriers. Intervirology. 1995;38(1–2):63–74. doi: 10.1159/000150415. [DOI] [PubMed] [Google Scholar]
- 71.Peyret H, Gehin A, Thuenemann EC, Blond D, El Turabi A, et al. Tandem fusion of Hepatitis B core antigen allows assembly of virus-like particles in bacteria and plants with enhanced capacity to accommodate foreign proteins. PloS One. 2015;10(4):e0120751. doi: 10.1371/journal.pone.0120751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walker A, Skamel C, Nassal M. Splitcore: an exceptionally versatile viral nanoparticle for native whole protein display regardless of 3d structure. Sci Rep. 2011;1:5. doi: 10.1038/srep00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C, Wang JC-Y, Pierson EE, Keifer DZ, Delaleau M, et al. Importin β can bind Hepatitis B Virus core protein and empty core-like particles and induce structural changes. PLoS Pathog. 2016 doi: 10.1371/journal.ppat.1005802. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen C, Wang JC-Y, Zlotnick A. A kinase chaperones Hepatitis B Virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 2011;7(11):e1002388. doi: 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selzer L, Kant R, Wang JC-Y, Bothner B, Zlotnick A. Hepatitis B Virus core protein phosphorylation sites affect capsid stability and transient exposure of the C-terminal domain. J Biol Chem. 2015;290(47):28584–93. doi: 10.1074/jbc.M115.678441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kann M, Gerlich WH. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of Hepatitis B Virus. J Virol. 1994;68(12):7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, et al. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating Hepatitis B Virus core protein. J Virol. 2002;76(16):8124–37. doi: 10.1128/JVI.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ludgate L, Ning X, Nguyen DH, Adams C, Mentzer L, Hu J. Cyclin-dependent kinase 2 phosphorylates S/T-P sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol. 2012;86(22):12237–50. doi: 10.1128/JVI.01218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basagoudanavar SH, Perlman DH, Hu J. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J Virol. 2007;81(4):1641–49. doi: 10.1128/JVI.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kock J, Kann M, Putz G, Blum HE, Von Weizsacker F. Central role of a serine phosphorylation site within duck Hepatitis B Virus core protein for capsid trafficking and genome release. J Biol Chem. 2003;278(30):28123–29. doi: 10.1074/jbc.M300064200. [DOI] [PubMed] [Google Scholar]
- 81.Gazina EV, Fielding JE, Lin B, Anderson DA. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck Hepatitis B Viruses. J Virol. 2000;74(10):4721–28. doi: 10.1128/jvi.74.10.4721-4728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci. 2006;103(46):17174–78. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perlmutter JD, Qiao C, Hagan MF. Viral genome structures are optimal for capsid assembly. eLife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chua PK, Tang F-M, Huang J-Y, Suen C-S, Shih C. Testing the balanced electrostatic interaction hypothesis of Hepatitis B Virus DNA synthesis by using an in vivo charge rebalance approach. J Virol. 2010;84(5):2340–51. doi: 10.1128/JVI.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Köck J, Nassal M, Deres K, Blum HE, von Weizsäcker F. Hepatitis B Virus nucleocapsids formed by carboxy-terminally mutated core proteins contain spliced viral genomes but lack full-size DNA. J Virol. 2004;78(24):13812–18. doi: 10.1128/JVI.78.24.13812-13818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newman M, Chua PK, Tang F-M, Su P-Y, Shih C. Testing an electrostatic interaction hypothesis of Hepatitis B Virus capsid stability by using an in vitro capsid disassembly/reassembly system. J Virol. 2009;83(20):10616–26. doi: 10.1128/JVI.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewellyn EB, Loeb DD. The arginine clusters of the carboxy-terminal domain of the core protein of Hepatitis B Virus make pleiotropic contributions to genome replication. J Virol. 2011;85(3):1298–1309. doi: 10.1128/JVI.01957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dhason MS, Wang JC-Y, Hagan MF, Zlotnick A. Differential assembly of Hepatitis B Virus core protein on single- and double-stranded nucleic acid suggest the dsDNA-filled core is spring-loaded. Virology. 2012;430(1):20–29. doi: 10.1016/j.virol.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chu T-H, Liou A-T, Su P-Y, Wu H-N, Shih C. Nucleic acid chaperone activity associated with the arginine-rich domain of human Hepatitis B Virus core protein. J Virol. 2014;88(5):2530–43. doi: 10.1128/JVI.03235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-beta bound to the ibb domain of importin-alpha. Nature. 1999;399(6733):221–29. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 91.Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 2010;285(18):13769–80. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H-C, Huang E-Y, Su P-Y, Wu S-Y, Yang C-C, et al. Nuclear export and import of human Hepatitis B Virus capsid protein and particles. PLoS Pathog. 2010;6(10):e1001162. doi: 10.1371/journal.ppat.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clark DN, Hu J. Hepatitis B Virus reverse transcriptase - target of current antiviral therapy and future drug development. Antiviral Res. 2015;123:132–37. doi: 10.1016/j.antiviral.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang JC-Y, Nickens DG, Lentz TB, Loeb DD, Zlotnick A. Encapsidated Hepatitis B Virus reverse transcriptase is poised on an ordered RNA lattice. Proc Natl Acad Sci. 2014;111(31):11329–34. doi: 10.1073/pnas.1321424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Das K, Xiong X, Yang H, Westland CE, Gibbs CS, et al. Molecular modeling and biochemical characterization reveal the mechanism of Hepatitis B Virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J Virol. 2001;75(10):4771–79. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tavis JE, Cheng X, Hu Y, Totten M, Cao F, et al. The Hepatitis B Virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLoS Pathog. 2013;9(1):e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abraham TM, Loeb DD. Base pairing between the 5′ half of epsilon and a cis-acting sequence, phi, makes a contribution to the synthesis of minus-strand DNA for human Hepatitis B Virus. J Virol. 2006;80(9):4380–87. doi: 10.1128/JVI.80.9.4380-4387.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lewellyn EB, Loeb DD. Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human Hepatitis B Virus. J Virol. 2007;81(12):6207–15. doi: 10.1128/JVI.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim H-Y, Park G-S, Kim E-G, Kang S-H, Shin H-J, et al. Oligomer synthesis by priming deficient polymerase in Hepatitis B Virus core particle. Virology. 2004;322(1):22–30. doi: 10.1016/j.virol.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 100.Tan Z, Pionek K, Unchwaniwala N, Maguire ML, Loeb DD, Zlotnick A. The interface between Hepatitis B Virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription. J Virol. 2015;89(6):3275–84. doi: 10.1128/JVI.03545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan Z, Maguire ML, Loeb DD, Zlotnick A. Genetically altering the thermodynamics and kinetics of Hepatitis B Virus capsid assembly has profound effects on virus replication in cell culture. J Virol. 2013;87(6):3208–16. doi: 10.1128/JVI.03014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68(7):4565–71. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chisari FV, Ferrari C. Hepatitis B Virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 104.Purcell RH, Gerin JL. Hepatitis Delta Virus. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, et al., editors. Fields Virology. Philadelphia: Lippincott - Raven; 1996. pp. 2819–29. [Google Scholar]
- 105.Bardens A, Doring T, Stieler J, Prange R. Alix regulates egress of hepatitis B virus naked capsid particles in an ESCRT-independent manner. Cell Microbiol. 2011;13:602–19. doi: 10.1111/j.1462-5822.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci. 2007;104:10205–10. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schädler S, Hildt E. HBV Life Cycle: Entry and Morphogenesis. Viruses. 2009;1:185–209. doi: 10.3390/v1020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stirk HJ, Thornton JM, Howard CR. A topological model for hepatitis B surface antigen. Intervirology. 1992;33:148–58. doi: 10.1159/000150244. [DOI] [PubMed] [Google Scholar]
- 109.Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. Large surface proteins of Hepatitis B Virus containing the pre-s sequence. J Virol. 1984;52(2):396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Short JM, Chen S, Roseman AM, Butler PJG, Crowther RA. Structure of Hepatitis B surface antigen from subviral tubes determined by electron cryomicroscopy. J Mol Biol. 2009;390(1):135–41. doi: 10.1016/j.jmb.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 111.Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the Hepatitis B Virus large envelope protein. EMBO J. 1994;13(10):2273–79. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eble BE, MacRae DR, Lingappa VR, Ganem D. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol Cell Biol. 1987;7:3591–601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lempp FA, Urban S. Inhibitors of Hepatitis B Virus attachment and entry. Intervirology. 2014;57(3–4):151–57. doi: 10.1159/000360948. [DOI] [PubMed] [Google Scholar]
- 114.Schulze A, Gripon P, Urban S. Hepatitis B Virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46(6):1759–68. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 115.Sureau C, Salisse J. A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved Hepatitis B Virus a-determinant. Hepatol. 2013;57(3):985–94. doi: 10.1002/hep.26125. [DOI] [PubMed] [Google Scholar]
- 116.Urban S. Entry and entry inhibition of Hepatitis B (HBV) and hepatitis delta virus (HDV) into hepatocytes. Hepatology. 2015;63:633. doi: 10.1002/hep.28308. [DOI] [PubMed] [Google Scholar]
- 117.Engelke M, Mills K, Seitz S, Simon P, Gripon P, et al. Characterization of a Hepatitis B and hepatitis delta virus receptor binding site. Hepatology. 2006;43(4):750–60. doi: 10.1002/hep.21112. [DOI] [PubMed] [Google Scholar]
- 118.Petersen J, Dandri M, Mier W, Lütgehetmann M, Volz T, et al. Prevention of Hepatitis B Virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol. 2008;26(3):335–41. doi: 10.1038/nbt1389. [DOI] [PubMed] [Google Scholar]
- 119.Volz T, Allweiss L, Ben MBarek M, Warlich M, Lohse AW, et al. The entry inhibitor myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with Hepatitis B Virus. J Hepatol. 2013;58(5):861–67. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 120.Bremer CM, Sominskaya I, Skrastina D, Pumpens P, El Wahed AA, et al. N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of Hepatitis B Virus. J Hepatol. 2011;55(1):29–37. doi: 10.1016/j.jhep.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 121.Roseman AM, Berriman JA, Wynne SA, Butler PJG, Crowther RA. A structural model for maturation of the Hepatitis B Virus core. Proc Natl Acad Sci. 2005;102(44):15821–26. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of Hepatitis B Virus envelope proteins interact with the core particle. Virology. 1997;228(1):115–20. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 123.Bruss V. A short linear sequence in the pre-S domain of the large Hepatitis B Virus envelope protein required for virion formation. J Virol. 1997;71(12):9350–57. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tan WS, Dyson MR, Murray K. Two distinct segments of the Hepatitis B Virus surface antigen contribute synergistically to its association with the viral core particles. J Mol Biol. 1999;286(3):797–808. doi: 10.1006/jmbi.1998.2525. [DOI] [PubMed] [Google Scholar]
- 125.Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of Hepatitis B Virus capsids required for envelopment and virion formation. J Virol. 2003;77(1):416–22. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dyson MR, Murray K. Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of Hepatitis B Virus. Proc Natl Acad Sci. 1995;92(6):2194–98. doi: 10.1073/pnas.92.6.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Katen SP, Tan Z, Chirapu SR, Finn MG, Zlotnick A. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify Hepatitis B Virus capsid tertiary and quaternary structure. Structure. 2013;21(8):1406–16. doi: 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Le Pogam S, Yuan TT, Sahu GK, Chatterjee S, Shih C. Low-level secretion of human Hepatitis B Virus virions caused by two independent, naturally occurring mutations (P5T and I60V) in the capsid protein. J Virol. 2000;74(19):9099–9105. doi: 10.1128/jvi.74.19.9099-9105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuan TT, Sahu GK, Whitehead WE, Greenberg R, Shih C. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human Hepatitis B Virus core antigen. J Virol. 1999;73(7):5731–40. doi: 10.1128/jvi.73.7.5731-5740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuan TT, Tai PC, Shih C. Subtype-independent immature secretion and subtype-dependent replication deficiency of a highly frequent, naturally occurring mutation of human Hepatitis B Virus core antigen. J Virol. 1999;73(12):10122–28. doi: 10.1128/jvi.73.12.10122-10128.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Böttcher B, Tsuji N, Takahashi H, Dyson MR, Zhao S, et al. Peptides that block Hepatitis B Virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 1998;17(23):6839–45. doi: 10.1093/emboj/17.23.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A theoretical model successfully identifies features of Hepatitis B Virus capsid assembly. Biochemistry. 1999;38(44):14644–52. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 133.Endres D, Zlotnick A. Model-based analysis of assembly kinetics for virus capsids or other spherical polymers. Biophys J. 2002;83(2):1217–30. doi: 10.1016/S0006-3495(02)75245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of Hepatitis B Virus capsids. Biochemistry. 2002;41(39):11525–31. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 135.Porterfield JZ, Zlotnick A. An Overview of Capsid Assembly Kinetics. In: Stockley PG, Twarock R, editors. Emerging Topics in Physical Virology. London: Imperial College Press; 2010. [Google Scholar]
- 136.Perlmutter JD, Hagan MF. Mechanisms of virus assembly. Annu Rev Phys Chem. 2015;66:217–39. doi: 10.1146/annurev-physchem-040214-121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, Zlotnick A. Regulating self-assembly of spherical oligomers. Nano Lett. 2005;5(4):765–70. doi: 10.1021/nl050274q. [DOI] [PubMed] [Google Scholar]
- 138.Tresset G, Le Coeur C, Bryche J-F, Tatou M, Zeghal M, et al. Norovirus capsid proteins self-assemble through biphasic kinetics via long-lived stave-like intermediates. J Am Chem Soc. 2013;135(41):15373–81. doi: 10.1021/ja403550f. [DOI] [PubMed] [Google Scholar]
- 139.Katen S, Zlotnick A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. J Biol Chem. 2003;278(20):18249–55. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 141.Hagan MF, Chandler D. Dynamic pathways for viral capsid assembly. Biophys J. 2006;91(1):42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJR. Subunit exchange rates in Hepatitis B Virus capsids are geometry- and temperature-dependent. Phys Chem Chem Phys. 2010;12(41):13368–71. doi: 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJR. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat Chem. 2011;3(2):126–32. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 144.Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited Biophys J. 1980;32(1):103–38. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Harms ZD, Mogensen KB, Nunes PS, Zhou K, Hildenbrand BW, et al. Nanofluidic devices with two pores in series for resistive-pulse sensing of single virus capsids. Anal Chem. 2011;83(24):9573–78. doi: 10.1021/ac202358t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, et al. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J Am Chem Soc. 2014;136(9):3536–41. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kukreja AA, Wang JC-Y, Pierson E, Keifer DZ, Selzer L, et al. Structurally similar woodchuck and human hepadnavirus core proteins have distinctly different temperature dependences of assembly. J Virol. 2014;88(24):14105–15. doi: 10.1128/JVI.01840-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pierson EE, Keifer DZ, Kukreja AA, Wang JC-Y, Zlotnick A, Jarrold MF. Charge detection mass spectrometry identifies preferred non-icosahedral polymorphs in the self-assembly of woodchuck hepatitis virus capsids. J Mol Biol. 2015 doi: 10.1016/j.jmb.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Perlmutter JD, Perkett MR, Hagan MF. Pathways for virus assembly around nucleic acids. J Mol Biol. 2014;426(18):3148–65. doi: 10.1016/j.jmb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zlotnick A, Porterfield JZ, Wang JC-Y. To build a virus on a nucleic acid substrate. Biophys J. 2013;104(7):1595–1604. doi: 10.1016/j.bpj.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cui X, Ludgate L, Ning X, Hu J. Maturation-associated destabilization of Hepatitis B Virus nucleocapsid. J Virol. 2013;87(21):11494–503. doi: 10.1128/JVI.01912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo J-T. Characterization of the intracellular deproteinized relaxed circular DNA of Hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007;81(22):12472–84. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- 154.Zlotnick A, Venkatakrishnan B, Tan Z, Lewellyn E, Turner W, Francis S. Core protein: a pleiotropic keystone in the HBV lifecycle. Antiviral Res. 2015;121:82–93. doi: 10.1016/j.antiviral.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Prevelige PE. Inhibiting virus-capsid assembly by altering the polymerisation pathway. Trends Biotechnol. 1998;16(2):61–65. doi: 10.1016/s0167-7799(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 156.Zlotnick A, Stray SJ. How does your virus grow? Understanding and interfering with virus assembly Trends Biotechnol. 2003;21(12):536–42. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 157.King RW, Ladner SK, Miller TJ, Zaifert K, Perni RB, et al. Inhibition of human Hepatitis B Virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (−)beta-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1998;42(12):3179–86. doi: 10.1128/aac.42.12.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weber O, Schlemmer K-H, Hartmann E, Hagelschuer I, Paessens A, et al. Inhibition of human Hepatitis B Virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res. 2002;54(2):69–78. doi: 10.1016/s0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 159.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 2007;76(2):168–77. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 160.Deres K, Schröder CH, Paessens A, Goldmann S, Hacker HJ, et al. Inhibition of Hepatitis B Virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299(5608):893–96. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 161.Billioud G, Pichoud C, Puerstinger G, Neyts J, Zoulim F. The main Hepatitis B Virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral Res. 2011;92(2):271–76. doi: 10.1016/j.antiviral.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 162.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, et al. Small-molecule effectors of Hepatitis B virus capsid assembly give insight into virus life cycle. J Virol. 2008;82(20):10262–70. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Delaney WE, Edwards R, Colledge D, Shaw T, Furman P, et al. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of Hepatitis B Virus in vitro. Antimicrob Agents Chemother. 2002;46(9):3057–60. doi: 10.1128/AAC.46.9.3057-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of Hepatitis B Virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol. 2010;5(12):1125–36. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. A heteroaryldihydropyrimidine activates and can misdirect Hepatitis B Virus capsid assembly. Proc Natl Acad Sci. 2005;102(23):8138–43. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stray SJ, Zlotnick A. Bay 41-4109 has multiple effects on Hepatitis B Virus capsid assembly. J Mol Recognit. 2006;19(6):542–48. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 167.Hacker HJ, Deres K, Mildenberger M, Schröder CH. Antivirals interacting with Hepatitis B Virus core protein and core mutations may misdirect capsid assembly in a similar fashion. Biochem Pharmacol. 2003;66(12):2273–79. doi: 10.1016/j.bcp.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 168.Hagan MF, Elrad OM. Understanding the concentration dependence of viral capsid assembly kinetics--the origin of the lag time and identifying the critical nucleus size. Biophys J. 2010;98:1065–74. doi: 10.1016/j.bpj.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Venkatakrishnan B, Katen SP, Francis S, Chirapu S, Finn MG, Zlotnick A. Hepatitis B Virus capsids have diverse structural responses to small-molecule ligands bound to the heteroaryldihydropyrimidine pocket. J Virol. 2016;90(8):3994–4004. doi: 10.1128/JVI.03058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bereszczak JZ, Rose RJ, van Duijn E, Watts NR, Wingfield PT, et al. Epitope-distal effects accompany the binding of two distinct antibodies to Hepatitis B Virus capsids. J Am Chem Soc. 2013;135(17):6504–12. doi: 10.1021/ja402023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Andoh Y, Yoshii N, Yamada A, Fujimoto K, Kojima H, et al. All-atom molecular dynamics calculation study of entire poliovirus empty capsids in solution. J Chem Phys. 2014;141(16):165101. doi: 10.1063/1.4897557. [DOI] [PubMed] [Google Scholar]
- 172.Phelps DK, Post CB. A novel basis of capsid stabilization by antiviral compounds. J Mol Biol. 1995;254(4):544–51. doi: 10.1006/jmbi.1995.0637. [DOI] [PubMed] [Google Scholar]
- 173.Machida A, Ohnuma H, Tsuda F, Yoshikawa A, Hoshi Y, et al. Phosphorylation in the carboxyl-terminal domain of the capsid protein of Hepatitis B Virus: evaluation with a monoclonal antibody. J Virol. 1991;65(11):6024–30. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yeh CT, Ou JH. Phosphorylation of Hepatitis B Virus precore and core proteins. J Virol. 1991;65(5):2327–31. doi: 10.1128/jvi.65.5.2327-2331.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gish RG, Yuen MF, Chan HL, Given BD, Lai CL, Locarnini SA, Lau JY, Wooddell CI, Schluep T, Lewis DL. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015;121:97–108. doi: 10.1016/j.antiviral.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 176.Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531(7594):386–9. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]