Abstract
Correct positioning of dendritic cells (DCs) is critical for efficient pathogen encounter and antigen presentation. Epstein-Barr virus–induced gene 2 (EBI2) has been identified as a chemoattractant receptor required for naïve CD4+DCIR2+ DC positioning in response to 7α,25-hydroxycholesterol (7α,25-HC). We now provide evidence that a second EBI2 ligand, 7α,27-HC, is involved in splenic DCIR2+ DC positioning and homeostasis. Cyp27a1, the enzyme uniquely required for 7α,27-HC synthesis, is expressed by stromal cells in the region of naïve DC localization. After activation, DCIR2+ DCs move into the T cell zone. We find that EBI2 is rapidly up-regulated in DCIR2+ DCs under certain activation conditions, and positioning at the B-T zone interface depends on EBI2. Under conditions of type I interferon induction, EBI2 ligand levels are elevated, causing activated DCIR2+ DCs to disperse throughout the T zone. Last, we provide evidence that oxysterol metabolism by Batf3-dependent DCs is important for EBI2-dependent positioning of activated DCIR2+DCs. This work indicates that 7α,27-HC functions as a guidance cue in vivo and reveals a multitiered role for EBI2 in DC positioning. Deficiency in this organizing system results in defective CD4+ T cell responses.
Introduction
Dendritic cells (DCs) play crucial roles in presenting antigens to T cells within lymphoid organs. In the spleen, classical DCs can be divided into two major subsets: cells that express CD4, CD11b, and DCIR2 and are dependent on interferon (IFN) regulatory factor 4 (IRF4) for their development (cDC2), and cells that express CD8α, DEC205, and XCR1 and require basic leucine zipper transcription factor activating transcription factor–like 3 (Batf3) for their development (cDC1) (1). The former DCs are important for presenting antigens to CD4+ T cells, whereas the latter are often crucial for cross-presentation of antigens to CD8+ T cells; however, both types of DCs can contribute to CD4+ and CD8+ T cell responses, depending on the type of antigen (1).
Within the spleen, DCIR2+ cDC2s are enriched in the blood-exposed marginal zone (MZ) bridging channels that extend between the T cell zone and the red pulp (RP) (2–4). XCR1+cDC1s are present in the MZ, RP, and T cell zone (5–9). After exposure to activating stimuli such as sheep red blood cells (SRBCs), lipopolysaccharide (LPS), or the double-stranded RNA mimetic polyinosinic:polycytidylic acid (poly I:C), splenic DCs move rapidly into the splenic T cell zone and, in some cases, position preferentially along the B-T zone interface (3, 10–16). Positioning at the B-T zone interface likely increases the amount of encounter with activated CD4+ T cells because they also favor this location (17, 18). DC movement into the T cell zone involves CCR7 up-regulation (3, 9, 11, 19). However, the factors that allow activated DCs to distinguish between the outer and the inner T cell zone are not defined.
Epstein-Barr virus–induced gene 2 (EBI2; also known as GPR183) is a Gαi-coupled chemoattractant receptor that is highly expressed by lymphocytes and DCs (20, 21). EBI2 in B cells functions together with CCR7 to distribute antigen-activated B cells along the B-T zone interface in lymphoid organs (22–24). After B cells have received T cell help and begun down-regulating CCR7, they position to inter- and outer-follicular regions in an EBI2-dependent manner (22–24). EBI2 function in B and T cells is important for mounting T cell–dependent antibody responses (18, 22, 23).
Using biochemical purification procedures, EBI2 ligands were identified as dihydroxylated forms of cholesterol (25, 26). In vitro studies established that 7α,25-HC was a potent EBI2 ligand. 7α,27-HC also had ligand activity but was about 10-fold less potent, whereas 25-HC and 27-HC had minimal activity (25, 26). 7α,25-HC is synthesized from cholesterol by the stepwise action of the enzymes Ch25h and Cyp7b1, whereas 7α,27-HC synthesis requires Cyp27a1 and Cyp7b1 (27). The enzyme Hsd3b7 metabolizes 7α,25-HC and 7α,27-HC into products that lack EBI2 ligand activity (28). In vivo studies established that mice lacking Ch25h suffered from defects in B and T cell positioning and antibody responses similar to mice lacking EBI2 (18, 28), suggesting that 7α,25-HC may be the sole ligand acting on EBI2 in lymphoid tissues.
In earlier studies, we and others established that EBI2 was critical for MZ bridging channel positioning of CD4+DCIR2+ DCs and for their homeostasis, with EBI2-deficient mice having about fourfold less of these splenic cells (3, 4). The defective DC homeostasis reflected a reduced ability of EBI2-deficient cells to access LTα1β2 on B cells. Ch25h- and Cyp7b1-deficient mice also showed defects in these DCs, establishing a critical role for 7α,25-HC in DC maintenance (3, 4).
When we further characterized Ch25h-deficient mice, we noted that the defects in their splenic DCIR2+ cDC2 compartment were less severe than those in EBI2-deficient mice. Specifically, we found that Ch25h-deficient mice retained more CD4+DCIR2+ DCs than EBI2-deficient mice and that the disruption in bridging channel positioning of these DCs was less complete. This led us to find that Cyp27a1 also contributes to the EBI2-dependent maintenance of splenic cDC2s. We show that both Cyp27a1 and Ch25h are expressed in MZ bridging channels, and we propose that these enzymes act cooperatively to control naïve (sentinel) DC positioning. We also found that cDC2s up-regulate EBI2 after activation and establish that their positioning in the splenic outer T cell zone depends on EBI2 and Ch25h. We show that EBI2 function in DCs after activation is suppressed by type I IFN, suggesting a mechanism whereby different innate stimuli lead to different T cell activating roles for DCs. Last, we provide evidence for cross-talk between DC subsets by showing that Batf3-dependent DCs promote outer T cell zone positioning of DCIR2+ DCs in a manner that depends on their expression of the oxysterol-metabolizing enzyme Hsd3b7.
Results
Cyp27a1 functions in positioning and homeostasis of DCIR2+ DCs
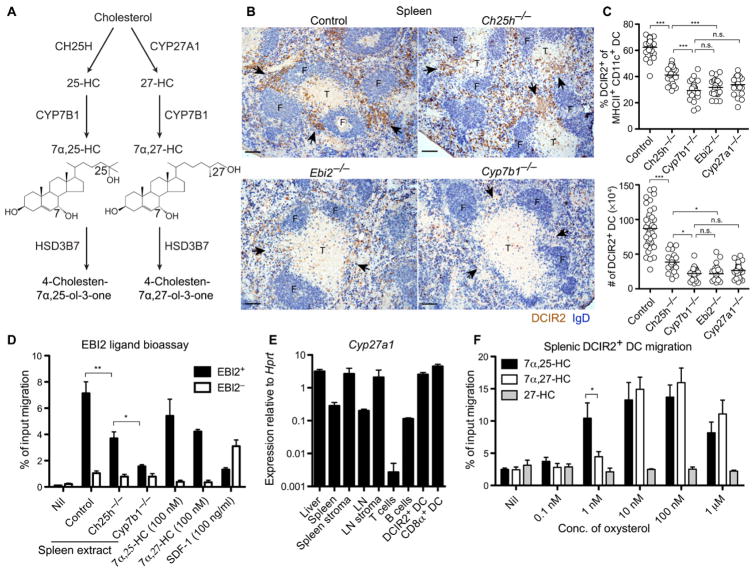
In the course of testing the enzyme requirements for EBI2 ligand production (Fig. 1A), we noticed that the defect in splenic DCIR2+ DC positioning in Ch25h-deficient mice was less severe than that in EBI2-deficient mice. Immunohistochemical staining of tissue sections revealed that Ch25h-deficient mice retained small clusters of DCIR2+ DCs in MZ bridging channels, whereas these were almost undetectable in EBI2- and Cyp7b1-deficient mice (Fig. 1B and fig. S1). Enumeration of splenic DCIR2+ cDCs by flow cytometry showed that their deficiency was less severe in Ch25h-deficient mice than in EBI2- and Cyp7b1-deficient mice (Fig. 1C). We used the DCIR2 marker in these studies because it uniquely allows identification of cDC2s in tissue sections and is expressed by most of the splenic cDC2s. Similar numerical deficiencies were obtained when we examined cDC2 frequencies using CD4, endothelial cell-selective adhesion molecule (ESAM), signal regulatory protein α (SIRPα), or CD11b (fig. S2). A comparison of spleen extracts for EBI2 ligand bioactivity using a subnanomolar-sensitive assay [(24); fig. S3A] revealed that Ch25h-deficient mice retained detectable EBI2 ligand, whereas ligand was almost undetectable in Cyp7b1-deficient mice (Fig. 1D). These observations and the in vitro evidence that 7α,27-HC has a potency on EBI2 within 10-fold of 7α,25-HC [(25); fig. S3A] led us to consider the possibility that Cyp27a1 acts as a second enzyme upstream of Cyp7b1 to generate EBI2 ligand activity in the spleen. Quantitative reverse transcription polymerase chain reaction analysis revealed that Cyp27a1 was highly expressed in the spleen and splenic stroma and also within the DCs themselves and was much less expressed in lymphocytes (Fig. 1E). In vitro migration analysis demonstrated that 7α,27-HC, although less potent than 7α,25-HC, was an efficacious attractant of DCs in the low nanomolar range (Fig. 1F). 7α,27-HC appeared to retain more activity at high (100 nM) concentrations than that observed for 7α,25-HC. This might reflect differences in the extent of receptor desensitization because 100 nM 7α,27-HC caused less internalization of EBI2 than 100 nM 7α,25-HC, although both ligands caused similar down-modulation at micromolar concentrations (fig. S3B).
Figure 1. Influences of Ch25h and Cyp27a1 on splenic DCIR2+ DCs.
(A) Pathway of 7α,25-HC and 7α,27-HC biosynthesis from cholesterol and metabolism by Hsd3b7. (B) Positioning of DCIR2+ DCs (brown) and B cells [immunoglobulin D (IgD), blue] in spleen sections from mice of the indicated genotype. Control indicates Ch25h+/−. Arrowheads indicate the location of MZ bridging channels. F, follicle; T, T cell zone. (C) Summary data for DCIR2+DC frequencies and numbers in mice of the indicated genotypes. Control indicates pooled data from Ch25h+/− and Cyp27a1+/− mice. (D) EBI2 ligand bioassay of spleen extracts from the indicated animals using a reporter cell line (M12) transduced with EBI2. Control indicates Ch25h+/− and Cyp27a1+/−mice. SDF-1, stromal cell-derived factor–1. Nil indicates migration media alone. (E) Cyp27a1 mRNA expression in tissue and cell subsets shown relative to Hprt. LN refers to mesenteric LN. (F) Migration of DCIR2+ DCs toward the indicated oxysterols in Transwell assays. Conc., concentration. Data in (B) to (E) are representative of at least two independent experiments with at least three mice per condition. Migration assay (F) contains data from three independent experiments. Scale bars, 100 μm. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant (P > 0.05) by one-way ANOVA with Bonferroni’s post hoc test for indicated comparisons.
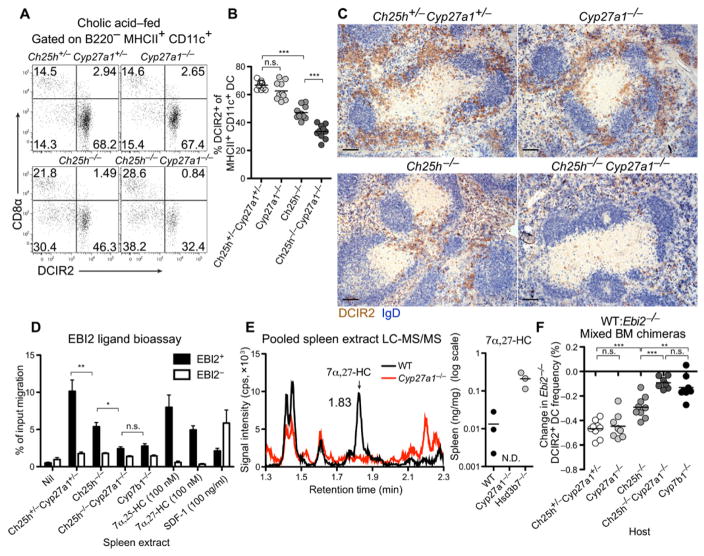
In initial experiments with Cyp27a1-deficient mice, we observed that they have a marked deficiency of DCIR2+ DCs (Fig. 1C). Unexpectedly, this deficiency exceeded that observed in Ch25h-deficient mice. Cyp27a1 has an important role in bile acid precursor synthesis in the liver, and mice lacking this enzyme have elevated plasma cholesterol and triglyceride concentrations (27). This is a consequence of elevated sterol response element–binding protein (SREBP-1 and SREBP-2) activity and reduced nuclear hormone receptor (farnesoid X receptor) activity in these mice because of their diminished bile acid pool and reduced absorption of dietary cholesterol (27). Previous work has shown that these phenotypes can be corrected by maintaining mice on a cholic acid–containing diet (29). When Cyp27a1 knockout (KO) mice were placed on this diet, their DCIR2+ DC frequencies became similar to those of controls (Fig. 2, A and B). One explanation for these observations is that Cyp27a1-deficient mice overproduce 7α,25-HC (30), and this disrupts the 7α,25-HC gradient needed for DC maintenance. Consistent with this notion, we found that EBI2 ligand bioactivity was elevated in spleen extracts from Cyp27a1 KO mice, but bioactivity was normal in spleen extracts from Cyp27a1 KO mice placed on a cholic acid–containing diet (fig. S3C). Similarly, bioactivity was present in plasma of Cyp27a1 KO mice on regular diet but was undetectable in plasma of mice maintained on a cholic acid–containing diet (fig. S3D).
Figure 2. Cyp27a1 Ch25h DKO mice reveal a role for 7α,27-HC in DCIR2+ DC positioning and maintenance.
Adult mice were fed 0.5% cholic acid diet for at least 2 weeks. (A to C) Flow cytometry plot (A), summary DCIR2+ DC frequency data (B), and positioning of DCIR2+ DCs (brown) and B cells (IgD, blue) in spleen sections (C) from the indicated mice. (D) EBI2 ligand bioassay of spleen extracts from the indicated mice. (E) LC-MS/MS detection of 7α,27-HC in spleen extracts pooled from 10 to 12 mice for WT and Cyp27a1−/− or from single Hsd3b7−/− mice per measurement. Left: Representative chromatogram (N.D., not detected; n = 2 sets of pooled mice for Cyp27a1−/−). cps, counts per second. (F) Percent change in Ebi2−/− DCIR2+ DCs in mice reconstituted with an equal mixture of WT and Ebi2−/− BM, calculated by comparing the chimerism of DCIR2+ DCs against CD8α+ DCs. Data in (A) to (D) and (F) are representative of at least two independent experiments with at least three mice per group. Scale bars, 100 μm. *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant (P > 0.05) by one-way ANOVA with Bonferroni’s post hoc test for indicated comparisons.
With these observations in mind, all further studies with Cyp27a1-deficient mice were performed with animals maintained on a cholic acid–containing diet. We speculated that the contribution of Cyp27a1 to EBI2-mediated DC positioning and homeostasis may normally be subdominant to the contribution of Ch25h. DCIR2+ DC frequencies in Ch25h and Cyp27a1 double-knockout (DKO) mice were reduced more substantially than in Ch25h-deficient mice and, to a similar extent, in EBI2- and Cyp7b1-deficient mice (Figs. 2, A and B, and 1C). This reduction in DCs was not due to selective loss of the DCIR2 marker because analysis of additional markers of splenic cDC2s (CD4, SIRPα, ESAM, and CD11b) revealed a similar deficiency in each case (fig. S2). Analysis of spleen sections revealed that the bridging channel DCIR2+ DC clusters detectable in Ch25h KO mice were no longer detectable in mice that also lacked Cyp27a1 (Fig. 2C and fig. S1). Instead, the remaining DCs were scattered in the RP as observed for EBI2-deficient mice (Fig. 2C and fig. S1). Consistent with these observations, spleen extracts from Ch25h and Cyp27a1 DKO mice had minimal EBI2 ligand bioactivity (Fig. 2D). Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of pooled soluble spleen extracts from multiple wild-type (WT) mice revealed that 7α,27-HC was present at a concentration of ~0.013 ng/mg tissue (~30 nM), whereas it was undetectable in Cyp27a1 KO spleen extracts (Fig. 2E). Because the splenocytes were removed without cell lysis during generation of these extracts, this measurement approximates what is found in the interstitial fluid. It is likely that local concentrations in tissue subregions are considerably higher. We also confirmed the importance of Hsd3b7 in metabolizing 7α,27-HC by observing that Hsd3b7 deficiency caused a marked increase in this oxysterol in the spleen (Fig. 2E).
To test whether the Ch25h and Cyp27a1 DKO mice retained any EBI2-dependent activity in vivo, we reconstituted irradiated DKO and control mice with an equal mixture of WT and EBI2-deficient bone marrow (BM). Upon reconstitution, we observed the expected twofold outcompetition of EBI2 KO DCs by WT DCs in control hosts (Fig. 2F). In Ch25h-deficient hosts, the WT DCs were deficient but still outcompeted the EBI2 KO DCs (Fig. 2F). However, in Ch25h and Cyp27a1 DKO hosts, the WT and EBI2 KO DCs were almost equally compromised, similar to the findings in Cyb7b1-deficient hosts (Fig. 2F). These data demonstrate that both Ch25h and Cyp27a1 contribute to the generation of EBI2 ligand activity controlling DCIR2+ DC homeostasis. These data also show that Cyp27a1 was required in radiation-resistant (stromal) cells and not intrinsically in the DCs to maintain DCIR2+ DC homeostasis (Fig. 2F).
Analysis of mesenteric lymph nodes (LNs) in cholic acid-fed Cyp27a1-deficient mice showed that migratory and resident DCs were present at normal frequencies and that DCIR2+ DCs were correctly distributed (fig. S4, A and B). Ch25h-deficient mice showed a reduction in DCIR2+ migratory (MHCIIhiCD11c+ CD11b+) and resident (MHCIIintCD11chi CD11b+) DCs that was comparable to the deficiency observed in EBI2-deficient mice (fig. S4, A and B). Ch25h and Cyp27a1 DKO mice showed the same deficiency observed in Ch25h KO mice (fig. S4, A and B) suggesting that Cyp27a1 may not be required for DC homeostasis in LNs. This was further established using the mixed BM chimera approach because the EBI2-deficient DCs were underrepresented to the same extent in WT and Cyp27a1 KO hosts (fig. S4C). Moreover, WT and EBI2-deficient cells were equally deficient in Ch25h KO hosts and further removing Cyp27a1 had no additional effect (fig. S4C). In tissue sections, DCIR2+ DCs were most concentrated in regions of the T cell zone near B cell follicles; this distribution was disrupted in EBI2-deficient and Ch25h-deficient mice but appeared unaffected in Cyp27a1-deficient mice (fig. S4D). Ch25h and Cyp27a1 DKO mice showed a similar disruption to that observed in Ch25h-deficient mice (fig. S4D). Thus, in contrast to the spleen, Cyp27a1 does not appear to be required for homeostasis of DCIR2+ DCs in mesenteric LNs despite the relatively high Cyp27a1 mRNA expression level in this tissue (Fig. 1E).
Patterns of enzyme expression in the spleen
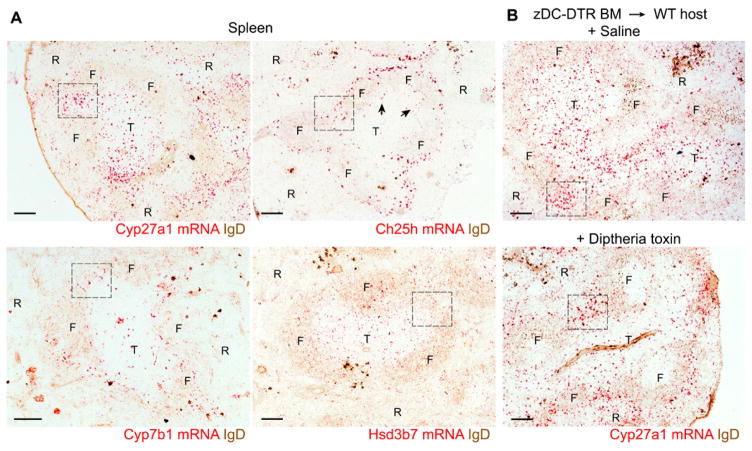
To understand how Cyp27a1 and Ch25h might work together to control DC positioning and homeostasis, it was critical to determine the pattern of enzyme expression. Attempts to stain for Cyp27a1 using rabbit antisera or detect transcript distribution using standard in situ hybridization techniques were unsuccessful. We therefore turned to the RNAscope procedure that uses multiple tagged DNA oligonucleotides and a secondary hybridization procedure to amplify the signal to detect mRNA distribution (31). This approach revealed that Cyp27a1 mRNA was particularly abundant in splenic MZ bridging channels and was also present in the T cell zone and at lower levels in B cell follicles (Fig. 3A, boxed regions highlight example bridging channels; see also fig. S5A). Because Cyp27a1 is expressed in DCs (Fig. 1E), we also examined diphtheria toxin (DT)–treated Zbtb46–DT receptor (DTR) (zDC-DTR) BM chimeric mice that lack DCs (fig. S5B). DC-deficient mice showed a pattern of Cyp27a1 expression in MZ bridging channels similar to saline-treated zDC-DTR mice, with considerably less signal in the T cell zone (Fig. 3B). These data suggest that Cyp27a1 is expressed by stromal cells in the MZ bridging channels but is expressed by both DCs and stromal cells in the T cell zone. Using the same method, we found that Ch25h was abundantly expressed by cells in the outer follicle and within interfollicular regions, including in the MZ bridging channel (Fig. 3A and fig. S5A). Ch25h expression was conspicuously low within B cell follicles, in accord with earlier laser capture microscopy analysis (28), and was also low within the T cell zone, although expression could be detected at the B-T zone interface (Fig. 3A, arrows). The specificity of the Ch25h probe was confirmed using tissue from Ch25h KO mice [fig. S3A; this control was not possible with Cyp27a KO mice because they retain most of the coding region (32) and express the mutated transcript]. The Ch25h mRNA detected by RNAscope was restricted to radiation-resistant cells because we observed no difference in the hybridization pattern in spleen tissue from mice reconstituted with Ch25h KO BM (fig. S5C). Cyp7b1 was more widely distributed than either Cyp27a1 or Ch25h, and expression could be detected in the MZ bridging channels, the T cell zone, and follicles (Fig. 3A and fig. S5A). The oxysterol-metabolizing enzyme, Hsd3b7, was particularly widely expressed (Fig. 3A and fig. S5A), and this included considerable expression by hematopoietic cells in the T cell zone (fig. S5C), most likely in DCs (28). RNAscope analysis of liver sections confirmed the very high expression of Cyp27a1, Cyp7b1, and Hsd3b7 and minimal expression of Ch25h in this bile-producing tissue (fig. S5D). The liver showed the expected sexual dimorphism in Cyp7b1 expression (33), a difference that is not observed in lymphoid tissues (fig. S5D). The presence of both Cyp27a1 and Ch25h in MZ bridging channels is consistent with these enzymes acting cooperatively to generate EBI2 ligand in this region of the spleen.
Figure 3. Expression pattern of Cyp27a1, Ch25h, Cyp7b1, and Hsd3b7 transcripts in the spleen determined using RNAscope.
(A) Distribution of Cyp27a1, Ch25h, Cyp7b1, and Hsd3b7 mRNA (red) in spleen sections co-stained for IgD (brown). Dashed box indicates staining within MZ bridging channel areas, and arrowheads highlight Ch25h signal at the B-T zone interface. T, T cell zone; F, follicle; R, red pulp. (B) Distribution of Cyp27a1 mRNA in spleen sections of zDC-DTR chimeric mice treated with either saline (top) or DT (bottom). Images are representative of at least five (A) or three (B) mice per condition. Scale bars, 100 μm.
EBI2 is required for positioning activated DCs in the outer T cell zone
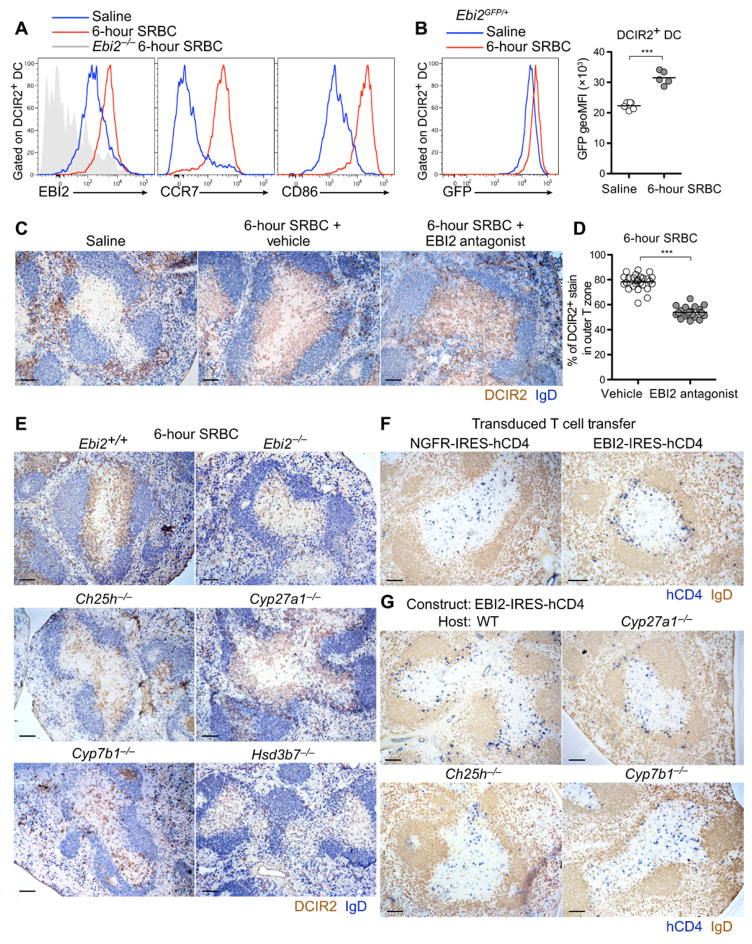
After activation, splenic DCIR2+ DCs rapidly relocalize from the MZ bridging channels, MZ, and RP and move into the T cell zone. In response to SRBC immunization and activation by missing self-recognition, the cells preferentially position in the outer T cell zone (3, 19). It seemed unlikely that EBI2 would have a role in this behavior, given its requirement for cell positioning in the bridging channels. However, studies in other contexts have shown that small shifts in responsiveness to competing cues can have major influences on cell localization (34). We therefore tested for changes in EBI2 expression in DCIR2+ DCs after SRBC-induced activation. Within 6 hours of activation, the cells not only had strongly up-regulated CCR7 and CD86 but also had increased their surface EBI2 expression (Fig. 4A). Analysis of green fluorescent protein (GFP) levels in immunized EBI2-GFP reporter mice established that this reflected an increase in transcript abundance in the cells (Fig. 4B). To test whether EBI2 was required for outer T cell zone positioning while avoiding the effects of chronic EBI2 deficiency on DC numbers, we took advantage of a recently described EBI2 antagonist (35). We first confirmed that the inhibitor was effective at inhibiting DC migration to EBI2 ligands in vitro while not affecting migration to control chemoattractants (fig. S6, A and B). Antagonist treatment resulted in a complete disruption in DCIR2+ DC outer T cell zone preference (Fig. 4C), a defect that was confirmed by quantitating the fraction of DCIR2 staining in the outer T cell zone across multiple sections (Fig. 4D and fig. S6C). This observation led us to reanalyze the distribution of the DCs remaining in EBI2-deficient mice; in the cases where the T cell zone contained sufficient DCs for comparative analysis, the KO cells were uniformly distributed throughout the T cell zone (Fig. 4E). Activated DCIR2+ DCs also showed little preference for the B-T zone interface in Ch25h-deficient mice, although, in this case, they were sometimes clustered within the T cell zone rather than being randomly dispersed (Fig. 4E). By contrast, in Cyp27a1-deficient mice, the activated DCs appeared to position normally to the B-T zone interface (Fig. 4E). In Cyp7b1- and Hsd3b7-deficient mice, the activated DCs were distributed uniformly throughout the T cell zone (Fig. 4E).
Figure 4. Activated DCIR2+ DCs up-regulate EBI2 and position at the B-T zone interface in an EBI2- and Ch25h-dependent manner.
(A and B) Surface expression of EBI2, CCR7, and CD86 in DCIR2+ DCs (A) and GFP expression in Ebi2gfp/+ DCIR2+ DCs (B) from mice 6 hours after intravenous immunization with saline (blue) or SRBCs (red). geoMFI, geometric mean fluorescence intensity. (C) Positioning of DCIR2+ DCs (brown) relative to B cells (blue) in mice 6 hours after treatment with saline, SRBCs plus vehicle (N-methyl-2-pyrrolidone), or SRBCs plus EBI2 antagonist (NIBR189). (D) Percentage of DCIR2+ DC staining in the outer T cell zone in the spleens of the type in (C). Each point represents a white pulp cord. (E) Positioning of DCIR2+ DCs in mice of the indicated genotypes 6 hours after SRBC immunization. (F and G) Positioning of activated T cells overexpressing EBI2 or a control receptor [truncated nerve growth factor receptor (NGFR)] 24 hours after transfer into WT mice (F) or mice of the indicated genotype (G). Sections were stained to detect transduced T cells (hCD4, blue) and B cells (IgD, brown). Data are representative of at least two independent experiments and at least four mice analyzed per condition. Scale bars, 100 μm. ***P < 0.001 by unpaired Student’s t test.
To determine whether EBI2 up-regulation on T cell zone tropic cells was sufficient to promote their positioning in the outer T cell zone, we examined the distribution of transferred T cells overexpressing EBI2. Flow cytometric analysis confirmed that EBI2 transduction led to higher EBI2 expression on the reporter-expressing cells than on the matched vector-transduced cells, and the EBI2hi cells responded more robustly to 7α,25-HC (fig. S6, D to F). The EBI2hi T cells showed a preference to position in the outer T cell zone, whereas control vector-transduced cells were distributed throughout the T cell zone (Fig. 4F). Consistent with the findings for activated DCIR2+ DCs, EBI2hi T cells continued to position in the outer T cell zone in Cyp27a1-deficient hosts but largely failed to do so in Ch25h-deficient hosts (Fig. 4G). In mice lacking Cyp7b1, the T cells were uniformly distributed throughout the T cell zone (Fig. 4G). These data indicate that Ch25h and Cyp7b1 contribute to EBI2 ligand production in the splenic outer T cell zone.
DC outer T zone positioning enhances T follicular helper cell responses
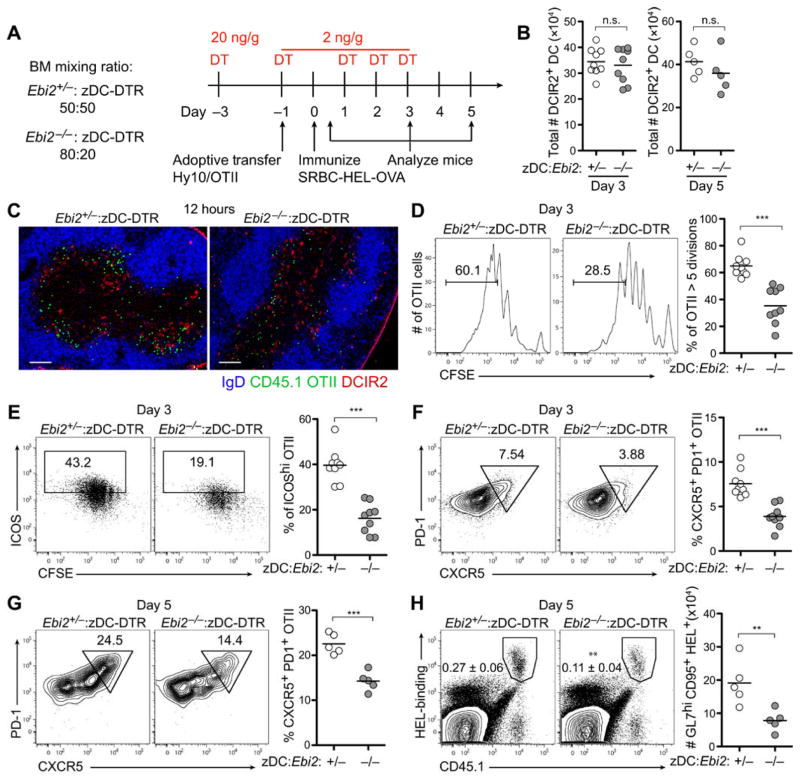
To test the importance of outer T cell zone positioning of DCs on the T cell response, we generated chimeric mice that had similar numbers of DCIR2+ DCs but where the DCs were either EBI2 HET (control) or KO. This was achieved by reconstituting mice with either a 50:50 ratio of EBI2 HET and zDC-DTR BM or an 80:20 ratio of EBI2 KO and zDC-DTR BM (the higher ratio is needed to compensate for the deficiency in EBI2 KO DCs) and then treating mice with DT (Fig. 5A and fig. S7A). After DT treatment, the two types of chimeric mice were found to have well-matched numbers of DCs (Fig. 5B). Carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled OTII T cells were transferred into these mice, and the animals were immunized with SRBC–ovalbumin (OVA). Analysis after 12 hours confirmed that the control and EBI2 KO DCIR2+ DCs became equally activated (fig. S7B). At this time point, WT DCIR2+DCs were colocalized with OTII cells in the outer T cell zone, whereas EBI2 KO DCIR2+ DCs were only partly overlapping with OTII T cells because the DCs remained uniformly distributed in the T cell zone (Fig. 5C). After 3 days, the OTII T cells in hosts that had EBI2 KO DCs were deficient in proliferation, up-regulation of the activation marker inducible costimulator (ICOS), and acquisition of T follicular helper (Tfh) cell markers CXCR5 and programmed cell death protein 1 (PD-1) (Fig. 5, D to F). In an adoptive cotransfer experiment using hen egg lysozyme (HEL)–specific Hy10 B cells and OTII T cells and immunization with SRBC-HEL-OVA, Tfh cell induction was again defective (Fig. 5G) and Hy10 cells were less able to give rise to germinal center B cells in mice that lacked EBI2 on DCs (Fig. 5H). These data support the conclusion that EBI2-dependent positioning of DCIR2+ DCs in the outer T cell zone augments their ability to support Tfh cell responses. The data do not exclude the possibility that EBI2 signaling in DCs may act in additional ways to promote Tfh cell responses.
Figure 5. EBI2-mediated DCIR2+ DC positioning in the outer T cell zone augments induction of Tfh cells.
(A) Time line of DT treatment, cell transfer, immunization, and analysis. OVA-specific OTII T cells were transferred alone (C to F) or together with HEL-specific Hy10 B cells (G and H) into mice containing matched frequencies of control (Ebi2+/−) or Ebi2−/− DCIR2+ DCs (B) Total DCIR2+ DC numbers in mice 3 or 5 days after immunization. (C) Immunofluorescence images of spleen sections showing distribution of CD45.1+ OTII T cells (green), DCIR2+ DCs (red), and B cells (IgD, blue) 12 hours after immunization. (D) Proliferation of OTII cells 3 days after immunization determined by CFSE dilution. (E and F) Up-regulation of ICOS (E) and acquisition of PD-1 and CXCR5 (F) by OTII cells 3 days after immunization. (G) Acquisition of PD-1 and CXCR5 by OTII cells after cotransfer with Hy10 B cells 5 days after immunization. (H) Frequency of total HEL-binding Hy10 cells (left plots) and total number of GL7hiFas+ HEL-binding B cells (graph on right) from the mice in (G). Images in (C) are representative of three mice, and data in (B), (D), (E), and (F) are representative of two independent experiments with at least three mice per group. Data in (G) and (H) are obtained from one experiment with five mice per condition. Scale bars, 100 μm. **P < 0.01; ***P < 0.001; n.s., not significant (P > 0.05) by unpaired Student’s t test.
Type I IFN signaling overrides DC outer T cell zone positioning
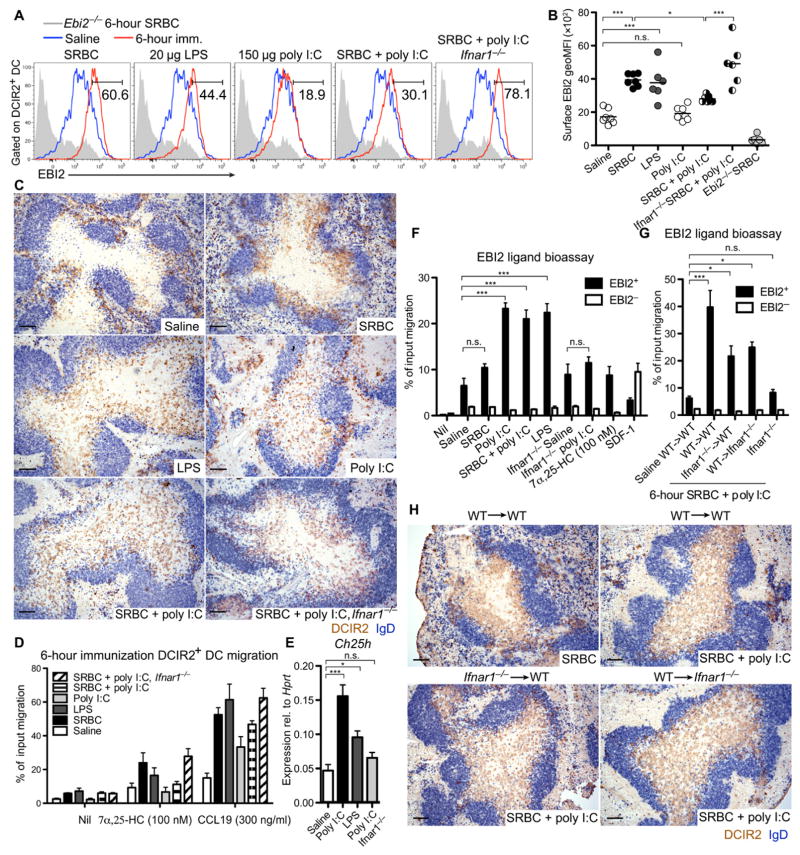
DCs also redistribute into the T cell zone after LPS or poly I:C treatment (10–13, 15), although whether these stimuli cause the cells to favor the outer T cell zone has been unclear. Analysis of EBI2 expression on DCIR2+ DCs after exposure to these activating stimuli revealed that LPS induced similar expression to SRBCs, whereas poly I:C induced inferior expression of both EBI2 and CCR7, despite up-regulation of CD86 to similar levels (Fig. 6, A and B, and fig. S8A). In accord with the differences in EBI2 surface expression, LPS caused DCIR2+ DCs to favor the B-T zone interface, whereas poly I:C did not (Fig. 6C and fig. S8B). In vitro migration assays confirmed that there was a close correspondence between the EBI2 surface levels and the ability of the DCIR2+ DCs to respond to 7α,25-HC (Fig. 6D). These data led us to examine whether poly I:C could have a suppressing effect on EBI2 function in DCIR2+ DCs. Analysis of mice treated with a mixture of SRBCs and poly I:C showed that, although DCIR2+ DCs were able to up-regulate CCR7 and CD86 (fig. S8A), there was an impaired surface expression of EBI2 (Fig. 6, A and B) and the DCs failed to position at the B-T zone interface (Fig. 6C and fig. S8B). Analysis of EBI2-GFP reporter expression showed that poly I:C did not suppress EBI2 at the transcriptional level, suggesting that the receptor was being posttranscriptionally regulated (fig. S8C). This observation brought our attention to the type I IFN inducibility of Ch25h (21). Ch25h transcripts were up-regulated in spleen tissue of poly I:C–treated mice (Fig. 6E), and there was a corresponding increase in EBI2 ligand bioactivity (Fig. 6F). The increase in bioactivity after poly I:C treatment was lost in IFN-α receptor (IFNAR) KO mice (Fig. 6F), and the inhibition of EBI2 surface expression and outer T cell zone positioning was rescued in these mice, establishing that these effects were mediated by type I IFN (Fig. 6, A to C, and fig. S8, A and B). Consistent with type I IFN–inducing Ch25h expression, induction of EBI2 ligand bioactivity by poly I:C was lost in Ch25h KO mice (fig. S8D). Immunization of IFNAR KO→WT and WT→IFNAR KO BM chimeras showed that IFN was acting on both radiation-sensitive (hematopoietic) and radiation-resistant (stromal) cells to promote EBI2 ligand production (Fig. 6G). In accord with this observation, IFNAR expression on either hematopoietic cells or stromal cells was sufficient to prevent DCIR2+ DC positioning in the outer T zone (Fig. 6H). In contrast to the findings for EBI2 ligand production, the modulating influence of poly I:C on surface EBI2 levels was dependent on IFNAR expression in hematopoietic cells (fig. S8E), and mixed BM chimeras showed that this dependency reflected a DC-intrinsic effect (fig. S8F). Analysis of BM chimeric mice in which DCs were selectively IFNAR-deficient also showed the recovery in surface EBI2 levels (fig. S8G). However, poly I:C treatment continued to cause dispersal of DCIR2+ DCs throughout the T cell zone in these mice (fig. S8H). Spleen extract bioactivity analysis showed that poly I:C caused elevations in EBI2 ligand levels in mice lacking IFNAR from DCs (fig. S8I). Together, these data indicate that type I IFN suppresses DCIR2+ DC positioning in the outer T zone and suggest that this effect occurs, at least in part, because of disruption of EBI2 ligand gradients as a result of increased 7α,25-HC production by both hematopoietic and nonhematopoietic cells.
Figure 6. Type I IFN suppresses EBI2 up-regulation and disrupts outer T cell zone positioning of DCIR2+ DCs.
(A) Surface EBI2 levels on DCIR2+ DCs in WT or Ifnar1−/− mice 6 hours after treatment with the indicated stimuli (red), plotted alongside the same saline-treated (blue) or Ebi2−/− (gray) control. Numbers indicate gate for the immunized (imm.) condition. (B) Summary MFI data from data of the type in (A). (C) Distribution of DCIR2+ DCs (brown) relative to B cells (blue) in the spleen of WT or Ifnar1−/− mice at 6 hours after immunization with the indicated stimuli. (D) Transwell migration of DCIR2+ DCs from WT or Ifnar1−/−mice 6 hours after treatment with the indicated stimuli toward 7α,25-HC or CCL19. (E) Relative expression of Ch25h in total spleen tissue 6 hours after the indicated immunization. (F and G) EBI2 ligand bioassay of spleen extracts from WT and Ifnar1−/− mice (F) or from WT→WT, Ifnar1−/−→WT, WT→Ifnar1−/− chimeric mice, and Ifnar1−/− nonchimeric mice (G) immunized as indicated. (H) Distribution of DCIR2+ DCs (brown) in the spleens of mice in (G) 6 hours after immunization with the indicated stimuli. Data are representative of at least three mice per condition, and migration assay (D) contains data from two independent experiments. Scale bars, 100 μm. *P < 0.05; ***P < 0.001; n.s., not significant (P > 0.05) by one-way ANOVA with Bonferroni’s post hoc test for indicated comparisons.
We note that LPS treatment caused a more diffuse distribution of DCIR2+ DCs in the outer T cell zone than induction by SRBCs (Fig. 6C and fig. S8B). Similar to poly I:C, LPS caused an elevation of Ch25h mRNA and EBI2 ligand bioactivity in total spleen (Fig. 6, E and F). However, EBI2 surface levels appeared similar to those induced by SRBCs (Fig. 6, A and B). We rationalize that LPS treatment may lead to a smaller increase in EBI2 ligand abundance specifically in the T cell zone compared with that occurring after poly I:C treatment, and this only partially disrupts the EBI2-mediated navigation of activated DCs to this region.
XCR1+ DCs help establish EBI2 ligand gradients used by DCIR2+ DCs
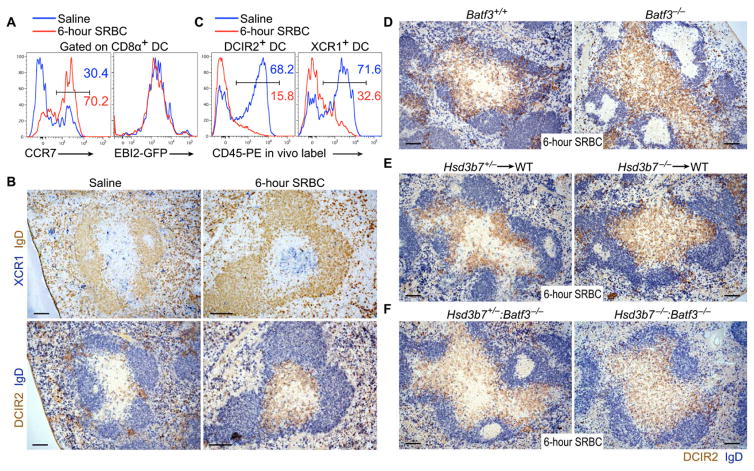
During our characterization of DCIR2+ DC positioning in response to the various stimuli, we observed that the T zone positioning of DCIR2+ DCs correlated with a distinct positioning pattern of activated Batf3-dependent XCR1+ DCs. XCR1 is expressed by ~85% of splenic CD8α+ DCs, and these cells are localized in the T cell zone, MZ, and RP in naïve mice (5–9). These DCs up-regulated CCR7 and CD86 but not EBI2 within 6 hours of SRBC immunization (Fig. 7A and fig. S9, A and B) and redistributed from MZ and RP locations into the central T cell zone (Fig. 7B and fig. S9C). In this location, the XCR1+ DCs were surrounded by DCIR2+DCs in the outer T cell zone (Fig. 7B and fig. S9C). In vivo pulse labeling of blood-exposed cells by intravenous injection of phycoerythrin (PE)–labeled antibody (36) showed that SRBC immunization caused redistribution of many DCIR2+ and XCR1+ DCs from a highly blood-exposed location (likely corresponding to the RP, MZ, and bridging channels) to a more secluded location, consistent with relocation into the white pulp (Fig. 7C). Redistribution of DCIR2+ and XCR1+ DCs into the white pulp could also be detected by in vivo CD45-PE labeling after immunization with LPS or poly I:C (fig. S9D).
Figure 7. Hsd3b7-expressing Batf3-dependent DCs are required for outer T cell zone positioning of activated DCIR2+ DCs.
(A) CCR7 and EBI2-GFP expression in CD8α+ DCs 6 hours after intravenous immunization with saline or SRBCs. (B) Serial spleen sections from saline or 6-hour SRBC-treated mice stained for XCR1 (upper, blue) or DCIR2 (lower, brown) and IgD (upper, brown; lower, blue). (C) In vivo PE labeling on DCIR2+(left) or XCR1+ (right) DCs in either saline-treated (blue line) or SRBC-treated (red line) mice injected with 1 μg of anti–CD45-PE for 5 min. (D to F) Positioning of DCIR2+ DCs in Batf3−/− and control mice (D), in mice lacking Hsd3b7 in BM-derived cells versus controls (E), or in Batf3−/−:Hsd3b7−/−mixed BM chimeric mice lacking Hsd3b7 in Batf3-dependent DCs versus controls (Batf3−/−:Hsd3b7+/− mixed BM chimeras) (F). Histograms and images are representative of at least three mice analyzed per condition. Scale bars, 100 μm.
The dichotomous relationship between XCR1+ and DCIR2+ DC positioning after activation, together with our earlier observation that CD8α+ DCs highly express the oxysterol-metabolizing enzyme Hsd3b7 [confirmed here for cells from naïve and SRBC-immunized mice (fig. S9E)] (28), led us to test whether XCR1+ DCs had a role in determining the distribution of DCIR2+ DCs. SRBC immunization of Batf3 KO mice, which are deficient in XCR1+ DCs (1), resulted in the inability of DCIR2+ DCs to position in the outer T cell zone (Fig. 7D and fig. S9F). Analysis of mice lacking Hsd3b7 specifically in hematopoietic cells revealed a defect in activated DCIR2+ DC positioning in the outer T cell zone (Fig. 7E), consistent with a requirement for Hsd3b7 in DCs. To test whether this requirement was within Batf3-dependent cells, we examined BM chimeric mice where only the Batf3-dependent DCs were fully Hsd3b7-deficient and also observed that activated DCIR2+ DCs failed to accumulate in the outer T cell zone (Fig. 7F). These observations provide evidence that XCR1+ DCs can promote EBI2 ligand turnover and thereby help establish EBI2 ligand gradients needed for activated DCIR2+ DC positioning. This property of XCR1+ DCs may be modulated under some conditions because poly I:C treatment down-regulated Hsd3b7 while up-regulating Ch25h and Cyp7b1 in these cells (fig. S9E).
Discussion
This study identifies a role for Cyp27a1 in DC positioning and homeostasis and provides evidence that this activity occurs through synthesis of a second EBI2 ligand, 7α,27-HC. Cyp27a1 is expressed in lymphoid stromal cells, and we propose that the enzyme functions together with Ch25h to generate paired gradients of 7α,27-HC and 7α,25-HC that act combinatorially to control naïve (sentinel) cDC2 positioning (fig. S10). We further demonstrate that EBI2 is up-regulated along with CCR7 in activated cDC2s, and the associated increase in responsiveness to EBI2 ligands is necessary for DC positioning in the outer T cell zone to promote efficient induction of Tfh cells (fig. S10). Type I IFN negatively regulates DC outer T cell zone positioning, an influence that is likely to alter the types of T cell responses that are favored (fig. S11). Last, we provide evidence that Batf3-dependent DCs can act as an EBI2 ligand sink, an activity that appears necessary for correct localization of cDC2s under some activation conditions, revealing population-level coordination of cDC1 and cDC2 distribution in lymphoid tissues (fig. S11).
Cyp27a1 is expressed in the T cell zone and at lower levels in follicles, as well as in splenic MZ bridging channels. Several factors beyond Cyp27a1 expression are likely at play to shape the 7α,27-HC gradient. Cyp7b1 is required downstream of Cyp27a1 for 7α,27-HC synthesis, although our previous work suggests that the two enzymes would not need to be expressed in the same cells (28). A key factor in determining local 7α,27-HC concentrations may be the density of Cyp27a1- and Cyp7b1-coexpressing cells, or the proximity of cells expressing the individual enzymes. It is also possible that the amounts of Cyp27a1 and Cyp7b1 or their enzymatic activities are modulated posttranscriptionally. Tissue-wide transcriptome analyses (e.g., Biogps.org) have shown that Cyp27a1 and Cyp7b1 are coexpressed in multiple tissues. 7α,27-HC may therefore be produced in a range of tissue types where the sterol could have actions, either together with or independently from 7α,25-HC, which influence the behavior of EBI2-expressing cells.
The expression pattern of Ch25h revealed by the RNAscope analyses agrees well with the distribution determined by less precise methods [laser capture microscopy and stromal cell sorting (28)] and demonstrates that this enzyme has a unique perifollicular expression pattern in the spleen. This pattern closely matches the established dependence of EBI2-mediated B cell positioning on Ch25h (20, 28) and is in accord with the EBI2 dependence of activated cDC2 positioning we describe here. The expression pattern in the outer follicle may correspond to marginal sinus–lining cells (37), although definitive statements will require the development of procedures for combining RNAscope with markers specific for these cells. The stromal cells expressing Ch25h at the B-T zone interface do not match a well-defined subset and may represent an uncharacterized subtype of fibroblastic reticular cell.
Our finding that EBI2 is needed for controlling distinct positioning events of naïve and activated cDC2s represents a new example of cell positioning that is determined by cooperative or balanced responsiveness to multiple cues (34). In naïve cDC2s where CCR7 expression is low, EBI2 likely guides the DCs to a position of high EBI2 ligand and low CCR7 ligand abundance (possibly cooperating with ligands for yet other receptors). In activated DCs, the increase in CCR7 makes the cells responsive to CCL21 and CCL19 that are abundant throughout the T cell zone, and EBI2 now cooperates with CCR7 to position the DCs in a zone where ligands for both receptors overlap, the outer T cell zone (fig. S10). The fact that DCIR2+DC positioning in this location augments CD4+ T cell responses is in agreement with the finding that activated CD4+ T cells undergo extensive interactions with DCs, preferentially in this region (17, 18).
SRBCs have been shown to cause cDC2 activation because of missing self-CD47 recognition (19). The physiologically analogous mode of cDC2 activation has not yet been determined, but it is speculated that any circulating endogenous cells with reduced CD47 may engage this pathway. The similar cDC2 distribution after LPS treatment indicates that multiple modes of activation are likely to cause EBI2 up-regulation and outer T zone preference by cDC2s. However, pathogens that also strongly induce type I IFN (as modeled here using poly I:C) may disrupt DC EBI2 function in a cell-extrinsic manner, likely because of increased EBI2 ligand production (and possibly reduced degradation) by various cell types in the inner T cell zone. Our data also showed that type I IFN had a DC-intrinsic posttranscriptional effect on EBI2 surface levels. Although further work will be needed to determine whether this reflects effects on EBI2 translation, internalization, or turnover, the findings suggest that EBI2 surface levels cannot be taken as a simple correlate of extracellular EBI2 ligand abundance. Marked changes in the expression of homeostatic lymphoid tissue organizers (the chemokines CCL21, CCL19, and CXCL13) have been reported in various infection models (37). It will be important in future studies to characterize how EBI2 expression and EBI2 ligand production are modulated in lymphoid tissues after various types of infection and to determine how this affects adaptive immune responses.
The finding that type I IFN antagonizes EBI2 function on activated cDC2s and favors their dispersal throughout the T cell zone raises questions about the different antigen-presenting roles of these DCs. Although they have been established to augment Tfh cell differentiation and antibody responses (1, 18, 38), they also support IFNγ-producing T cell responses (39, 40). Because type I IFN is most prominently induced during infections with intracellular pathogens, perhaps dispersal throughout the T cell zone allows the cDC2s to present antigen in a manner that more effectively induces a T helper 1 cell fate. In accord with this notion, cDC2s were shown to depend on interleukin-12 from cDC1s for induction of IFNγ-producing cells (40). By up-regulating Ch25h expression, type I IFN increases not only EBI2 ligand but also 25-HC abundance (21). Because 25-HC can have multiple actions on cells, including repression of cholesterol biosynthesis and antagonism of viral replication, it is possible that the actions of 7α,25-HC and 25-HC on DCs are coordinated to match the response to the type of invader.
Although some studies had suggested that CD8α+ DCs may mostly be in the splenic T cell zone, other observations showed that they were more widely distributed (15, 41, 42) and their presence in the RP and MZ, as well as in the T cell zone, was more clearly established using the XCR1 marker (5–9). These DCs move into the T cell zone after activation (8, 9, 42). Our finding that CD8α+ DCs up-regulate CCR7 but not EBI2 under the activation conditions tested provides a possible explanation for why they were not enriched in the outer T cell zone. We establish that Batf3-dependent DCs help exclude EBI2hi DCIR2+ DCs from the central T cell zone through their high expression of Hsd3b7 and presumed efficient metabolism of 7α,25-HC. We do not exclude the possibility that Hsd3b7-deficient Batf3-dependent DCs are altered in additional ways that affect T cell zone organization. Future work is also needed to determine how much the cDC1 influence on cDC2 positioning affects T cell responses to various immunogens. The cross-talk between DC populations that we describe here likely represents an example of a broader set of population-level interplays that help fine-tune tissue reorganization events during the immune response.
Materials and Methods
Study design
The aim of this study was to characterize the oxysterol requirements for EBI2-dependent positioning of naïve and activated DCs in lymphoid tissues and to assess how this positioning affects Tfh cell responses. Most of the experiments consisted of enumeration of population frequencies by flow cytometry, assessment of cell or enzyme distribution using immunohistochemistry, quantitation of EBI2 ligand abundance using bioassay, and testing Tfh cell responses via adoptive transfer of antigen-specific T or B cells. Littermate comparisons were used for all mouse studies unless otherwise indicated. Control and experimental treatments were administered to age- and sex-matched mice. The investigators were not blinded. Experimental replication is indicated in the figure legends.
Animals
C57BL/6NCr (code 556) and B6-Ly5.1/Cr (code 564) mice of 6 weeks of age were purchased from the National Cancer Institute (NCI) at Charles River (CRV). Cyp27a1−/−, Batf3−/−, Ifnar1−/−, Zbtb46-DTR (zDC-DTR) and OVA-specific OTII TCR-transgenic mice were purchased from Jackson Laboratories. Ebi2−/−, Cyp7b1−/−, Ch25h−/−, Hsd3b7−/−, and HEL-specific Hy10 knock-in mice have been described ((23, 28) and references therein). To generate BM chimeras, mice were lethally irradiated by exposure to 900–1100 rads of γ-irradiation in two doses 3 hr apart followed by injection of donor BM (5x106 cells). Chimeric mice were analyzed 6–10 weeks after reconstitution. Where indicated, adult mice or chimeric mice were fed rodent diet supplemented with 0.5% cholic acid (TD.04046, Envigo) for at least two weeks prior to analysis.
Animals were housed in a pathogen-free environment in the Laboratory Animal Research Center at the University of California, San Francisco, and all experiments conformed to ethical principles and guidelines approved by the Institutional Animal Care and Use Committee.
Immunizations and adoptive transfer
For 6-hour immunizations, 2 × 108 SRBCs (Colorado Serum Company), 20 μg of LPS (0111:B4, Sigma), 150 μg of poly I:C (GE Healthcare), or saline was injected retro-orbitally into mice. For immunization of mice with both SRBC and poly I:C, SRBCs were resuspended directly in corresponding doses of poly I:C.
For analysis of CD4+ T cell responses, pooled spleen and LN cells containing 106CD45.1+CD4+Vα2+ OTII were adoptively transferred into mice. For analysis of B cell responses, spleen cells containing 105 HEL-binding Hy10 B cells and 104 OTII T cells were cotransferred. One day after cell transfer, recipients were immunized intraperitoneally with HEL-OVA conjugated to SRBCs (SRBC-HEL-OVA; 2 × 108 cells).
RNAscope in situ hybridization
RNA in situ hybridization was performed using the RNAscope RED 2.5HD manual assay kit (Advanced Cell Diagnostics) (31). Using this kit was necessary because we could not detect signal using digoxigenin-labeled probes and conventional in situ hybridization. The RNAscope probes used were: Ch25h (NM_009890.1, targeting bp 115-1240), Cyp27a1 (NM_024264.5, targeting bp 555-1440), Cyp7b1 (NM_007825.4, targeting bp 179-1182), and Hsd3b7 (NM_133943.2, targeting bp 330-1192). Tissues were frozen in OCT and stored at −80°C. Within 24 hr of freezing, cryosections of 14 μm were cut and slides were dried at −20°C for 1 hr and at −80°C for 30 min. Slides were fixed for 15 min with ice-cold 4% paraformaldehyde and washed in 50%, 70%, and 100% ethanol for 5 min each. After drying for 5 min, slides were treated with hydrogen peroxide (from kit) for 8 min and protease IV (from kit) for 15 min. Probes were allowed to hybridize in a humidified chamber at 40°C for 3.5 hr. The following incubation times for the amplification steps were used: Amp 1, 40 min; Amp 2, 25 min; Amp 3, 40 min; Amp 4, 25 min; Amp 5, 40 min; Amp 6, 25 min. Slides were then developed with FastRed (from kit) for 20 minutes, washed in PBS, and counterstained for IgD using goat anti-mouse IgD (Cedarlane Laboratories) and HRP-conjugated donkey anti-goat IgG (Jackson Immunoresearch).
Conjugation of HEL-OVA to SRBCs
For SRBC-HEL-OVA conjugation, OVA protein (Sigma-Aldrich) was first cross-linked to HEL using glutaraldehyde. This HEL-OVA conjugate was further conjugated to SRBCs. 1 ml of SRBCs was washed with PBS three times and resuspended in 1mL of conjugation buffer (0.35 M mannitol & 0.01 M NaCl). 50μg of HEL-OVA was mixed with the SRBCs for 10 min and crosslinked using 100μL of 100 mg/ml EDCI (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, Fisher) for 30 minutes at 4°C while rocking. This was followed by three washes in PBS to remove free protein. Conjugation was confirmed by flow cytometry by staining the SRBCs with HyHEL9 mAb. This produces 8 doses of SRBC-conjugates and was scaled according to the number of mice requiring immunization. To visualize cell proliferation, OTII cells were labeled with 5μM CFSE (Invitrogen) for 10 min at 37°C in PBS with 1% FBS prior to transfer.
Dendritic Cell Ablation
For DC ablation in zDC-DTR:EBI2 mixed BM chimeras, mice were injected i.v. with 20ng/g of diphtheria toxin (DT) (Sigma-Aldrich) 2 days before cell transfer of antigen specific B and/or T cells. These mice also received 2ng/g DT i.p. on the day of cell transfer as well as 1 day, 2 days, and 4 days after immunization to maintain ablation. DC ablation in zDC-DTR:IFNAR mixed chimeras and full zDC-DTR chimeras was performed using an initial injection of 20ng/g DT 4 days before analysis and a second injection of 4ng/g DT 2 days before analysis.
EBI2 Antagonist Treatment
EBI2 antagonist (NIBR189, Avanti Polar Lipids (35)) was administered retro-orbitally 30 min before and 3 hr after immunization with SRBCs (1.5mg/kg/dose). Mice were analyzed 6 hr after immunization for DC positioning. Each dose of antagonist was prepared by diluting a 10mg/mL stock solution into 50μl of saline. Control mice were given equivalent injections of vehicle (N-Methyl-2-pyrrolidone, NMP) diluted in 50μl of saline.
In vivo labeling of dendritic cells
In vivo labeling of blood-exposed DCs was performed as described (36) with minor modifications. Mice were injected retro-orbitally with 1 μg PE-conjugated anti-CD45.2 (clone 104, Biolegend) diluted in 100μL of saline, 5 min before sacrifice. Spleens were digested for 10 minutes at 37°C, mashed through a 70μm filter and processed for flow cytometric analysis.
Flow cytometry and cell sorting
For EBI2 surface staining, cells were incubated with 4% normal mouse serum (NMS) and 4% normal donkey serum (NDS) for 10 min on ice, a 1:20 dilution of goat polyclonal antibody against EBI2 (A-20, Santa Cruz Biotechnology) for 1 hr at RT, a 1:200 dilution of biotin-conjugated donkey anti-goat IgG (Jackson Immunoresearch) for 30 min on ice, and a 1:200 dilution of PE-conjugated streptavidin (BD Biosciences) for 25 min on ice. Staining for CCR7 was performed using biotin-conjugated anti-CCR7 (4B12, Biolegend) at a dilution of 1:50 for 30 min at RT, followed by 1:200 PE-conjugated streptavidin. OTII-derived Tfh cell staining was performed using a 1:100 dilution of BV605-conjugated anti-CXCR5 (L138D7, Biolegend) for 1 hr at RT, followed by PE-conjugated anti-PD-1 (29F.1A12, Biolegend), PE-Cy7-conjugated anti-CD4 (RM4-5, Biolegend), Percp-Cy5.5-conjugated TCR Vα2 (B20.1, Biolegend), and APC-conjugated CD45.1 (A20, Biolegend) for 20 min on ice. Staining of Hy10-derived GCB cells was performed using HEL conjugated to A647, PE-Cy7-conjugated anti-CD95 (Jo2, BD Biosciences), Pacific Blue-conjugated anti-GL7 (GL7, Biolegend), Percp-Cy5.5-conjugated anti-B220 (RA3-6B2, Tonbo Biosciences), and BV605-conjugated CD45.1 (A20, Biolegend). Staining of DCs was performed using Alexa 647-conjugated anti-DCIR2 (33D1, Biolegend), PE-Cy7-conjugated anti-CD11c (N418, Tonbo Biosciences), FITC-conjugated anti-I-Ab (AF6-120.1, BD Biosciences), Pacific Blue-conjugated anti-CD8α (53-6.7, Biolegend), and PerCP-Cy5.5-conjugated anti-B220 (RA3-6B2, Tonbo Biosciences) to exclude B cells. Spleens (cut into 6–7 pieces) and mesenteric LNs were digested in RPMI 1640 media supplemented with 2% fetal bovine serum, 10mM HEPES, 1mg/mL collagenase type IV (Worthington Biochemical), and 20ug/mL DNAse I (Sigma), for 15 min in a 37°C incubator while rotating. After digestion, cells were isolated by mashing through a 70 μm cell strainer. DCs were stained in PBS containing 2% FBS and 0.1% sodium azide without EDTA. EDTA-free staining buffer is required since binding of anti-DCIR2 clone 33D1 is cation dependent. For sorting of DCs, DCs were first pre-enriched using anti-CD11c microbeads (Miltenyi Biotec) and sorted on a BD FACS Aria III. See Fig. S12 for sort gating strategy and purity. Flow cytometry data were analyzed using Flowjo (ver 9.7.6). Histograms for EBI2 and CCR7 surface levels were plotted using biexponential display.
Immunohistochemistry
Tissues were frozen in OCT. Cryosections of 7μm were dried for 1 hr at −20°C, fixed in 4°C acetone for 10 min, and dried for 1 hr at RT. For staining of DCIR2+ DCs, a 1:100 dilution of biotin-conjugated anti-DCIR2 (33D1, Biolegend) and a 1:100 dilution of goat anti-mouse IgD (GAM/IGD(FC)/7S, Cedarlane Laboratories) along with 1% NMS and NDS was incubated with the slides overnight at 4°C. Next, a tyramide signal amplification kit was used (TSA Biotin System; Perkin Elmer), in which the sections were first stained with streptavidin-HRP (1:100 dilution, Jackson Immunoresearch) for 45 min at RT, washed, then incubated with tyramide (1:50 dilution) for 4 min at RT. This was followed by staining with AP-conjugated donkey anti-goat IgG (1:100 dilution, Jackson Immunoresearch) and HRP-conjugated streptavidin (1:100 dilution, from TSA kit) for 2 hours at RT. Staining for XCR1 did not require signal amplification and was performed using biotin-conjugated anti-XCR1 (1:100 dilution, clone ZET) and AP-conjugated streptavidin (Jackson Immunoresearch). For immunofluorescence, slides were prepared for DCIR2+ DC staining as described above with the addition of anti-mouse CD45.1-FITC (clone A20, biolegend), followed by AMCA-conjugated donkey anti-goat IgG (Jackson Immunoresearch), Alexa 488-conjugated donkey anti-FITC and Alexa 647-conjugated streptavidin (Invitrogen). Images were captured with a Zeiss AxioObserver Z1 microscope.
Quantification of IHC images
Images of immunohistochemical stains of DCIR2+ DC positioning were quantified using ImageJ (version 1.49). All images were captured at the same magnification using a Zeiss AxioObserver Z1 inverted microscope. Images were loaded into ImageJ and the freehand selection tool was used to outline the B-T zone interface. This selection was scaled to 60% of its size, centered, and saved in the ImageJ ROI manager. The outer T cell zone area was defined as the area between this scaled selection and original B-T zone interface. The ImageJ IHC Image Analysis Toolbox was then used to isolate DCIR2+ staining and create a binary black and white image (DCIR2+ staining in black). The total amount of DCIR2+ staining (total amount of black pixels) was quantified. Then, the previously defined selection from the ROI manager was applied to this binary image, and the black pixels within the selection (the “inner T-cell zone”) were erased. The total amount of remaining black pixels (in the “outer T-cell zone”) was quantified, and the proportion of DCIR2+ staining in the outer T-cell zone was calculated. See Suppl. Fig. S6C for an example.
Transwell migration assay and M12 EBI2 ligand Bioassay
For splenic DC migration assays, spleens were digested for 10 min in a 37°C incubator using 1mg/mL collagenase type IV (Worthington Biochemical; Lakewood, NJ) and 20μg/mL DNAse I (Sigma) in migration media (RPMI 1640 containing 0.5% fatty acid free BSA and 10mM HEPES) and mashed through a 70μm filter. The cells were RBC-lysed, washed, and resensitized in migration assay media at 37°C for 20 min. 106 cells were placed into 5 μm transwell filters (Corning Costar), allowed to migrate for 3 hr at 37°C, and migrated cells were enumerated by flow cytometry. 7α,25-HC and 7α,27-HC was purchased from Avanti Polar Lipids and reconstituted as a 10mM solution in DMSO. For bioassays, spleen extracts were prepared by mashing spleens into migration media (1:10, weight by volume) through a 70μm filter and centrifuging at 500g and 4000g for 10 minutes to remove cells. Extracts were frozen at −80°C and re-centrifuged at 4000g prior to use. All extracts were further diluted 1:20 for use in transwell migration assays. EBI2-transduced M12 cells were grown to confluency in T75 flasks, washed three times in migration media, and resensitized for 20 min at 37°C in migration media prior to use in migration assays. 5x105 M12 cells were added to each transwell and allowed to migrate for 3 hr towards spleen extracts. Data are shown as % of input cells that migrated. For spleen extract data, each mouse was plotted as a mean of technical duplicate.
EBI2 Receptor Internalization Assay
Splenocytes were prepared as for migration assay. A titration of 7α,25-HC and 7α,27-HC in migration medium was prepared in a 96-well round bottom plate, with each well receiving 10 μl of oxysterol. 90 μl of cells (106 cells total) were added by multichannel to the 96-well plate and the plate was immediately placed in a 37°C incubator for 30 min. Afterwards, the plate was placed on ice and cells were washed three times before staining for EBI2 surface levels.
Retroviral transduction of T cells
T cells were enriched from spleen and LNs of wild type mice using MACS manual cell separation LS columns (Miltenyi Biotec). Enrichment was performed by negative selection using biotin-conjugated anti-B220 and streptavidin-conjugated-MACS beads. T cells were cultured in 24 well plates (106 cells/2ml/well) and activated using a final concentration of 3μg/ml anti-CD3ε (LEAF clone 2C11, Biolegend) and 1μg/ml anti-CD28 (LEAF clone 37.51, Biolegend) diluted in RPMI 1640 containing 10% FBS, 10mM HEPES, 55μM 2-mercaptoethanol, 2mM glutamine, and 50 IU penicillin/streptomycin. 24 hr after activation, the plate was centrifuged and the culture supernatant was saved. Retrovirus encoding a MSCV-EBI2-IRES-hCD4 plasmid (23) or control plasmid (truncated nerve growth factor receptor, NGFR) was produced using the PLAT-E packaging cell line (43) and added to the activated T cells. The T cells were spinfected at 2400rpm for 2 hr at RT, the viral supernatant was aspirated, and the original culture supernatant was returned to the cells. This spinfection was repeated a second time 24 hr later. 24 hr after the second spinfection, cells were washed twice and adoptively transferred into mice. Positioning of transduced T cells was analyzed 16–24 hr later in tissue sections by staining for hCD4.
Quantitative RT-PCR
Total RNA from tissues or sorted cells was extracted using an RNeasy kit (Qiagen) and reverse-transcribed. Quantitative PCR was performed as described (28). Data were analyzed using the comparative CT (2−ΔΔCt) method using Hprt as the reference.
LC-MS/MS
Spleen extracts were prepared as for the M12 bioassay. 10–12 spleen extracts from WT or Cyp27a1−/− mice were pooled per sample. Hsd3b7−/− spleen extracts were generated from single mice. The entire volume of spleen extract for each sample was extracted for LC-MS/MS analysis. 7α,27-HC was quantitated using a SCIEX API 5000 mass spectrometer coupled to a Shimadzu LC-20XR high performance liquid chromatograph as previously described (44). The measured amount of 7α,27-HC for each sample was divided by the total mg of spleen tissue used to generate the extract in order to determine ng per mg of spleen tissue.
Statistical analysis
Prism software (GraphPad, ver. 5.0a) was used for all statistical analyses. Two-tailed, unpaired Student’s t tests were performed when comparing only two groups, and one-way analysis of variance (ANOVA) using Bonferroni’s post hoc test for the indicated comparisons was performed when comparing one variable across multiple groups. P values less than 0.05 were considered significant. In summary graphs, points indicate individual mice, and horizontal lines indicate means. In bar graphs, bars indicate means, and error bars indicate SEM.
Supplementary Material
Acknowledgments
We thank J. An and Y. Xu for expert technical assistance, B. M. Thompson and K. M. Eckert for technical assistance with LC-MS/MS, D. Russell for enzyme-deficient mice and Cyp27a1 antisera, T. Yi and J. Muppidi for helpful discussions, and L. Rodda and J. Wu for critical reading of the manuscript. Funding: E.L. was supported by the University of California San Francisco (UCSF) Biomedical Sciences Graduate Program and NSF grant 1144247. E.V.D. was supported by the UCSF Medical Scientist Training Program and NIH F30 grant F30AI120527. J.G.C. is an investigator of the Howard Hughes Medical Institute. This work was supported in part by NIH grant AI040098, and the mass spectrometry work was supported by grant HL20948. Author contributions: E.L. and J.G.C. designed the study, analyzed the data, and wrote the paper. E.L. performed the experiments. E.V.D. performed the initial experiments identifying the DC defect in Cyp27a1 single-KO mice on control diet and edited the paper. J.G.M. provided LC-MS/MS data for 7α,27-HC in pooled spleen extracts. E.L. performed the statistical analysis. Competing interests: The authors declare that they have no competing interests.
Footnotes
This manuscript has been accepted for publication in Science Immunology. This version has not undergone final editing. Please refer to the complete version of record at http://immunology.sciencemag.org. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Refer to Web version on PubMed Central for supplementary material.
References
- 1.Durai V, Murphy KM. Functions of Murine Dendritic Cells. Immunity. 2016;45:719–736. doi: 10.1016/j.immuni.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinman RM, Pack M, Inaba K. Dendritic cells in the T-cell areas of lymphoid organs. Immunol Rev. 1997;156:25–37. doi: 10.1111/j.1600-065x.1997.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 3.Yi T, Cyster JG. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. Elife. 2013;2:e00757. doi: 10.7554/eLife.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatto D, Wood K, Caminschi I, Murphy-Durland D, Schofield P, Christ D, Karupiah G, Brink R. The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat Immunol. 2013 doi: 10.1038/ni.2555. [DOI] [PubMed] [Google Scholar]
- 5.Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Guttler S, Hutloff A, Mages HW, Ranke K, Schaefer M, Jack RS, Henn V, Kroczek RA. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 2009;31:823–833. doi: 10.1016/j.immuni.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Bachem A, Hartung E, Guttler S, Mora A, Zhou X, Hegemann A, Plantinga M, Mazzini E, Stoitzner P, Gurka S, Henn V, Mages HW, Kroczek RA. Expression of XCR1 Characterizes the Batf3-Dependent Lineage of Dendritic Cells Capable of Antigen Cross-Presentation. Front Immunol. 2012;3:214. doi: 10.3389/fimmu.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazaki C, Sugiyama M, Ohta T, Hemmi H, Hamada E, Sasaki I, Fukuda Y, Yano T, Nobuoka M, Hirashima T, Iizuka A, Sato K, Tanaka T, Hoshino K, Kaisho T. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J Immunol. 2013;190:6071–6082. doi: 10.4049/jimmunol.1202798. [DOI] [PubMed] [Google Scholar]
- 8.Alexandre YO, Ghilas S, Sanchez C, Le Bon A, Crozat K, Dalod M. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J Exp Med. 2016;213:75–92. doi: 10.1084/jem.20142350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calabro S, Liu D, Gallman A, Nascimento MS, Yu Z, Zhang TT, Chen P, Zhang B, Xu L, Gowthaman U, Krishnaswamy JK, Haberman AM, Williams A, Eisenbarth SC. Differential Intrasplenic Migration of Dendritic Cell Subsets Tailors Adaptive Immunity. Cell Rep. 2016;16:2472–2485. doi: 10.1016/j.celrep.2016.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reis e Sousa C, Germain RN. Analysis of adjuvant function by direct visualization of antigen presentation in vivo: endotoxin promotes accumulation of antigen-bearing dendritic cells in the T cell areas of lymphoid tissue. J Immunol. 1999;162:6552–6561. [PubMed] [Google Scholar]
- 11.Gunn MD, Kyuwa S, Tam C, Kakiuchi T, Matsuzawa A, Williams LT, Nakano H. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J Exp Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Becker G, Moulin V, Pajak B, Bruck C, Francotte M, Thiriart C, Urbain J, Moser M. The adjuvant monophosphoryl lipid A increases the function of antigen-presenting cells. Int Immunol. 2000;12:807–815. doi: 10.1093/intimm/12.6.807. [DOI] [PubMed] [Google Scholar]
- 13.De Trez C, Pajak B, Brait M, Glaichenhaus N, Urbain J, Moser M, Lauvau G, Muraille E. TLR4 and Toll-IL-1 receptor domain-containing adapter-inducing IFN-beta, but not MyD88, regulate Escherichia coli-induced dendritic cell maturation and apoptosis in vivo. J Immunol. 2005;175:839–846. doi: 10.4049/jimmunol.175.2.839. [DOI] [PubMed] [Google Scholar]
- 14.Asselin-Paturel C, Brizard G, Chemin K, Boonstra A, O’Garra A, Vicari A, Trinchieri G. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J Exp Med. 2005;201:1157–1167. doi: 10.1084/jem.20041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idoyaga J, Suda N, Suda K, Park CG, Steinman RM. Antibody to Langerin/CD207 localizes large numbers of CD8alpha+ dendritic cells to the marginal zone of mouse spleen. Proc Natl Acad Sci U S A. 2009;106:1524–1529. doi: 10.1073/pnas.0812247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czeloth N, Schippers A, Wagner N, Muller W, Kuster B, Bernhardt G, Forster R. Sphingosine-1 phosphate signaling regulates positioning of dendritic cells within the spleen. J Immunol. 2007;179:5855–5863. doi: 10.4049/jimmunol.179.9.5855. [DOI] [PubMed] [Google Scholar]
- 17.Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of specific B and T lymphocyte interactions in the lymph node. Science. 1998;281:96–99. doi: 10.1126/science.281.5373.96. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Lu E, Yi T, Cyster JG. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 2016;533:110–114. doi: 10.1038/nature17947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi T, Li J, Chen H, Wu J, An J, Xu Y, Hu Y, Lowell CA, Cyster JG. Splenic Dendritic Cells Survey Red Blood Cells for Missing Self-CD47 to Trigger Adaptive Immune Responses. Immunity. 2015;43:764–775. doi: 10.1016/j.immuni.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatto D, Brink R. B cell localization: regulation by EBI2 and its oxysterol ligand. Trends Immunol. 2013;14:446–453. doi: 10.1016/j.it.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Cyster JG, Dang EV, Reboldi A, Yi T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat Rev Immunol. 2014;14:731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- 22.Gatto D, Paus D, Basten A, Mackay CR, Brink R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 2009;31:259–269. doi: 10.1016/j.immuni.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–1126. doi: 10.1038/nature08226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly LM, Pereira JP, Yi T, Xu Y, Cyster JG. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J Immunol. 2011;187:3026–3032. doi: 10.4049/jimmunol.1101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, Knochenmuss R, Noel S, Gessier F, Kelly LM, Vanek M, Laurent S, Preuss I, Miault C, Christen I, Karuna R, Li W, Koo DI, Suply T, Schmedt C, Peters EC, Falchetto R, Katopodis A, Spanka C, Roy MO, Detheux M, Chen YA, Schultz PG, Cho CY, Seuwen K, Cyster JG, Sailer AW. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Yang XV, Wu J, Kuei C, Mani NS, Zhang L, Yu J, Sutton SW, Qin N, Banie H, Karlsson L, Sun S, Lovenberg TW. Oxysterols direct B-cell migration through EBI2. Nature. 2011;475:519–523. doi: 10.1038/nature10226. [DOI] [PubMed] [Google Scholar]
- 27.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 28.Yi T, Wang X, Kelly LM, An J, Xu Y, Sailer AW, Gustafsson JA, Russell DW, Cyster JG. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37:535–548. doi: 10.1016/j.immuni.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Repa JJ, Lund EG, Horton JD, Leitersdorf E, Russell DW, Dietschy JM, Turley SD. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. J Biol Chem. 2000;275:39685–39692. doi: 10.1074/jbc.M007653200. [DOI] [PubMed] [Google Scholar]
- 30.Soroosh P, Wu J, Xue X, Song J, Sutton SW, Sablad M, Yu J, Nelen MI, Liu X, Castro G, Luna R, Crawford S, Banie H, Dandridge RA, Deng X, Bittner A, Kuei C, Tootoonchi M, Rozenkrants N, Herman K, Gao J, Yang XV, Sachen K, Ngo K, Fung-Leung WP, Nguyen S, de Leon-Tabaldo A, Blevitt J, Zhang Y, Cummings MD, Rao T, Mani NS, Liu C, McKinnon M, Milla ME, Fourie AM, Sun S. Oxysterols are agonist ligands of RORgammat and drive Th17 cell differentiation. Proc Natl Acad Sci U S A. 2014;111:12163–12168. doi: 10.1073/pnas.1322807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen H, Reshef A, Maeda N, Lippoldt A, Shpizen S, Triger L, Eggertsen G, Bjorkhem I, Leitersdorf E. Markedly reduced bile acid synthesis but maintained levels of cholesterol and vitamin D metabolites in mice with disrupted sterol 27-hydroxylase gene. J Biol Chem. 1998;273:14805–14812. doi: 10.1074/jbc.273.24.14805. [DOI] [PubMed] [Google Scholar]
- 33.Li-Hawkins J, Lund EG, Turley SD, Russell DW. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J Biol Chem. 2000;275:16536–16542. doi: 10.1074/jbc.M001811200. [DOI] [PubMed] [Google Scholar]
- 34.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 35.Gessier F, Preuss I, Yin H, Rosenkilde MM, Laurent S, Endres R, Chen YA, Marsilje TH, Seuwen K, Nguyen DG, Sailer AW. Identification and characterization of small molecule modulators of the Epstein-Barr virus-induced gene 2 (EBI2) receptor. J Med Chem. 2014;57:3358–3368. doi: 10.1021/jm4019355. [DOI] [PubMed] [Google Scholar]
- 36.Cinamon G, Zachariah M, Lam O, Cyster JG. Follicular shuttling of marginal zone B cells facilitates antigen transport. Nat Immunol. 2008;9:54–62. doi: 10.1038/ni1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller SN, Germain RN. Stromal cell contributions to the homeostasis and functionality of the immune system. Nat Rev Immunol. 2009;9:618–629. doi: 10.1038/nri2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin C, Han JA, Choi B, Cho YK, Do Y, Ryu S. Intrinsic features of the CD8alpha(−) dendritic cell subset in inducing functional T follicular helper cells. Immunol Lett. 2016;172:21–28. doi: 10.1016/j.imlet.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Neubert K, Lehmann CH, Heger L, Baranska A, Staedtler AM, Buchholz VR, Yamazaki S, Heidkamp GF, Eissing N, Zebroski H, Nussenzweig MC, Nimmerjahn F, Dudziak D. Antigen delivery to CD11c+CD8− dendritic cells induces protective immune responses against experimental melanoma in mice in vivo. J Immunol. 2014;192:5830–5838. doi: 10.4049/jimmunol.1300975. [DOI] [PubMed] [Google Scholar]
- 40.Soares H, Waechter H, Glaichenhaus N, Mougneau E, Yagita H, Mizenina O, Dudziak D, Nussenzweig MC, Steinman RM. A subset of dendritic cells induces CD4+ T cells to produce IFN-gamma by an IL-12-independent but CD70-dependent mechanism in vivo. J Exp Med. 2007;204:1095–1106. doi: 10.1084/jem.20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLellan AD, Kapp M, Eggert A, Linden C, Bommhardt U, Brocker EB, Kammerer U, Kampgen E. Anatomic location and T-cell stimulatory functions of mouse dendritic cell subsets defined by CD4 and CD8 expression. Blood. 2002;99:2084–2093. doi: 10.1182/blood.v99.6.2084. [DOI] [PubMed] [Google Scholar]
- 42.Qiu CH, Miyake Y, Kaise H, Kitamura H, Ohara O, Tanaka M. Novel subset of CD8{alpha}+ dendritic cells localized in the marginal zone is responsible for tolerance to cell-associated antigens. J Immunol. 2009;182:4127–4136. doi: 10.4049/jimmunol.0803364. [DOI] [PubMed] [Google Scholar]
- 43.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 44.McDonald JG, Smith DD, Stiles AR, Russell DW. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J Lipid Res. 2012;53:1399–1409. doi: 10.1194/jlr.D022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.