Abstract
Expression of activating transcription factor 5 (ATF5) negatively correlates with patient survival for different types of cancers. Here, we illustrate ATF5 is important for survival and proliferation of cancer and can be targeted to selectively trigger apoptosis of cancer cells, but sparing normal cells. Cell-penetrating peptides combined with a dominant negative ATF5 cargo have shown efficacy in brain, breast, melanoma, and prostate cancers.
Keywords: CP (Cell Penetrating Peptide), ATF5, CP-d/n-ATF5 (Cell penetrating peptide dominant negative ATF5), apoptosis, and cancer
Forum
Transcription factors activate or repress genes involved in development, proliferation, differentiation, homeostasis, and in cancer. Targeting transcription factors has the advantage of direct control of gene regulation to promote a desired cell response. Classical pharmaceuticals achieve their function through direct binding to receptors to initiate or inhibit signaling cascades leading to gene activation/repression. The receptor mediated transcriptional therapeutic effects often coexist with unwanted irrelevant-transcriptional and non-transcriptional side effects.
To date, apart from nuclear hormone receptors, transcription factors proteins have not been shown to be good accessible drug targets for small organic molecule drugs. Small protein/peptides that behave as dominant negative counterparts to their targeted transcription factors, namely activating transcription factor 5 (ATF5), have shown promise of blocking proliferation and survival of cancer cells. Blocking ATF5 triggers selective apoptosis in cancer with the advantage of sparing normal cells, which has not been demonstrated with other transcription factors.
ATF5 Biology
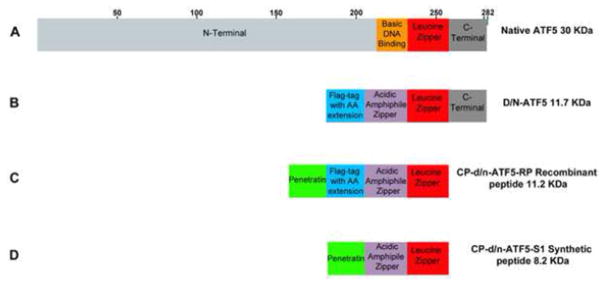
ATF5 belongs to the family of cAMP-response-element binding proteins/activating transcription factors (CREBs)/ATFs) that serve in neural synaptic plasticity and memory, endoplasmic reticulum and cellular stress, maintenance of neural progenitors, and cell proliferation. Like each family member, ATF5 possesses an N-terminal amino acid sequence for transactivation, a basic DNA binding domain, a leucine zipper composed of a sequence of heptad repeats of leucine involved in homodimerization or binding to other leucine zipper transcription factor binding partners, and a C-terminal end (Figure 1A) [1]. Two mRNAs distinct on their 5′-untranslated ends, ATF5α and less abundant ATF5β translate the same ATF5 protein [1]. The ATF5α transcript has two open reading frame sites. The first initiates ATF5, while the second inhibits translation. There are two putative Kozak translational start sites initiating expression of a longer 30 KDa or a shorter 22 KDa ATF5 protein. Expression of the two lengths variants is specific to cell type, but functional roles regarding their lengths are not known. [1].
Figure 1.
ATF5 and dominant negative ATF5 constructs. (A) Human wild type native full length ATF5 contains an N-terminal, basic DNA binding, leucine zipper, and C-terminal within 282 amino acid length. (B) D/N ATF5 protein has the Flag-tagged reporter with amino acid extension, acidic amphipathic α-helical sequence, wild type leucine zipper and C-terminal domain. (C) The bacteria recombinant cell penetrating d/n-ATF5 has the Penetratin motif N-terminal to the Flag-tagged reporter with amino acid extension, acidic amphipathic α-helical sequence, wild type leucine zipper, but C-terminally truncated. (D) The chemical synthetic form has penetratin N-terminal to acidic amphipathic α-helical sequence, wild type leucine zipper, but lacks the Flag-tagged reporter with amino acid extension, and C-terminus. The removal of the C-terminal domain reduces peptide aggregation during bacterial or chemical manufacturing, but does not affect drug mechanism of action, and pharmacological properties. The removal of Flag with amino acid extension for the chemical synthetic CP-d/n-ATF5 eliminates Flag antibody detection required for detection in tissues, but there are no differences in drug mechanism, and pharmacological properties. Elimination of Flag-tag with amino acid extension increases ease of manufacturing with cost reduction for preclinical studies.
ATF5 is expressed in a variety of tissue types that include olfactory neurons, neural progenitor cells, with the highest expression in the liver. ATF5 serves in cell survival, proliferation, and inhibition of neural differentiation. ATF5 upregulates both Egr1 [2] and cycD3 genes [1] that are involved in proliferation. Egr-1 also has a role in survival along with ATF5-transcriptionally regulated anti-apoptotic genes, BCL2 and MCL1 [3, 4]. Deubiquitinase USP9X expression is also governed by ATF5, and USP9X post-translationally stabilizes both BCL2, MCL1, and MCL1-stability protein, Bag3 [5]. ATF5 itself is stabilized by binding to heat shock protein 70 [6], but is degraded by binding to nucleophosmin in hepatocellular carcinomas [6]. Under cellular stress, ATF5 is either transcriptionally induced as in amino acid starvation or in cytotoxicity, [1, 7], or translationally induced by heat shock, endoplasmic reticulum stress, or arsenite exposure [1]. The latter promotes phosphorylation of eukaryotic initiation factor eIF2 leading to translation of ATF5 using the first opening frame site [1]. ATF5-mediated rescue of cells from stress is achieved by upregulation of chaperone heat shock proteins [1]. Given these roles, it is apparent that ATF5 may encourage proliferation in cancer and aid in the rescue of cancer cells from stress.
High ATF5 tumor specific expression is seen in glioblastomas, astrocytomas, adenocarcinomas, transitional cell carcinomas, squamous cell carcinomas, and metastatic carcinomas [1]. Surrounding cells in the tumor microenvironment are ATF5 negative [1]. One exception is hepatic carcinomas where a worse prognosis is associated with lower levels of ATF5 [6].
Development of ATF5 peptide drugs
The ATF5 antagonist (dominant negative ATF5; d/n-ATF5) was designed as a N-terminally truncated construct, with the DNA binding domain replaced with an acidic amphipathic α-helical sequence encompassing mostly heptad repeats of leucine, and an unchanged leucine zipper and C-terminal [8] (Figure 1B).
Transfection of d/n-ATF5 transgene triggered massive apoptosis in glioma, breast, ovarian, and pancreatic cancer cell lines (Table 1) [1, 9, 10]. Infection of rat brain glioma allografts with d/n-ATF5 retrovirus resulted in massive apoptosis of tumor cells [1] but not of normal astrocytes. Induction of transgenic d/n-ATF5 in a mouse model which gliomas were created from resident neural progenitors resulted in tumor prevention or complete eradication (Table 1) [11]. In all cases, normal cells were spared [11]. To migrate from a transgene to a therapeutic deliverable drug, d/n-ATF5 was N-terminally fused to the cell penetrating peptide motif penetratin. This fusion was manufactured in bacteria as Cell Penetrating-d/n-ATF5 recombinant peptide (CP-d/n-ATF5-RP), or as a chemically synthesized peptide (CP-d/n-ATF5-S1). Both forms included truncation of 25 C-terminal amino acids, and exclusion of the C-terminus domain, to reduce aggregation without compromising efficacy (Figure 1C & D) [12]. In vitro, CP-d/n-ATF5-RP and CP-d/n-ATF5-S1 promoted apoptosis of cancer cell lines. In fact, CP-d/n-ATF5-S1 was shown to promote apoptosis in a broad spectrum of tumors (Table 1) [5, 12]. CP-d/n-ATF5-S1 induces apoptosis by disrupting the expression of prosurvival proteins, ATF5 itself, deubiquitinase USP9X, BCL2, BAG3, and MCL1 [5]. Loss of USP9X leads to greater degradation of Bag3, MCL1, and BCL2 through their enhanced ubiquitination. The addition of CP-d/n-ATF5-S1 with TRAIL or the BH3-mimetic ABT263 to cancer cell lines revealed synergism by increased apoptosis compared to CP-d/n-ATF5-S1, MCL1, or ABT263 added separately. In fact, the loss of MCL1 and Bag3 induced by CP-d/n-ATF5 is thought to promote this synergism including cancer cells more resistant to TRAIL or ABT263 [5].
Table 1.
Tumors responsive toward ATF5 antagonism.
| Tumor Type | Tumor origin | d/n-ATF5 Antagonism Method | Outcomes |
|---|---|---|---|
|
| |||
| Glioma | In vitro | Transgene transfection [1] | Apoptosis[1] |
| In vivo | |||
| Allograft | Retrovirus [1] | Apoptosis [1] | |
| Induced Gliomas | Transgenic [11] | Prevention/Eradication [11] | |
| Orthotopic | CP-d/n-ATF5-RP [12] | Growth Attenuation [12] | |
| Orthotopic | CP-d/n-ATF5-S1 [5] | Growth Attenuation [5] | |
| Xenograft | CP-d/n-ATF5-S1 [5] | Growth Attenuation [5] | |
|
| |||
| Epithelial Ovarian | In vitro | Transgene transfection [9] | Apoptosis [9] |
|
| |||
| Breast | In vitro | Transgene transfection [1] | Apoptosis [1] |
| In vivo | |||
| Orthotopic | CP-d/n-ATF5-S1 [5] | Regression [5] | |
|
| |||
| Pancreatic | In vitro | Transgene transfection [10] | Apoptosis [10] |
| In vivo | |||
| Xenograft | CP-d/n-ATF5-S1 [5, 10] | No Growth Attenuation [5] | |
|
| |||
| Prostate (hormone-refractory) | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
| In Vivo | |||
| Xenograft | CP-d/n-ATF5-S1 [5] | Growth Attenuation [5] | |
|
| |||
| Lung (non-small cell) | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
|
| |||
| Colorectal | In vivo/Xenograft | CP-d/n-ATF5-S1 [5] | No Growth Attenuation [5] |
|
| |||
| Melanoma (BRAF mutated) | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
| In vivo | |||
| Orthotopic | CP-d/n-ATF5-S1 [5] | Growth Attenuation [5] | |
|
| |||
| Myeloid Leukemia | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
|
| |||
| B-Cell Lymphoma | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
|
| |||
| Chronic Myeloid Leukemia | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
|
| |||
| Raji Burkitt Lymphoma Cells | In vitro | CP-d/n-ATF5-S1 [5] | Apoptosis [5] |
Intraperitoneal or subcutaneous systemic drug injection of mice with PDGF-B/sh-p53 induced gliomas showed that CP-d/n-ATF5-RP crosses the blood brain barrier and is incorporated in glioma and normal brain cells with retention up to 72 hours. The gliomas were eradicated after two succeeding treatments of CP-d/n-ATF5. There was no regrowth or recurrence of gliomas from 6 to 13 months after CP-d/n-ATF5-RP treatment [12]. In a separate study, using intracranial human glioma orthotopic xenografts as a model for human gliomas in mice, CP-d/n-ATF5-RP and CP-d/n-ATF5-S1 suppressed glioma growth with increased survival time for one glioma line [5, 12]. Likewise, CP-d/n-ATF5-S1 significantly reduced the growth of melanoma, prostate, and glioma subcutaneous xenografts, and caused regression of orthotopic triple negative breast cancer [5] (Table 1). CP-d/n-ATF5-S1 was not effective in pancreatic and colorectal cancers [5]. Both forms of the cell-penetrable d/n-ATF5 were not toxic to normal cells [5, 12] or tissues, and the kidney and liver function was not affected in exposed animals [12].
In summary, interfering with ATF5 function selectively triggers massive apoptosis and regression of tumors. Future efforts are underway to identify transcription factors that directly bind to ATF5 and serve as partners in regulation of prosurvival genes. The multi-targeting of ATF5 and its binding partners could both increase the efficacy and prolong the disruption of prosurvival genes to maximize cancer treatment. These findings open doors for the pursuit of drug candidates that block the function of downstream prosurvival proteins.
Concluding Remarks
The concept of selectively targeting transcription factors as a regimen for cancer therapeutics is on the path to realization. In this case, a cell penetrable dominant negative form (that shares the leucine zipper of ATF5) blocks ATF5 function. It is well-documented that in the absence of neoplasia, ATF5 function is cell or tissue type dependent. By contrast, ATF5 appears to govern a battery of the same prosurvival proteins across a broad spectrum of rare and more common cancers. Finally, these pioneering efforts opens the doors of targeting ATF5 by different therapeutic strategies.
Acknowledgments
Source of funding:
NIH- R01NS083795 (JMA)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greene LA, et al. The transcription factor ATF5: role in neurodevelopment and neural tumors. J Neurochem. 2009;108(1):11–22. doi: 10.1111/j.1471-4159.2008.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu DX, et al. p300-Dependent ATF5 Acetylation Is Essential for Egr-1 Gene Activation and Cell Proliferation and Survival. Molecular and cellular biology. 2011;31(18):3906–16. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dluzen D, et al. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. The Journal of biological chemistry. 2011;286(9):7705–13. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng Z, et al. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16(6):671–7. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karpel-Massler G, et al. A synthetic cell-penetrating dominant-negative ATF5 peptide exerts anti-cancer activity against a broad spectrum of treatment resistant cancers. Clin Cancer Res. 2016;22(18):4698–4711. doi: 10.1158/1078-0432.CCR-15-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, et al. Nucleophosmin (NPM1/B23) interacts with activating transcription factor 5 (ATF5) protein and promotes proteasome- and caspase-dependent ATF5 degradation in hepatocellular carcinoma cells. The Journal of biological chemistry. 2012;287(23):19599–609. doi: 10.1074/jbc.M112.363622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pei Z, et al. Transcriptomic and functional pathways analysis of ascorbate-induced cytotoxicity and resistance of Burkitt lymphoma. Oncotarget. 2016;7(39):63950–63959. doi: 10.18632/oncotarget.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moll JR, et al. Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J Biol Chem. 2000;275(44):34826–32. doi: 10.1074/jbc.M004545200. [DOI] [PubMed] [Google Scholar]
- 9.Chen A, et al. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2012;31(6):532–7. doi: 10.1097/PGP.0b013e31824df26b. [DOI] [PubMed] [Google Scholar]
- 10.Hu M, et al. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer research. 2012;32(10):4385–94. [PubMed] [Google Scholar]
- 11.Arias A, et al. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene. 2012;31(6):739–51. doi: 10.1038/onc.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cates CC, et al. Regression/Eradication of gliomas in mice by a systemically-deliverable ATF5 dominant-negative peptide. Oncotarget. 2016;15(11):12718–12730. doi: 10.18632/oncotarget.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]