Abstract Abstract
Orestias Valenciennes, 1839 is a genus of freshwater fish endemic to the South American Altiplano. Cytogenetic studies of these species have focused on conventional karyotyping. The aim of this study was to use classical and molecular cytogenetic methods to identify the constitutive heterochromatin distribution and chromosome organization of four classes of repetitive DNA sequences (histone H3 DNA, U2 snRNA, 18S rDNA and 5S rDNA) in the chromosomes of O. ascotanensis Parenti, 1984, an endemic species restricted to the Salar de Ascotán in the Chilean Altiplano. All individuals analyzed had a diploid number of 48 chromosomes. C-banding identified constitutive heterochromatin mainly in the pericentromeric region of most chromosomes, especially a GC-rich heterochromatic block of the short arm of pair 3. FISH assay with an 18S probe confirmed the location of the NOR in pair 3 and revealed that the minor rDNA cluster occurs interstitially on the long arm of pair 2. Dual FISH identified a single block of U2 snDNA sequences in the pericentromeric regions of a subtelocentric chromosome pair, while histone H3 sites were observed as small signals scattered in throughout the all chromosomes. This work represents the first effort to document the physical organization of the repetitive fraction of the Orestias genome. These data will improve our understanding of the chromosomal evolution of a genus facing serious conservation problems.
Keywords: Orestias, molecular cytogenetics, multigene families
Introduction
Cytogenetic analysis is a useful tool for describing evolutionary patterns and the histories of closely-related species or species complexes. Orestias Valenciennes, 1839 is a genus of freshwater fish endemic to the South American Altiplano. The genus includes 45 species, grouped into four complexes: O. cuvieri, O. mulleri, O. gilsoni and O. agassii (Costa 1999, Parenti 1984). Conventional karyotyping studies involving the seven species of O. agassii complex found in the Chilean Altiplano (17°and 22°S) have revealed variations in the chromosome number (2n=48-55) and the presence/absence of microchromosomes, suggesting that Robertsonian rearrangements may play a role in the karyotypic evolution of these species (Arratia 1982, Vila et al. 2007, 2010, 2011, Habit et al. 2006, Villwock and Sienknecht 1996).
The most commonly-used approaches for comparative cytogenetic analysis of fish include characterizing the distribution and composition of constitutive heterochromatin and fluorescence in situ hybridization (FISH) mapping of molecular landmarks such as 18S and 5S ribosomal DNA. New markers of repeated elements such as histone H1, H3 and H4 genes and the U2 snRNA gene have recently been incorporated into these studies (Hashimoto et al. 2011, Utsunomia et al. 2014a, Silva et al. 2015, Utsunomia et al. 2016). The repetitive nature of these sequences makes them useful markers for chromosomal mapping as they provide insight into the structure and organization of the genome and facilitate detection of karyotype rearrangements (Kavalco et al. 2013). However, studies involving chromosomal mapping of repetitive sequences in fish are scarce and typically focus exclusively on the location of ribosomal DNA sites. Studies involving physical mapping of histone genes and mobile elements are also limited, and data is available for only a few species (Pendas et al. 1994, Hashimoto et al. 2011, Ferreira et al. 2011).
Orestias ascotanensis Parenti, 1984 is an endemic species restricted to the small isolated freshwater springs of the Salar de Ascotán. This fish is on the Chilean Endangered Species List (MINSEGPRES, 2008). Major threats to conservation of this species include global climate change and intense regional mining activity. Both situations contribute to a gradual lowering of the water level in the springs, potentially making the salinity of the water incompatible with life for these populations (Vila et al. 2007, Morales et al. 2011). The O. ascotanensis karyotype consists of 48 chromosomes, which is the most common diploid number among species in the order Cyprinodontiformes. The chromosomal formula is (2M + 4SM + 4 ST + 38T) (Vila et al. 2010).
The aim of this study was to identify for the first time the constitutive heterochromatin distribution and chromosome organization of four classes of repetitive DNAs (histone H3 DNA, U2 snRNA and 18S and 5S rDNA) in the chromosomes of O. ascotanensis. This data will shed light on the physical organization of the repetitive fraction of the genome of O. ascotanensis, a species endemic to the Chilean Altiplano that is facing serious conservation problems. In addition, application of these cytogenetic tools will allow for comparisons among Orestias species, facilitating the identification of genomic modifications underlying the chromosomal variations observed in these species.
Materials and methods
Sampling and mitotic chromosome isolation
Eight O. ascotanensis individuals, 3 male and 5 female, were obtained from Salar de Ascotán (21°31'S 68°15'W), Region de Antofagasta, Chile, under Scientific Collection Permit Number 1103 issued by SERNAPESCA. The fish were transported to the laboratory and maintained alive in aquaria until processing. Mitotic chromosomes were obtained from kidney cell suspensions according to a modified version of the protocol established by Foresti et al. (1993). Approximately 20 metaphase spreads from different individuals were analyzed to confirm the diploid number and karyotype structure of O. ascotanensis. The chromosomes were measured and classified as metacentric (m), submetacentric (sm), subtelocentric (st) or telocentric (t) (Levan et al. 1964), and the karyotype was arranged according to Vila et al. (2010). The images were captured with a digital camera (Nikon D60) attached to an epifluorescence photomicroscope (Nikon Optiphot). Karyotype mounting and image brightness and contrast adjustments were performed in Adobe Photoshop CS6.
Chromosome banding: C- banding and CMA3
The constitutive heterochromatin (HC) distribution pattern was visualized according to a modified version of the protocol established by Sumner (1972); briefly, chromosomes were subjected to hydrolysis with HCL 0.2 N for 45 min at room temperature, denatured with 5% barium hydroxide at 60°C for 8 min and incubated in saline buffer 2× SSC, and stained with propidium iodide (50 ug/mL) (Lui et al. 2009). Chromomycin A3 staining was then performed using the method described by Sola et al. (1992). Metaphase plates were observed using a Nikon (Optiphot) microscope with the appropriate filter.
Repetitive sequence probes and FISH experiments
U2 snRNA, 5S rDNA, 18S rDNA and histone H3 DNA probes were obtained from the genomic DNA of O. ascotanensis. DNA was collected from a piece of fin tissue with the Wizard Genomic DNA Purification Kit (Promega) according to manufacturer instructions, using previously-described primers (Table 1). The U2 snRNA and 5S rDNA probes were labeled by PCR with biotin-16-dUTP, and the 18S rDNA and histone H3 DNA probes were labeled by PCR with digoxigenin-11-dUTP. FISH was performed under high-stringency conditions using the method described by Pinkel et al. (1986). Slides were incubated with RNAse (50μg/ml) for 1 h at 37°C. Next, the chromosomal DNA was denatured in 70% formamide/2× SSC for 5 min at 70°C, and the slides were taken through an ice-cold ethanol series (70°-80°-100°). For each slide, 30μl of hybridization solution containing 200 ng of each labeled probe, 50% formamide, 2× SSC and 10% dextran sulfate was denatured for 10 min at 95°C, dropped onto the slides and hybridized overnight at 37°C in a 2× SSC moist chamber. After hybridization, slides were washed in 0.2× SSC/15% formamide for 20 min at 42°C, followed by a second wash in 0.1× SSC for 15 min at 60°C and a final wash at room temperature in 4× SSC/0.5% Tween for 10 min. Probe detection was carried out with Avidin-FITC (Sigma) or anti-digoxigenin-rhodamine (Roche). Chromosomes were counterstained with DAPI (4’,6-diamidino-2-phenylindole, Vector Laboratories).
Table 1.
Primers used to PCR amplification for gene fragments 5S rDNA, 18S rDNA, U2 snRNA and Histone H3.
| Gene | Primers sequences | References |
|---|---|---|
| 5S rDNA | 5SA 5’-TCAACCAACCACAAAGACATTGGCAC-3’ | Pendás et al. 1994 |
| 5SB 5’-TAGACTTCTGGGTGGCCAAAGGAATCA-3’ | ||
| 18S rDNA | 18SF 5’-GTAGTCATATGCTTGTCTC-3’ | White et al. 1990 |
| 18SR 5’-TCCGCAGGTTCACCTACGGA-3’ | ||
| U2snRNA | U2 F 5’-ATCGCTTCTCGGCCTTATG-3’ | Bueno et al. 2013 |
| U2 R 5’-TCCCGGCGGTACTGCAATA-3’ | ||
| Histone H3 | H3F 5’- ATATCCTTRGGCATRATRGTGAC-3’ | Colgan et al. 1998 |
| H3R 5’- ATGGCTCGTACCAAGCAGACVGC-3’ |
Results
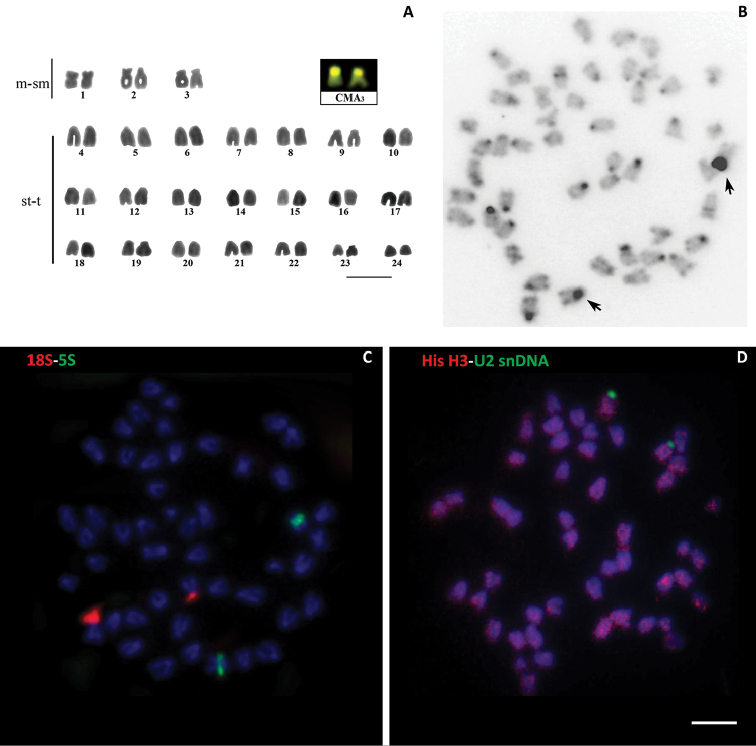
All O. ascotanensis individuals analyzed had a diploid number of 48 chromosomes, consistent with the chromosome formula defined by Vila et al. (2010) (Fig. 1A). No morphologically differentiated sex chromosomes were found when metaphase plates from males and females were compared. C-banding revealed that the constitutive heterochromatin was mainly distributed in the pericentromeric regions of most chromosomes (Fig. 1B). Submetacentric pair 3 was noteworthy due to the presence of conspicuous HC blocks extending along the entire short arm. An interstitial C-band was present on the long arm of chromosome pair 2, proximal to the centromeric region. Additionally, in all observed metaphases, CMA3-banding revealed that the short arm of pair 3 was strongly stained, reflecting a greater abundance of GC bases in this heterochromatic region (see box, Fig. 1A).
Figure 1.
Karyotype of O. ascotanensis (female), 2n=48. A Giemsa-stained karyotype and CMA3 positive bands (box) B C-banded somatic metaphase C metaphases counterstained with DAPI after FISH treatment using 5S and 18S rDNA probes D metaphases counterstained with U2 snDNA/histone H3 DNA probes. The arrows show a block of HC on the short arm of pair 3. Bar = 10µm.
Dual FISH detected 18S and 5S rDNA probes on different chromosome pairs (Fig. 1C). The major rDNA cluster (18S) was located on the short arm of pair 3, with a size polymorphism between the bearing arms of these sequences. 5S rDNA sequences were detected in the region proximal to the centromere of the long arm of pair 2, coincident with the HC band described above. Dual FISH (Fig. 1D) identified a single block of U2 snDNA sequences in the pericentromeric region of a subtelocentric chromosome pair, while histone H3 sites were detected as scattered signals throughout the O. ascotanensis chromosomes.
Discussion
Previous cytogenetic studies involving the seven species of O. agassii complex of the Chilean Altiplano were limited to characterizing the chromosome number and morphology of the species. The diploid number has been reported to vary between 48 and 55 chromosomes and the fundamental number of chromosome arms (FN) between 54 and 56 (Arratia 1982, Vila et al. 2010, Vila et al. 2011)
Characterization of the repetitive fraction of the genome is a useful tool for identifying recent genomic changes during the evolutionary process as well as possible hotspots associated with chromosomal rearrangements (Valente et al. 2011, Ozouf-Costaz et al. 2004, Yano et al. 2014). The organization of the repetitive fraction of the genome in Cyprinodontiformes fish has remained relatively unexplored, with prior studies focusing primarily on the distribution and composition of constitutive heterochromatin and physical mapping of 18S rDNA genes. Noteworthy studies include reports on: Fundulus (Lacépède, 1803) (Kornfield 1981); Austrolebias Costa, 1998 (García et al. 1993, 1995, 2001, 2014, 2015); Aphanius Nardo, 1827 (Vitturi et al. 2005, Gaffaroglu et al. 2014) and Hypsolebias Costa, 2006 (Do Nascimento et al. 2014), with constitutive heterochromatin found to be distributed mainly in centromeric, telomeric and interstitial regions. In addition, in some species of Chromaphyosemion Myers, 1924, conspicuous blocks of HC have been identified in the short arm of bi-armed chromosomes (Völker et al. 2005, Völker et al. 2006, Volker et al. 2007, Völker et al. 2008).
In O. ascotanensis, the C-band regions were found mainly in the pericentromeric regions, unlike other Cyprinodontiformes that have been studied. CMA3 also revealed that the conspicuous blocks of HC found in the short arm of pair 3 have a higher proportion of GC bases than previously-analyzed fish. Moreover, the presence of 18S rDNA sequences in this chromosome arm defines this pair as the carrier of the NOR. An association between 18S and 28S rDNA sequences and heterochromatin has been found in other fish, such as salmonids (Fujiwara et al. 1998, Pendas et al. 1994), species of the genera Epinephelus Bloch, 1793 (Sola et al. 2000), Imparfinis Eigenmann & Norris, 1900 and Pimelodella Eigenmann & Eigenmann, 1888 (Gouveia et al. 2013) and sturgeon species (Fontana et al. 2003), suggesting that the repeated HC sequences play an important role and exercise diverse functions in the eukaryotic genome (Grewal and Jia 2007). It has even been postulated that heterochromatin is involved in maintaining the structure of the nucleolus and the integrity of ribosomal DNA repeats (McStay and Grummt 2008).
The single 18S rDNA sequence-bearing chromosome pair in O. ascotanensis (Fig. 1C) is a feature observed in most teleosts (Pisano and Ghigliotti 2009, Gornung 2013). However, varied numbers of chromosomes carrying the major ribosomal DNA cluster have been reported in Cyprinodontiformes, with findings ranging from one to seven pairs of chromosomes (Völker et al. 2005, Völker et al. 2006, Volker et al. 2007, Völker et al. 2008). Data on the chromosomal location of the minor ribosomal sites are almost non-existent for Cyprinodontiformes. In O. ascotanesis, pair 2 is the 5S-bearing pair, with submetacentric morphology (Fig. 1C). The hybridization signal was detected on the long arm, proximal to the centromere region, associated with the interstitial heterochromatic band of this pair. 5S and 18S rDNA are typically localized on different chromosomes in vertebrates, including teleosts (Scacchetti et al. 2015, Sánchez-Romero et al. 2015). However, in the Cyprinodontiform Lebias fasciata (Valenciennes, 1821), FISH mapping has shown that the 28S and 5S ribosomal DNA probes co-localize on a pair of telocentric chromosomes, conserving the 5S locus on the medial position of the chromosome (Tigano et al. 2004). In general, these sequences vary among teleosts in relation to the chromosomal distribution due to their association with transposable elements, typically within the internal spacer regions (Martins and Galetti 2001, Cabral-de-Mello et al. 2011, Scacchetti et al. 2012, Sene et al. 2015).
Data on the physical location of U2 snRNA sites in fish is also scarce. Two general configurations are recognized: (I) clustered on a single pair of chromosomes, as in the present case and (II) scattered throughout the genome (Ubeda-Manzanaro et al. 2010, Utsunomia et al. 2014, Scacchetti et al. 2015, Silva et al. 2015). According to Medrano et al. (1988), teleosts show low levels of genomic compartmentalization, suggesting that the configuration observed for the U2 snRNA, 5S rDNA and 18S rDNA in O. ascotanensis represents a relatively simple genomic organization.
The finding of scattered histone H3 sites distributed throughout the O. ascotanensis chromosomes diverges strongly from data reported for other fish, such as Characiformes (Hashimoto et al. 2011, Pansonato-Alves et al. 2013a, Silva et al. 2015), Siluriformes (Hashimoto et al. 2013, Pansonato-Alves et al. 2013b) and Perciformes (Lima-Filho et al. 2012), which generally have large blocks of these sequences in specific chromosome pairs. The histone H3 DNA site distribution found in O. ascotanensis chromosomes is similar to the organization described for Synbranchus marmoratus Bloch, 1795, suggesting that H3 sequences may be organized in small, abundant copies throughout the genome, as has been proposed by Utsunomia et al. (2014b). Further studies are necessary to confirm that this scattered distribution of H3 DNA is conserved among Orestias species.
To understand the relationship of these repeated genomic elements to the chromosomal evolution of these fish and to historical changes in the fishes’ environment, further studies are needed to physically map the repetitive DNA in other Orestias representatives. These findings would enhance our understanding of native wildlife species facing serious conservation problems.
Acknowledgments
This research work was financed by projects Fondecyt 1080390 and Fondecyt 1140543.
Citation
Araya-Jaime C, Lam N, Pinto IV, Méndez MA, Iturra P (2017) Chromosomal organization of four classes of repetitive DNA sequences in killifish Orestias ascotanensis Parenti, 1984 (Cyprinodontiformes, Cyprinodontidae). Comparative Cytogenetics 11(3): 463–475. https://doi.org/10.3897/CompCytogen.v11i3.11729
References
- Arratia G. (1982) Peces del altiplano de Chile. In: Veloso A, Bustos-Obregon E. (Eds) El Ambiente Natural y las Poblaciones Humanas de los Andes del Norte Grande de Chile. UNESCO, MAB-6, La Vegetación y los Vertebrados Inferiores de los Pisos Altitudinales entre Arica y Lago Chungará. Oficina Regional de Ciencia y Técnología de la UNESCO para América Latina y el Caribe, Uruguay, Montevideo, 93–133.
- Bueno D, Palacios-Gimenez OM, Cabral-de-Mello DC. (2013) Chromosomal Mapping of Repetitive DNAs in the Grasshopper Abracris flavolineata Reveal Possible Ancestry of the B Chromosome and H3 Histone Spreading. In: Wutz A (Ed.) PLoS ONE 8: e66532. https://doi.org/10.1371/journal.pone.0066532 [DOI] [PMC free article] [PubMed]
- Cabral-de-Mello DC, Oliveira S, Moura R, Martins C. (2011) Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genetics 12: 88. https://doi.org/10.1186/1471-2156-12-88 [DOI] [PMC free article] [PubMed]
- Colgan D, McLauchlan A, Wilson G, Livingston S, Edgecombe G, Macaranas J, Cassis G, Gray M. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Australian Journal of Zoology 46: 419–437. https://doi.org/10.1071/ZO98048 [Google Scholar]
- Costa WJEM. (1999) Phylogeny and Classification of the Cyprinodontiformes (Euteleostei: Atherinomorpha): A Reappraisal. In: Reis RE, Vari RP, Lucena ZM, Lucena CAS (Eds) Phylogeny and Classification of Neotropical Fishes. Edipucrs, Porto Alegre, 603 pp. [Google Scholar]
- Do Nascimento WS, Bezerra JG, Lima-Filho PA, Yamamoto ME, Chellappa S, Molina WF. (2014) Karyotype patterns of Hypsolebias antenori (Cyprinodontiformes: Rivulidae): an endangered killifish of the semiarid region of Brazil. The Scientific World Journal 2014: 862434. https://doi.org/10.1155/2014/862434 [DOI] [PMC free article] [PubMed]
- Ferreira DC, Oliveira C, Foresti F. (2011) Chromosome Mapping of Retrotransposable Elements Rex1 and Rex3 in Three Fish Species in the Subfamily Hypoptopomatinae (Teleostei, Siluriformes, Loricariidae). Cytogenetic and Genome Research 132: 64–70. https://doi.org/10.1159/000319620 [DOI] [PubMed] [Google Scholar]
- Fontana F, Lanfredi M, Congiu L, Leis M, Chicca M, Rossi R. (2003) Chromosomal mapping of 18S-28S and 5S rRNA genes by two-colour fluorescent in situ hybridization in six sturgeon species. Genome 46: 473–7. https://doi.org/10.1139/g03-007 [DOI] [PubMed] [Google Scholar]
- Foresti F, Oliveira C, de Almeida-Toledo L. (1993) A method for chromosome preparations from large fish specimens using in vitro short-term treatment with colchicine. Experientia 9: 810–813. https://doi.org/10.1007/BF01923555 [Google Scholar]
- Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida MC. (1998) Chromosomal localization and heterochromatin association of ribosomal RNA gene loci and silver-stained nucleolar organizer regions in salmonid fishes. Chromosome research 6: 463–71. https://doi.org/10.1023/A:1009200428369 [DOI] [PubMed] [Google Scholar]
- Gaffaroglu M, Ayata MK, Unal S, Ozkan M. (2014) Karyological Analysis of Some Species of Aphanius (Osteichthyes: Cyprinodontidae) From Anatolia. Pakistan Journal of Zoology 45: 1271–1275. [Google Scholar]
- García G, Gutiérrez V, Ríos N, Turner B, Santiñaque F, López-Carro B, Folle G. (2014) Burst speciation processes and genomic expansion in the neotropical annual killifish genus Austrolebias (Cyprinodontiformes, Rivulidae). Genetica 142: 87–98. https://doi.org/10.1007/s10709-014-9756-7 [DOI] [PubMed] [Google Scholar]
- García G, Lalanne AI, Aguirre G, Cappetta M. (2001) Chromosome evolution in the annual killifish genus Cynolebias and mitochondrial phylogenetic analysis. Chromosome research 9: 437–48. [DOI] [PubMed] [Google Scholar]
- García G, Ríos N, Gutiérrez V. (2015) Next-generation sequencing detects repetitive elements expansion in giant genomes of annual killifish genus Austrolebias (Cyprinodontiformes, Rivulidae). Genetica 143: 353–360. https://doi.org/10.1007/s10709-015-9834-5 [DOI] [PubMed] [Google Scholar]
- García G, Scvortzoff E, Hernández A. (1995) Karyotypic Heterogeneity in South American Annual Killifishes of the Genus Cynolebias (Pisces, Cyprinodontiformes Rivulidae). Cytologia 103–110. https://doi.org/10.1508/cytologia.60.103
- García G, Scvortzoff E, Máspoli M, Vaz-Ferreira R. (1993) Analysis of Karyotypic Evolution in Natural Populations of Cynolebias (Pisces: Cyprinodontiformes, Rivulidae) Using Banding Techniques. Cytologia 58: 85–94. https://doi.org/10.1508/cytologia.58.85 [Google Scholar]
- Gornung E. (2013) Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenetic and Genome Research 141: 90–102. https://doi.org/10.1159/000354832 [DOI] [PubMed] [Google Scholar]
- Gouveia JG, de Moraes VPO, Sampaio TR, Da Rosa R, Dias AL. (2013) Considerations on karyotype evolution in the genera Imparfinis Eigenmann and Norris 1900 and Pimelodella Eigenmann and Eigenmann 1888 (Siluriformes: Heptapteridae). Reviews in Fish Biology and Fisheries 23: 215–227. https://doi.org/10.1007/s11160-012-9286-2 [Google Scholar]
- Grewal SIS, Jia S. (2007) Heterochromatin revisited. Nature reviews. Genetics 8: 35–46. https://doi.org/10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Habit E, Dyer B, Vila I. (2006) Estado de conocimiento de los peces dulceacuícolas de Chile. Gayana 70: 111–113. https://doi.org/10.4067/S0717-65382006000100016 [Google Scholar]
- Hashimoto DT, Ferguson-Smith MA, Rens W, Foresti F, Porto-Foresti F. (2011) Chromosome mapping of H1 histone and 5S rRNA gene clusters in three species of Astyanax (Teleostei, Characiformes). Cytogenetic and Genome Research 134: 64–71. https://doi.org/10.1159/000323512 [DOI] [PubMed] [Google Scholar]
- Hashimoto DT, Ferguson-Smith MA, Rens W, Prado FD, Foresti F, Porto-Foresti F. (2013) Cytogenetic mapping of H1 histone and ribosomal RNA genes in hybrids between catfish species Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum. Cytogenetic and Genome Research 139: 102–6. https://doi.org/10.1159/000345299 [DOI] [PubMed] [Google Scholar]
- Kavalco KF, Pazza R, Brandão KO, de Almeida-Toledo LF. (2013) Biogeographic patterns in the chromosomal distribution of a satellite DNA in the banded tetra Astyanax fasciatus (Teleostei: Characiformes). Organisms Diversity & Evolution 13: 67–76. https://doi.org/10.1007/s13127-012-0100-8 [Google Scholar]
- Kornfield I. (1981) Distribution of Constitutive Heterochromatin and the Evolution of Sex Chromosomes in Fundulus Copeia: 916–918. https://doi.org/10.2307/1444204
- Levan A, Fredga K, Sandberg A. (1964) Nomenclature for centromeric position on chromosomes. Hereditas 2: 201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x [Google Scholar]
- Lima-Filho P, Cioffi M, Bertollo L, Molina WF. (2012) Chromosomal and morphological divergences in Atlantic populations of the frillfin goby Bathygobius soporator (Gobiidae, Perciformes). Journal of Experimental Marine Biology and Ecology 434–435: 63–70. dx.doi.org/10.1016/j.jembe.2012.08.004
- Lui RL, Blanco DR, Margarido VP, Orl Filho OM, Filho OM. (2009) First description of B chromosomes in the family Auchenipteridae, Parauchenipterus galeatus (Siluriformes) of the São Francisco River basin (MG, Brazil). Micron, Oxford, England, 40: 552–559. https://doi.org/10.1016/j.micron.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Martins C, Galetti PM Jr. (2001) Two 5S rDNA arrays in neotropical fish species: is it a general rule for fishes? Genetica 111: 439–46. https://doi.org/10.1023/A:1013799516717 [DOI] [PubMed]
- McStay B, Grummt I. (2008) The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology. Annual Review of Cell and Developmental Biology 24: 131–157. https://doi.org/10.1146/annurev.cellbio.24.110707.175259 [DOI] [PubMed] [Google Scholar]
- Medrano L, Bernardi G, Couturier J, Dutrillaux B. (1988) Chromosome banding and genome compartmentalization in fishes. Chromosoma 96: 178–183. https://doi.org/10.1007/BF00331050 [Google Scholar]
- MINSEGPRES (2008) Decreto Supremo N 51/2008. Aprueba y oficializa nómina para el tercer proceso de clasificación de especies según su estado de conservación. Santiago, Chile: Ministerio Secretaría General de la Presidencia, Gobierno de Chile. Diario Oficial de la República de Chile, 30 de junio de 2008.
- Morales P, Vila I, Poulin E. (2011) Genetic structure in remnant populations of an endangered cyprinodontid fish, Orestias ascotanensis, endemic to the Ascotán salt pan of the Altiplano. Conservation Genetics: 1639–1643. https://doi.org/10.1007/s10592-011-0245-6
- Ozouf-Costaz C, Brandt J, Körting C, Pisano E, Bonillo C, Coutanceau J, Volff J. (2004) Genome dynamics and chromosomal localization of the non-LTR retrotransposons Rex1 and Rex3 in Antarctic fish. Antarctic Science 16: 51–57. https://doi.org/10.1017/S0954102004001816 [Google Scholar]
- Pansonato-Alves JC, Hilsdorf AWS, Utsunomia R, Silva DMZA, Oliveira C, Foresti F. (2013a) Chromosomal mapping of repetitive DNA and cytochrome C oxidase I sequence analysis reveal differentiation among sympatric samples of Astyanax fasciatus (Characiformes, Characidae). Cytogenetic and Genome Research 141: 133–42. https://doi.org/10.1159/000354885 [DOI] [PubMed] [Google Scholar]
- Pansonato-Alves JC, Serrano ÉA, Utsunomia R, Scacchetti PC, Oliveira C, Foresti F. (2013b) Mapping five repetitive DNA classes in sympatric species of Hypostomus (Teleostei: Siluriformes: Loricariidae): analysis of chromosomal variability. Reviews in Fish Biology and Fisheries 23: 477–489. https://doi.org/10.1007/s11160-013-9303-0 [Google Scholar]
- Parenti LR. (1984) A taxonomic revision of the Andean killifish genus Orestias (Cyprinodontiformes, Cyprinodontidae). Bulletin of the AMNH; v. 178, article 2. Bulletin of the Americam Museum of the Natural History 178: 110–214. [Google Scholar]
- Pendas A, Moran P, Freije J, Garcia-Vazquez E. (1994) Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenetic and Genome Research 67: 31–36. https://doi.org/10.1159/000133792 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray J. (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proceedings of the National Academy of Sciences 83: 2934–2938. https://doi.org/10.1073/pnas.83.9.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano E, Ghigliotti L. (2009) Ribosomal genes in notothenioid fishes: Focus on the chromosomal organisation. Marine Genomics 2: 75–80. https://doi.org/10.1016/j.margen.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Sánchez-Romero O, Abad CQ, Cordero PQ, de Sene VF, Nirchio M, Oliveira C. (2015) First description of the karyotype and localization of major and minor ribosomal genes in Rhoadsiaaltipinna Fowler, 1911 (Characiformes, Characidae) from Ecuador Comparative Cytogenetics 9(2): 271–80. http://doi.org/10.3897/CompCytogen.v9i2.4504 [DOI] [PMC free article] [PubMed]
- Sene VF, Pansonato-Alves JC, Ferreira DC, Utsunomia R, Oliveira C, Foresti F. (2015) Mapping of the Retrotransposable Elements Rex1 and Rex3 in Chromosomes of Eigenmannia (Teleostei, Gymnotiformes, Sternopygidae). Cytogenetic and Genome Research. https://doi.org/10.1159/000441465 [DOI] [PubMed]
- Scacchetti PC, Alves JCP, Utsunomia R, Claro FL, de Toledo LFA, Oliveira C, Foresti F. (2012) Molecular characterization and physical mapping of two classes of 5S rDNA in the genomes of Gymnotus sylvius and G. inaequilabiatus (Gymnotiformes, Gymnotidae). Cytogenetic and Genome Research 136: 131–7. https://doi.org/10.1159/000335658 [DOI] [PubMed] [Google Scholar]
- Scacchetti PC, Utsunomia R, Pansonato-Alves JC, Da Silva GJC, Vicari MR, Artoni RF, Oliveira C, Foresti F. (2015) Repetitive DNA Sequences and Evolution of ZZ/ZW Sex Chromosomes in Characidium (Teleostei: Characiformes). Plos One 10: e0137231. https://doi.org/10.1371/journal.pone.0137231 [DOI] [PMC free article] [PubMed]
- Silva DMZA, Utsunomia R, Pansonato-Alves JC, Oliveira C, Foresti F. (2015) Chromosomal Mapping of Repetitive DNA Sequences in Five Species of Astyanax (Characiformes, Characidae) Reveals Independent Location of U1 and U2 snRNA Sites and Association of U1 snRNA and 5S rDNA. Cytogenetic and Genome Research. https://doi.org/10.1159/000438813 [DOI] [PubMed]
- Sola L, de Innocentils S, Gornung E, Papalia S, Rossi AR, Marino G, de Marco P, Cataudella S. (2000) Cytogenetic analysis of Epinephelus marginatus (Pisces: Serranidae), with the chromosome localization of the 18S and 5S rRNA genes and of the (TTAGGG)n telomeric sequence. Marine Biology 35: 47–51. https://doi.org/10.1007/s002270000334 [Google Scholar]
- Sola L, Rossi AR, Iaselli V, Rasch EM, Monaco PJ. (1992) Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenetics and cell genetics 60: 229–35. https://doi.org/10.1159/000133346 [DOI] [PubMed] [Google Scholar]
- Sumner A. (1972) A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75: 304–306. https://doi.org/10.1016/0014-4827(72)90558-7 [DOI] [PubMed] [Google Scholar]
- Tigano C, Rocco L, Ferrito V, Costagliola D, Pappalardo AM, Stingo V. (2004) Chromosomal mapping and molecular characterization of ribosomal RNA genes in Lebias fasciata (Teleostei, Cyprinodontidae). Genetica 121: 95–100. https://doi.org/10.1023/B:GENE.0000019931.89458.dc [DOI] [PubMed] [Google Scholar]
- Ubeda-Manzanaro M, Merlo M, Palazón J, Cross I, Sarasquete C, Rebordinos L. (2010) Chromosomal mapping of the major and minor ribosomal genes, (GATA)n and U2 snRNA gene by double-colour FISH in species of the Batrachoididae family. Genetica 138: 787–94. https://doi.org/10.1007/s10709-010-9460-1 [DOI] [PubMed] [Google Scholar]
- Utsunomia R, Scacchetti PC, Pansonato-Alves JC, Oliveira C, Foresti F. (2014a) Comparative chromosome mapping of U2 snRNA and 5S rRNA genes in Gymnotus species (Gymnotiformes, Gymnotidae): evolutionary dynamics and sex chromosome linkage in G . pantanal. Cytogenetic and Genome Research 142: 286–92. https://doi.org/10.1159/000362258 [DOI] [PubMed] [Google Scholar]
- Utsunomia R, Pansonato-Alves JC, Scacchetti PC, Oliveira C, Foresti F. (2014b) Scattered organization of the histone multigene family and transposable elements in Synbranchus. Genetics and Molecular Biology 37: 30–36. https://doi.org/10.1590/S1415-47572014000100007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomia R, de Silva DMZA, Ruiz-Ruano FJ, Araya-Jaime C, Pansonato-Alves JC, Scacchetti PC, Hashimoto DT, Oliveira C, Trifonov VA, Porto-Foresti F, Camacho JPM, Foresti F. (2016) Uncovering the Ancestry of B Chromosomes in Moenkhausia sanctaefilomenae (Teleostei, Characidae). Plos One 11: e0150573. https://doi.org/10.1371/journal.pone.0150573 [DOI] [PMC free article] [PubMed]
- Valente GT, Mazzuchelli J, Ferreira IA, Poletto AB, Fantinatti BEA, Martins C. (2011) Cytogenetic Mapping of the Retroelements Rex1, Rex3 and Rex6 among Cichlid Fish: New Insights on the Chromosomal Distribution of Transposable Elements. Cytogenetic and Genome Research 133: 34–42. https://doi.org/10.1159/000322888 [DOI] [PubMed] [Google Scholar]
- Vila I, Mendez M, Scott S, Morales P, Poulin E. (2007) Threatened fishes of the world: Orestias ascotanensis Parenti, 1984 (Cyprinodontidae). Environmental Biology of Fishes 80: 491–492. https://doi.org/10.1007/s10641-006-9150-0 [Google Scholar]
- Vila I, Scott S, Lam N, Iturra P, Méndez MA. (2010) Karyological and morphological analysis of divergence among species of the killifi sh genus Orestias (Teleostei: Cyprinodontidae) from the southern Altiplano. In: Nelson JS, Schultze H, Wilson MHV (Eds) Origin and Phylogenetic Interrelationships of Teleosts. Verlag Dr. Friedrich Pfeil, Munchen, 480.
- Vila I, Scott S, Mendez M, Iturra P, Valenzuela F, Poulin E. (2011) Orestias gloriae, a new species of cyprinodontid fish from saltpan spring of the southern high Andes (Teleostei: Cyprinodontidae). Ichthyological Exploration of Freshwaters 22: 345–353. [Google Scholar]
- Villwock W, Sienknecht U. (1996) Contribución al conocimiento e historia de los peces chilenos. Los Cyprinodóntidos del género Orestias Val. 1839 (Teleostei: Cyprinodontidae) del altiplano chileno. Medio Ambiente 1: 119–126. [Google Scholar]
- Vitturi R, Colomba M, Vizzini S, Libertini A, Barbieri R. (2005) Chromosomal location polymorphism of major rDNA sites in two Mediterranean populations of the killifish Aphanius fasciatus (Pisces: Cyprinodontidae). Micron 3: 243–246. dx.doi.org/10.1016/j.micron.2004.11.006 [DOI] [PubMed]
- Völker M, Ráb P, Kullmann H. (2005) Karyotype Differentiation in Chromaphyosemion Killifishes (Cyprinodontiformes, Nothobranchiidae). I: Chromosome Banding Patterns of C. alpha, C. kouamense and C. lugens. Genetica 125: 33–41. https://doi.org/10.1007/s10709-005-4267-1 [DOI] [PubMed] [Google Scholar]
- Völker M, Rab P, Kullmann H. (2008) Karyotype differentiation in Chromaphyosemion killifishes (Cyprinodontiformes, Nothobranchiidae): patterns, mechanisms, and evolutionary implications. Biological Journal of the Linnean Society: 143–153. https://doi.org/10.1111/j.1095-8312.2008.00967.x
- Völker M, Sonnenberg R, Ráb P, Kullmann H. (2006) Karyotype differentiation in Chromaphyosemion killifishes (Cyprinodontiformes, Nothobranchiidae). II: cytogenetic and mitochondrial DNA analyses demonstrate karyotype differentiation and its evolutionary direction in C. riggenbachi. Cytogenetic and Genome Research 115: 70–83. https://doi.org/10.1159/000094803 [DOI] [PubMed] [Google Scholar]
- Völker M, Sonnenberg R, Ráb P, Kullmann H. (2007) Karyotype differentiation in Chromaphyosemion killifishes (Cyprinodontiformes, Nothobranchiidae). III: extensive karyotypic variability associated with low mitochondrial haplotype differentiation in C. bivittatum. Cytogenetic and Genome Research 116: 116–26. https://doi.org/10.1159/000097429 [DOI] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J. (Eds) PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc., New York, 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
- Yano CF, Yano A, Poltronieri J, Bertollo LAC, Artoni RF, Liehr T, de Cioffi MB. (2014) Chromosomal mapping of repetitive DNAs in Triportheus trifurcatus (Characidae, Characiformes): insights into the differentiation of the Z and W chromosomes. Plos One 9: e90946. https://doi.org/10.1371/journal.pone.0090946 [DOI] [PMC free article] [PubMed]