Abstract
What is known and objective
OATP1B1 mediates the transport of a diverse range of amphiphilic organic compounds that includes bile acids, steroid conjugates, and hormones. This retrospective pharmacogenetic study was conducted to assess the impact of the OATP1B1 c.521T>C single nucleotide polymorphism (SNP) on the pharmacokinetics of the steroidal aromatase inhibitor drug exemestane in healthy volunteers.
Methods
Exemestane (25 mg) was administered orally to 14 healthy postmenopausal women. All of the 14 subjects were sampled for pharmacokinetic (PK) analyses and retrospectively genotyped for OATP1B1 c.521T>C (rs 4149056).
Results and discussion
Of the 14 subjects enrolled in the study, 5 were carriers of the minor C allele (OATP1B1 c.521TC+CC), and the remaining 9 were carriers of the OATP1B1 c.521TT genotype. PK was assessed over 8 hours post-dosing. Our results showed statistically significant differences (P = 0.04) in the plasma exemestane AUC0–8 between the OATP1B1 genotype groups. Our data also showed statistically significant differences (P = 0.04) in the plasma AUC0–8 of 17-hydroexemestane (the major biologically active metabolite) between the OATP1B1 genotype groups.
What is new and conclusion
Our data suggest that the OAPTP1B1 c.521T>C SNP may influence exemestane pharmacokinetics in humans.
Keywords: Exemestane, Pharmacokinetics, Pharmacogenetics, OATP1B1, Healthy Volunteers
CURRENT KNOWLEDGE AND OBJECTIVE
Estrogen-dependent breast cancers constitute the most common type of breast cancer1–5 and a leading cause of cancer-related mortality among women worldwide.6 The steroidal aromatase inhibitor exemestane, which works by inhibiting aromatase (the rate-limiting enzyme responsible for the conversion of androgens into estrogens in postmenopausal women), is one of the standard treatments of postmenopausal women with estrogen-dependent breast cancers.7–11 Significant interindividual variability has been observed in the clinical response to exemestane.12–16
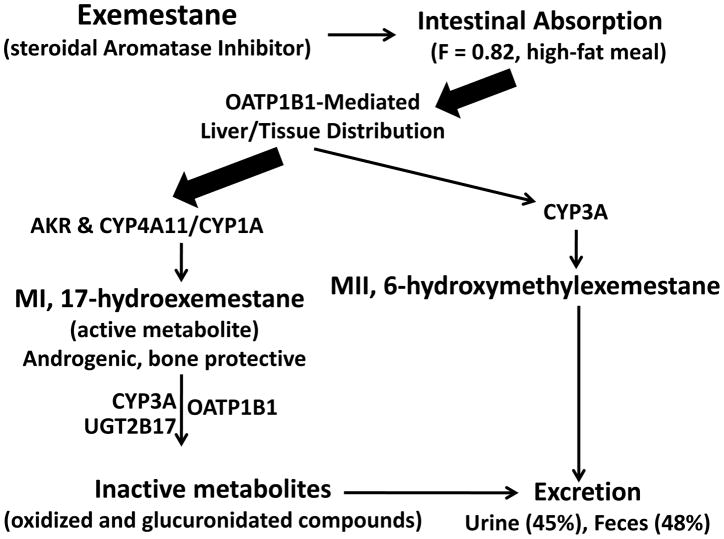
To better understand the mechanisms behind the interindividual variability in exemestane response, the metabolic pathways of exemestane were characterized, and specific enzymes responsible for the drug’s disposition were identified (Fig. 1). These findings suggest that, after absorption, exemestane is shunted to two different primary metabolic processes.10,17–19 One route involves its oxidation to 6-hydroxymethylexemestane primarily by CYP3A4. The other route involves its reduction by aldoketoreductases (AKR) and CYP4A11/CYP1A into an active (anti-estrogenic) and bone-protective androgenic metabolite, 17-hydroexemestane, which is the major metabolite of exemestane in humans in vivo. This bone-protective androgenic metabolite is secondarily deactivated by two main enzymes, CYP3A4 and UGT2B17.10,17–20
Figure 1.
Exemestane Pharmacokinetics
In this study, we hypothesize that other factors contributing to drug disposition, such as transporters (e.g., organic anion-transporting polypeptides/OATP), may play a role in the pharmacokinetics of the steroidal aromatase inhibitor exemestane. The role of steroid hormones as substrates of members of the OATP family is well documented.21–24 OATPs are a superfamily of transporter proteins that facilitate the cellular uptake of a diverse range of endogenous compounds and drugs, and are expressed in organs of importance in drug disposition, such as the liver, kidney and intestine.25–28 OATP proteins have a structure comprising 12 transmembrane domains,29,30 and the specific members OATP1B1 and OATP1B3 mediate the sodium-independent transport of a diverse range of amphiphilic organic compounds, including bile acids, steroid conjugates, thyroid hormones, anionic peptides, numerous drugs and other xenobiotic substances.29,30
Although many single-nucleotide polymorphisms (SNPs) have been identified in OATP1B1/3, only a few (OATP1B1 rs4149056, OATP1B1 rs2306283, OATP1B3 rs4149117, and OATP1B3 rs7311358) are known to have functional effects30–32 and clinical significance.
To test the hypothesis that OATP1B1/3 SNPs influence the pharmacokinetics of exemestane, we retrospectively genotyped DNA samples from 14 healthy postmenopausal women for the most common SNPs within OATP1B1/3 and correlated the genotyping results with previously published exemestane pharmacokinetic parameters.20
METHODS
Study design and subjects
This was a retrospective, non-randomized, open-label, single-dose pharmacokinetic study. Study subjects were enrolled with written informed consent under a protocol that was reviewed and approved by the Institutional Review Board at Harding University. All participants were healthy postmenopausal female volunteers who were judged to be in good medical health based on their medical history, a physical exam, and blood and urine chemistry, as reported previously.20 In preparation for the study, the subjects were asked to abstain from consuming alcoholic beverages, garlic, citrus products (especially grapefruit and grapefruit juice) apples and grapes for at least one week prior to the study. The healthy volunteers were also required to stop all over-the-counter medications, caffeinated beverages, and herbal or dietary supplements two days prior to exemestane dosing. On the study day, baseline blood samples were collected via upper extremity peripheral venipuncture, and urine samples were collected prior to dosing. The subjects were then given a single dose of exemestane orally (25 mg). Blood samples were taken via intravenous catheterization at 0.5, 1, 2, 4, 6, and 8 hours after drug administration. The catheters were removed and the subjects were allowed to return home. Blood samples in tubes were placed on ice immediately after collection and centrifuged at high speed for 10 minutes within 15 minutes of collection. The plasma samples were removed and stored at −80°C until liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
Compliance with ethical standards
All procedures performed in the studies that involved human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
LC-MS/MS and pharmacokinetics
Plasma concentrations of exemestane and its major metabolite 17-hydroexemestane were quantified using a previously described LC-MS/MS methodology.20 The LC-MS/MS instrument consisted of an Agilent 1100 HPLC pump (Tokyo, Japan), a CTC Analytics HTC-PAC autosampler (Zwingen, Switzerland), and an API 4000 QTRAP MS/MS system (Toronto, Canada). The separation system included a Kinetex 2.6 μm 150 × 4.6 mm column (Phenomenex, Torrance, California), a security 4 × 3 mm guard column, and a mobile phase consisting of 50% acetonitrile and 50% water at a flow rate of 500 μL/min. The mass spectrometer was operated using electrospray ionization with an ion spray voltage of 5000 V and the temperature was set at 700°C. The positive ion multiple reaction monitoring (MRM) mode was used for the analysis and was performed using nitrogen as the collision gas. The nebulizer gas (GS1) and the turbo gas (GS2) were set at 20 V and 50 V, respectively. The declustering potential was 50 V. A dwell time of 200 milliseconds and a pause time of 5 milliseconds between the scans were used to monitor precursor/product ion pairs. Two positive transitions (a quantifier MRM and qualifier MRM) were used for each analyte. Plasma concentrations of exemestane and 17-hydroexemestane were quantified using the ratio of the peak areas of exemestane and 17-hydroexemestane to that of the internal standard and calibration curves, which were constructed by spiking blank plasma with known amounts of exemestane and 17-hydroexemestane. Data collection and processing were performed using the Analyst software package (version 1.6, AB SCIEX, Ontario, Canada).
Chemicals
17-hydroexemestane was obtained from Toronto Research Chemicals Incorporation (Toronto, Canada). Exemestane and norgestrel (internal standard) were purchased from Sigma-Aldrich (St. Louis, Missouri). All other reagents were of HPLC grade or of the highest grade commercially available.
Genotyping
Genomic DNA was extracted from whole blood using a Promega Maxwell biorobot (Madison, Wisconsin). DNA quantity and quality/purity were determined photometrically at 260 nm and 280 nm using the Thermo Scientific NanoDrop 2000C spectrophotometer (Wilmington, Delaware) according to the manufacturer’s instructions. Genotyping was performed for 4 polymorphic loci in OATP1B1 and OATP1B3 transporter genes that are known to have functional effects30–32 and clinical significance. The 4 polymorphic loci [OATP1B1 rs4149056T>C, OATP1B1 rs2306283G>A, OATP1B3 rs4149117G>T, and OATP1B3 rs7311358A>G] were genotyped on the MassArray® System by CD Genomics (Shirley, NY, USA). The primers used for genotyping are described in Table 1. PCR amplifications were carried out using the GeneAmp PCR System 9700 (Applied Biosystems, Incorporation) with the following conditions: 94°C for 15 min (denaturation), followed by 45 cycles of amplification at 94°C for 20 sec (denaturation), 56°C for 30 sec (annealing) and 72°C for 1 min (extension). The PCR was completed with a final extension at 72°C for 3 min.
Table 1.
Primer sequences for PCR
| SNP | Primer sequence (Forward) | Primer sequence (Reverse) |
|---|---|---|
| OATP1B1 rs4149056T>C | ACGTTGGATGAATCTGGGTCATACATGTGG | ACGTTGGATGTATGGGAGTCTCCCCTATTC |
| OATP1B1 rs2306283G>A | ACGTTGGATGATTAAACAAGTGGATAAGG | ACGTTGGATGGATGTTCTTACAGTTACAGG |
| OATP1B3 rs4149117G>T | ACGTTGGATGGGTTGTCTCCTTATGGGAAC | ACGTTGGATGGGCTCAGAGCTGTTTAACAC |
| OATP1B3 rs7311358A>G | ACGTTGGATGTGGTCCAGTCATTGGCTTTG | ACGTTGGATGCTTACTCAGATCTACATATCC |
Statistical analysis
Pharmacokinetic and statistical analyses were performed using Microsoft Excel (version 2013) and Prism (version 6, GraphPad Software, San Diego, California), respectively. The AUC0–8 was calculated by the linear trapezoidal method for the observed values. The demographics (BMI and age) and the AUC0–8 were compared with the subjects’ genotypes using a 2-sided unpaired nonparametric Mann-Whitney U Test. Initially, two types of binary groupings were assigned to each SNP genotype: (AA + AB vs. BB) or (AA vs. AB + BB). To simplify the presentation, only one genotyping group (the heterozygous AB grouped with either AA or BB homozygous genotypes) is given: the group yielding the lowest P value for the association of that locus with the AUC0–8. A value of P ≤ 0.05 was considered to indicate statistical significance.
RESULTS AND DISCUSSION
Clinical characteristics and OATP1B1/3 genotyping data
Fourteen healthy postmenopausal women participated in this retrospective, open-label, non-randomized pharmacokinetic study. The subject characteristics of these healthy female volunteers are described in Table 2. Body mass index values ranged from 18.6 to 34.6 kg/m2, with a mean value of 27.13 kg/m2 and a standard deviation of 0.16. Ages ranged from 45 to 65 years, with a mean age of 55.5 and a standard deviation of 5.08 years. No statistically significant differences were observed between body mass index (BMI) and the OATP1B1 rs4149056T>C (P = 0.5), OATP1B1 rs2306283G>A (P = 0.9), OATP1B3 rs4149117G>T (P = 0.9), or OATP1B3 rs7311358A>G (P = 0.9) genotype groups (Table 2). Additionally, no statistically significant differences were observed between age and the OATP1B1 rs4149056T>C (P = 0.14), OATP1B3 rs4149117G>T (P = 0.3), or OATP1B3 rs7311358A>G (P = 0.28) genotype groups. However, we found a statistically significant difference (P = 0.04) between age and the OATP1B1 rs2306283G>A genotype group.
Table 2.
Clinical characteristics of the study participants
| OATP1B1 Genotype | OATP1B3 Genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OATP1B1 rs4149056T>C | OATP1B1 rs2306283G>A | OATP1B3 rs4149117G>T | OATP1B3 rs7311358A>G | |||||||||
| Demogra-phics | CC +CT | TT | P | AA +AG | GG | P | GT | GG | P | AA | GA | P |
| BMI kg/m2 (SD) | 28.4 (5.2) | 26.4 (5.2) | 0.5 | 26.9 (4.9) | 28.2 (7.7) | 0.9 | 27.7 (4.9) | 26.9 (6.1) | 0.9 | 27.7 (4.9) | 26.9 (6.1) | 0.9 |
| Age, years (SD) | 53 (5.5) | 56.9 (4.5) | 0.14 | 56.6 (4.2) | 48.5 (4.9) | 0.04 | 53.7 (2.8) | 56.2 (5.7) | 0.3 | 53.7 (2.8) | 56.2 (5.7) | 0.28 |
The areas under the plasma concentration-time curves (AUC0–8) of exemestane and 17-hydroexemestane (17-HEXE) were calculated based on plasma concentrations at predose and at 0.5, 1, 2, 4, 6, and 8 hours after exemestane administration using the linear trapezoidal rule. Plasma concentrations beyond 8 hours after exemestane administration were not calculated because they were below the lowest limit of quantification.
Table 3 describes the pharmacokinetic characteristics of exemestane (AUC0–8) and 17-HEXE (AUC0–8) based on the OATP1B1 and OATP1B3 genotype groups. The impact of OATP1B1 and OATP1B3 SNPs, namely, rs4149056T>C, rs2306283G>A, rs4149117G>T, and rs7311358A>G, on exemestane and 17-HEXE pharmacokinetics was assessed using a 2-sided unpaired nonparametric Mann-Whitney U Test.
Table 3.
Pharmacokinetics (PK) of exemestane (exemestane) and 17-hydroexemestane (17-HEXE)
| OATP1B1 Genotype | OATP1B3 Genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OATP1B1 rs4149056T>C | OATP1B1 rs2306283G>A | OATP1B3 rs4149117G>T | OATP1B3 rs7311358A>G | |||||||||
| PK | CC +CT | TT | P | AA +AG | GG | P | GT | GG | P | AA | GA | P |
| exemestane | ||||||||||||
| AUC0–8, ng × h/mL (SD) | 82.5 (82.5) | 29.04 (10.8) | 0.04 | 35.2 (16) | 125.3 (143.7) | 0.65 | 43.5 (22.5) | 49.9 (63) | 0.6 | 43.5 (22.5) | 49.9 (63) | 0.6 |
| 17-HEXE | ||||||||||||
| AUC0–8, ng × h/mL (SD) | 3.09 (1.2) | 1.83 (0.3) | 0.04 | 3.03 (1.9) | 2.15 (0.84) | 0.92 | 2.5 (1.4) | 2.3 (0.86) | 0.9 | 2.5 (1.4) | 2.3 (0.86) | 0.9 |
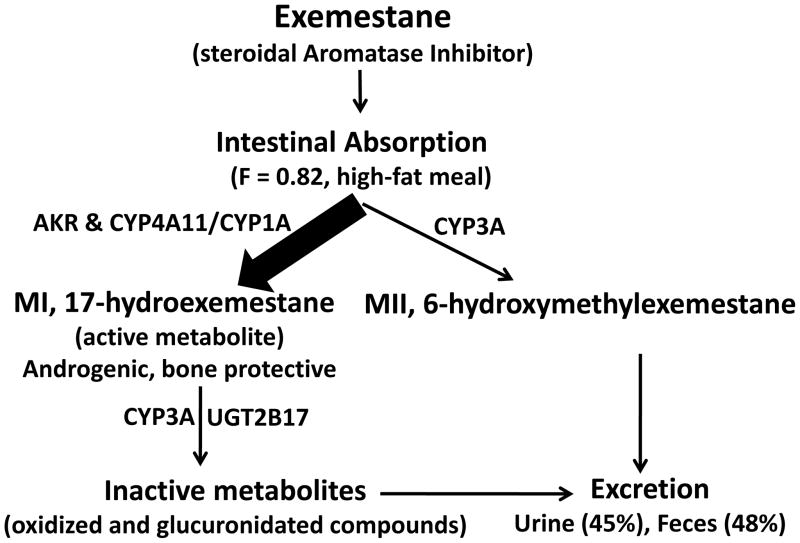
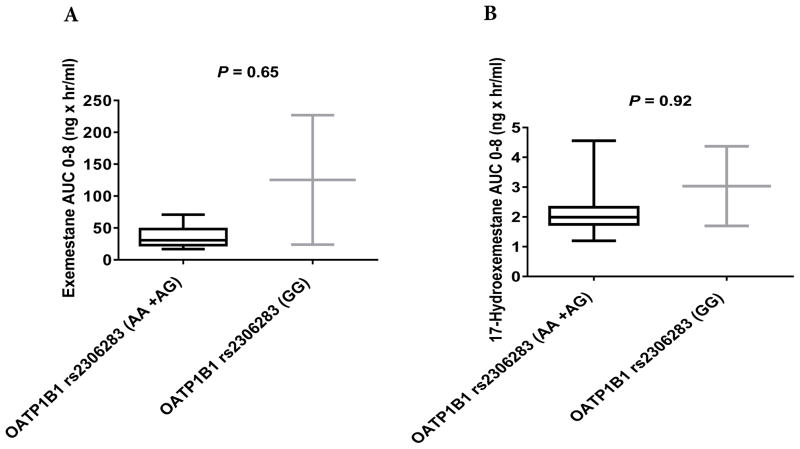
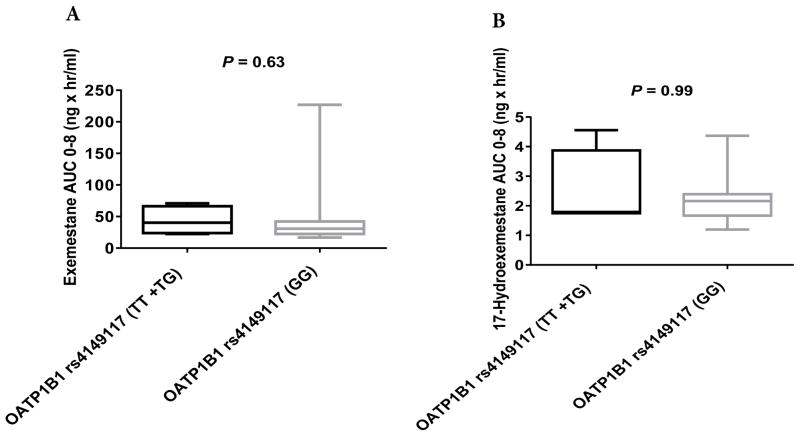
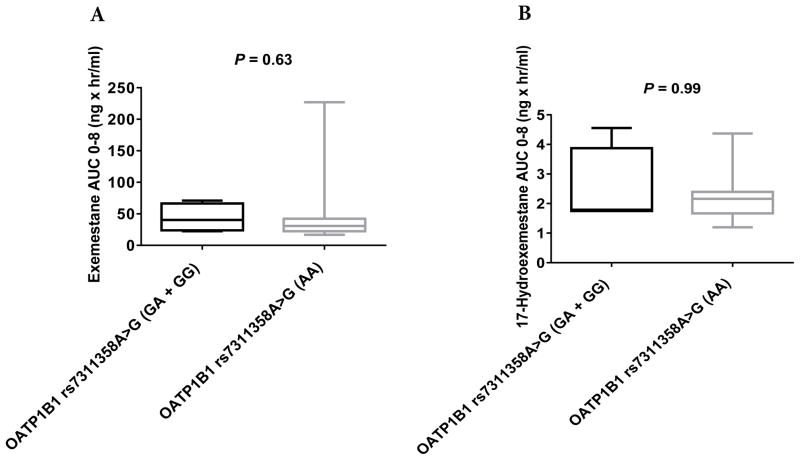
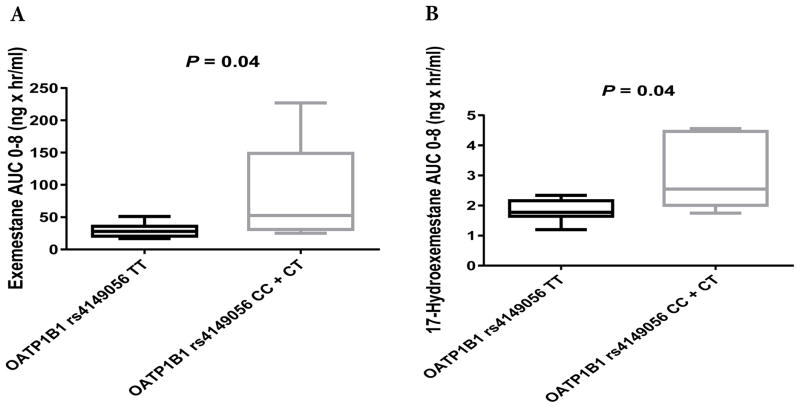
No significant differences were observed in the pharmacokinetics of exemestane (AUC0–8) or 17-HEXE (AUC0–8) between the OATP1B1 rs2306283G>A, OATP1B3 rs4149117G>T, and OATP1B3 rs7311358A>G genotype groups (Table 3, Figs. 2A and 2B, 3A and 3B, and 4A and 4B). However, statistically significant differences were observed in the exemestane AUC0–8 and the 17-HEXE AUC0–8 between the OATP1B1 rs4149056T>C (CC + CT vs. TT) genotype groups (exemestane AUC0–8, 82.5 vs. 29.04 ng × hr/mL, P = 0.04; 17-HEXE AUC0–8, 3.09 vs. 1.83 ng × hr/mL, P = 0.04) (Table 3, Fig. 5A and 5B).
Figure 2.
AUC0–8 values of exemestane (A) and 17-hydroexemestane (B) stratified by OATP1B1 rs2306283G>A genotype.
Figure 3.
AUC0–8 values of exemestane (A) and 17-hydroexemestane (B) stratified by OATP1B3 rs4149117G>T genotype.
Figure 4.
AUC0–8 values of exemestane (A) and 17-hydroexemestane (B) stratified by OATP1B3 rs7311358A>G genotype.
Figure 5.
AUC0–8 values of exemestane (A) and 17-hydroexemestane (B) stratified by OATP1B1 rs4149056T>C genotype.
The impact of common OATP1B1 and OATP1B3 SNPs on exemestane and 17-hydroexemestane pharmacokinetics
This retrospective, open-label, pharmacogenetic study shows for the first time that there is an association between the OATP1B1 rs4149056C>T SNPs and exemestane metabolism in humans in vivo. In our study of 14 healthy postmenopausal female volunteers, results showed statistically significant differences in the total plasma exemestane AUC0–8 and the 17-HEXE AUC0–8 between the OATP1B1 rs4149056C>T genotype groups. We did not find any statistically significant differences in the total plasma exemestane AUC0–8 or the 17-HEXE AUC0–8 between the OATP1B1 rs2306283G>A, OATP1B3 rs4149117G>T, and OATP1B3 rs7311358A>G genotype groups. These findings suggest that the OATP1B1 rs4149056C>T SNP influences the pharmacokinetics of exemestane and 17-HEXE in humans.
Our updated exemestane pharmacokinetic pathway (Fig. 6) now suggests that, after absorption, exemestane is first transported by the polymorphic OATP1B1 protein and then shunted to two different primary metabolic processes.10,17–19 One route involves its oxidation to 6-hydroxymethylexemestane primarily by CYP3A4. The other route involves its reduction by aldoketoreductases (AKR) and CYP4A11/CYP1A into an active (anti-estrogenic) and bone-protective androgenic metabolite, 17-hydroexemestane, which is the major metabolite of exemestane in humans in vivo. This bone-protective androgenic metabolite is then transported into the liver by the polymorphic OATP1B1 protein and then secondarily deactivated by two main enzymes, CYP3A4 and UGT2B17.10,17–20
Figure 6.
Updated Pharmacokinetics of Exemestane
OATP1B1, the organic anion transporting polypeptide 1B1, is one of the main hepatic uptake transporters. It is localized on the basolateral part of hepatocytes and is involved in the active cellular influx of diverse endogenous substrates, such as bile acids, bilirubin, conjugates of steroid hormones and drugs such as HMG-CoA reductase inhibitors or statins.25–28 The OATP1B1 protein (691 amino acid residues) is encoded by the SLCO1B1 gene, which consists of 15 exons (including non-coding exon 1) and spans approximately 109 kb on chromosome 12p12.2-p12.1. Similar to other OATP family members, this transporter is predicted to have 12 transmembrane domains.29,33 Although many SNPs have been identified in OATP1B1, only a few are known to have functional effects.32 The common OATP1B1 c.521T>C variant, rs4149056, produces a p.V174A substitution and is the predominant OATP genetic predictor of drug disposition.30 The minor rs4149056 C allele has been associated with decreased transport function in vitro and decreased clearance for a number of drugs in vivo.30 Among all currently available drugs, HMG-CoA reductase inhibitors or statins are believed to be the most clinically relevant substrates of OATP1B1. Several single-dose pharmacokinetic studies have shown AUCs of the statins simvastatin acid, pitavastatin, atorvastatin, pravastatin, and rosuvastatin that were 221%, 162–191%, 144%, 57–130%, and 62–117% higher, respectively, in rs4149056 CC homozygotes than in rs4149056 TT homozygotes.27,32
Hormone-dependent breast cancers constitute the most common type of breast cancer and the second leading cause of mortality among women in the United States.2–4 Consequently, long-term adjuvant endocrine therapy in cancer-free (healthy) patients is commonly included in treatment for at least 5 years, with evidence of improved survival and reduced metastatic disease.7,34–36 Unfortunately, emerging breast cancer outcome data suggest that up to 50% of women diagnosed with breast cancer and receiving endocrine therapy experience little or no clinical benefit from treatment (non-responders)37 and that up to 70% of women who begin one hormonal therapy drug will be forced to discontinue and/or switch to another hormonal therapy drug due to intolerable and undesirable toxic side effects (toxic responders).13,38–40 The molecular basis of non-response and toxic responses could be linked to genetic variations within the organic anion transporting polypeptides (OATPs). Our data support the strong role of OATP1B1 in the disposition of exemestane. Our findings suggest that exemestane could be even more clinically relevant than statins because our single-dose healthy volunteer pharmacokinetic study showed that the plasma AUC values of exemestane were 284% higher in rs4149056 CC + CT (82.5 ng × hr/mL) individuals than in TT (29.04 ng × hr/mL) homozygous individuals.
Several limitations of our study should be considered. First, this was a retrospective study that was not powered to detect a difference between the OATP1B1 rs4149056 (CC + CT) and (TT) genotype groups. Of the 14 subjects enrolled in this study, 5 were carriers of the minor C allele (OATP1B1 c.521 TC+CC), and the remaining 9 were carriers of the OATP1B1 c.521TT genotype. A post hoc calculation using the Mann-Whitney statistical test and G*Power software version 3.1.9.2 gave a power of 51%. Second, in our study, the healthy postmenopausal female volunteers only received a single 25 mg dose of exemestane. Finally, the women were asked to abstain from the use of potential confounders such as co-medication and also herbal and dietary supplements. All of these limitations suggest that caution should be used in interpreting the results of our study and that future prospective pharmacogenetic studies are needed to validate the involvement of OATP1B1 in the disposition of the hormonal breast cancer agent exemestane.
NEW INFORMATION AND CONCLUSION
Our data suggest that the OAPTP1B1 c.521T>C SNP may influence exemestane pharmacokinetics in humans.
Acknowledgments
We are very grateful to the study volunteers, students, faculty, and staff of the Harding Community for their participation and contributions.
Sources of funding
This work was supported by the Arkansas INBRE Program, supported by grant funding from the National Institute of Health (NIH) National Institute of General Medical Sciences (NIGMS) (P20GM103429).
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Cauley JA, Lucas FL, Kuller LH, Stone K, Browner W, Cummings SR. Elevated serum estradiol and testosterone concentrations are associated with a high risk for breast cancer. Study of Osteoporotic Fractures Research Group. Ann Intern Med. 1999;130:270–277. doi: 10.7326/0003-4819-130-4_part_1-199902160-00004. [DOI] [PubMed] [Google Scholar]
- 2.Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 5.Joslyn SA. Hormone receptors in breast cancer: racial differences in distribution and survival. Breast Cancer Res Treat. 2002;73:45–59. doi: 10.1023/a:1015220420400. [DOI] [PubMed] [Google Scholar]
- 6.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 7.Buzdar AU, Coombes RC, Goss PE, Winer EP. Summary of aromatase inhibitor clinical trials in postmenopausal women with early breast cancer. Cancer. 2008;112:700–709. doi: 10.1002/cncr.23193. [DOI] [PubMed] [Google Scholar]
- 8.Coombes RC, Kilburn LS, Snowdon CF, et al. Survival and safety of exemestane versus tamoxifen after 2–3 years’ tamoxifen treatment (Intergroup Exemestane Study): a randomised controlled trial. Lancet. 2007;369:559–570. doi: 10.1016/S0140-6736(07)60200-1. [DOI] [PubMed] [Google Scholar]
- 9.Keating GM. Letrozole: a review of its use in the treatment of postmenopausal women with hormone-responsive early breast cancer. Drugs. 2009;69:1681–1705. doi: 10.2165/10482340-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Robinson A. A review of the use of exemestane in early breast cancer. Ther Clin Risk Manag. 2009;5:91–98. [PMC free article] [PubMed] [Google Scholar]
- 11.Sanford M, Plosker GL. Anastrozole: a review of its use in postmenopausal women with early-stage breast cancer. Drugs. 2008;68:1319–1340. doi: 10.2165/00003495-200868090-00007. [DOI] [PubMed] [Google Scholar]
- 12.Cheung AM, Tile L, Cardew S, et al. Bone density and structure in healthy postmenopausal women treated with exemestane for the primary prevention of breast cancer: a nested substudy of the MAP.3 randomised controlled trial. Lancet Oncol. 2012;13:275–284. doi: 10.1016/S1470-2045(11)70389-8. [DOI] [PubMed] [Google Scholar]
- 13.Cheung AM, Tomlinson G, Goss PE. Bone loss with exemestane: is the jury still out? J Clin Oncol. 2005;23:9433–9434. doi: 10.1200/JCO.2005.04.1376. author reply 9433–9435. [DOI] [PubMed] [Google Scholar]
- 14.Chien AJ, Goss PE. Aromatase inhibitors and bone health in women with breast cancer. J Clin Oncol. 2006;24:5305–5312. doi: 10.1200/JCO.2006.07.5382. [DOI] [PubMed] [Google Scholar]
- 15.Goss PE, Qi S, Cheung AM, Hu H, Mendes M, Pritzker KP. Effects of the steroidal aromatase inhibitor exemestane and the nonsteroidal aromatase inhibitor letrozole on bone and lipid metabolism in ovariectomized rats. Clin Cancer Res. 2004;10:5717–5723. doi: 10.1158/1078-0432.CCR-04-0438. [DOI] [PubMed] [Google Scholar]
- 16.Lonning PE, Geisler J, Krag LE, et al. Effects of exemestane administered for 2 years versus placebo on bone mineral density, bone biomarkers, and plasma lipids in patients with surgically resected early breast cancer. J Clin Oncol. 2005;23:5126–5137. doi: 10.1200/JCO.2005.07.097. [DOI] [PubMed] [Google Scholar]
- 17.Kamdem LK, Flockhart DA, Desta Z. In vitro cytochrome P450-mediated metabolism of exemestane. Drug Metab Dispos. 2011;39:98–105. doi: 10.1124/dmd.110.032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landry K, David F, Zeruesenay D. 17-hydroexemestane: a potent inhibitor of CYP19 (aromatase) and substrate of CYP3A. J Drug Metab Toxicol. 2014;5:2. [Google Scholar]
- 19.Sun D, Chen G, Dellinger RW, Sharma AK, Lazarus P. Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics. 2010;20:575–585. doi: 10.1097/FPC.0b013e32833b04af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SM, Atchley DH, Murphy MA, Gurley BJ, Kamdem LK. Impact of UGT2B17 gene deletion on the pharmacokinetics of 17-hydroexemestane in healthy volunteers. J Clin Pharmacol. 2016;56:875–884. doi: 10.1002/jcph.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Y, Konig J, Leier I, Buchholz U, Keppler D. Hepatic uptake of bilirubin and its conjugates by the human organic anion transporter SLC21A6. J Biol Chem. 2001;276:9626–9630. doi: 10.1074/jbc.M004968200. [DOI] [PubMed] [Google Scholar]
- 22.Keppler D, Konig J, Cui Y. The human hepatocyte-specific organic anion transporter encoded by the SLC21A8 gene. Gastroenterology. 2002;122:1545–1546. doi: 10.1053/gast.2002.33377. [DOI] [PubMed] [Google Scholar]
- 23.Green SM, Kaipainen A, Bullock K, et al. Role of OATP transporters in steroid uptake by prostate cancer cells in vivo. Prostate Cancer Prostatic Dis. 2017;20:20–27. doi: 10.1038/pcan.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Pu Z, Ge J, Shen J, Yuan X, Xie H. Association of CYP2D6*10, OATP1B1 A388G, and OATP1B1 T521C polymorphisms and overall survival of breast cancer patients after tamoxifen therapy. Med Sci Monit. 2015;21:563–569. doi: 10.12659/MSM.893473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung JY, Cho JY, Yu KS, et al. Effect of OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of pitavastatin in healthy volunteers. Clin Pharmacol Ther. 2005;78:342–350. doi: 10.1016/j.clpt.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Maeda K, Ieiri I, Yasuda K, et al. Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin Pharmacol Ther. 2006;79:427–439. doi: 10.1016/j.clpt.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 28.Treiber A, Schneiter R, Hausler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug Metab Dispos. 2007;35:1400–1407. doi: 10.1124/dmd.106.013615. [DOI] [PubMed] [Google Scholar]
- 29.Hagenbuch B, Meier PJ. The superfamily of organic anion transporting polypeptides. Biochim Biophys Acta. 2003;1609:1–18. doi: 10.1016/s0005-2736(02)00633-8. [DOI] [PubMed] [Google Scholar]
- 30.International Transporter Consortium. Giacomini KM, Huang SM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boivin AA, Cardinal H, Barama A, Pichette V, Hebert MJ, Roger M. Organic anion transporting polypeptide 1B1 (OATP1B1) and OATP1B3: genetic variability and haplotype analysis in white Canadians. Drug Metab Pharmacokinet. 2010;25:508–515. doi: 10.2133/dmpk.dmpk-10-sh-046. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96:423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamai I, Nezu J, Uchino H, et al. Molecular identification and characterization of novel members of the human organic anion transporter (OATP) family. Biochem Biophys Res Commun. 2000;273:251–260. doi: 10.1006/bbrc.2000.2922. [DOI] [PubMed] [Google Scholar]
- 34.Bertelli G, Gangadhara S. Exemestane in postmenopausal women with early or advanced breast cancer: a review. Expert Opin Pharmacother. 2010;11:1933–1942. doi: 10.1517/14656566.2010.495945. [DOI] [PubMed] [Google Scholar]
- 35.Bertelli G, Hall E, Ireland E, et al. Long-term endometrial effects in postmenopausal women with early breast cancer participating in the Intergroup Exemestane Study (IES)--a randomised controlled trial of exemestane versus continued tamoxifen after 2–3 years tamoxifen. Ann Oncol. 2010;21:498–505. doi: 10.1093/annonc/mdp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisen A, Trudeau M, Shelley W, Messersmith H, Pritchard KI. Aromatase inhibitors in adjuvant therapy for hormone receptor positive breast cancer: a systematic review. Cancer Treat Rev. 2008;34:157–174. doi: 10.1016/j.ctrv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Anderson H, Bulun S, Smith I, Dowsett M. Predictors of response to aromatase inhibitors. J Steroid Biochem Mol Biol. 2007;106:49–54. doi: 10.1016/j.jsbmb.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Hershman DL, Shao T, Kushi LH, et al. Early discontinuation and non-adherence to adjuvant hormonal therapy are associated with increased mortality in women with breast cancer. Breast Cancer Res Treat. 2011;126:529–537. doi: 10.1007/s10549-010-1132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kadakia KC, Snyder CF, Kidwell KM, et al. Patient-reported outcomes and early discontinuation in aromatase inhibitor-treated postmenopausal women with early stage breast cancer. Oncologist. 2016;21:539–546. doi: 10.1634/theoncologist.2015-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy CC, Bartholomew LK, Carpentier MY, Bluethmann SM, Vernon SW. Adherence to adjuvant hormonal therapy among breast cancer survivors in clinical practice: a systematic review. Breast Cancer Res Treat. 2012;134:459–478. doi: 10.1007/s10549-012-2114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]