Abstract
The chromatin structure of DNA determines genome compaction and activity in the nucleus. On the basis of in vitro structures and electron microscopy (EM) studies, the hierarchical model is that 11-nanometer DNA-nucleosome polymers fold into 30- and subsequently into 120- and 300-to 700-nanometer fibers and mitotic chromosomes. To visualize chromatin in situ, we identified a fluorescent dye that stains DNA with an osmiophilic polymer and selectively enhances its contrast in EM. Using ChromEMT (ChromEM tomography), we reveal the ultrastructure and three-dimensional (3D) organization of individual chromatin polymers, megabase domains, and mitotic chromosomes. We show that chromatin is a disordered 5- to 24-nanometer-diameter curvilinear chain that is packed together at different 3D concentration distributions in interphase and mitosis. Chromatin chains have many different particle arrangements and bend at various lengths to achieve structural compaction and high packing densities.
In 1953, Watson and Crick determined that DNA forms a double helix, which provided a structural basis for how our genetic information is stored and copied (1). However, the double helix captures only the first-order structure of DNA. In the nucleus, DNA is assembled into chromatin structures that determine the activity and inheritance of human genomic DNA. A 147–base pair (bp)–length of DNA is wrapped around an octamer of histones H2A, H2B, H3, and H4 into an 11-nm DNA–core nucleosome particle (2). Each DNA-nucleosome particle is separated by 20 to 75 bp of DNA that can bind to histone H1 (3). However, to fit 2 m of human genomic DNA into the nucleus, a further level of structural compaction is thought to be necessary. The long-standing model in most textbooks is that primary DNA-nucleosome polymers progressively fold into discrete higher-order chromatin fibers and, ultimately, mitotic chromosomes (Fig. 1A) (4, 5). However, the hierarchical folding model is based on chromatin structures that are formed in vitro by reconstituting purified DNA and histones (6–9) or in permeabilized cells from which other components had been extracted (10, 11). Thus, a remaining question is, what is the local chromatin polymer structure and three-dimensional (3D) organization of human genomic DNA in the nucleus of interphase and mitotic cells in situ?
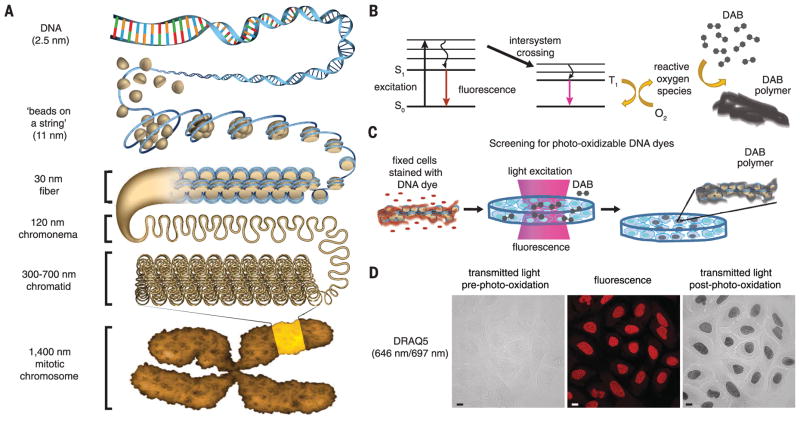
Fig. 1. A fluorescent DNA-binding dye that catalyzes local DAB polymerization on chromatin in the nucleus.
(A) Hierarchical chromatin-folding model. (B) Excited fluorophores that undergo intersystem crossing generate reactive oxygen species that catalyze DAB polymerization. S0, ground state; S1, excited singlet state; T1, excited triplet state. (C) Schema for cell-based screen for DNA-binding dyes that photo-oxidize DAB. (D) U2OS cells were fixed with glutaraldehyde and stained with DRAQ5. Cells were incubated with DAB and excited by continuous epifluorescence illumination for 5 min. DAB photo-oxidation was identified by the appearance of dark DAB precipitates in the nucleus. Fluorescence (middle), transmitted-light images pre– (left panel) and post–photo-oxidation (right panel). Scale bar, 10 μm. See Movie 1 for photo-oxidation of DAB by DRAQ5.
In vitro reconstituted purified nucleosomes and DNA in low salt form “beads-on-a-string” structures, 2.5-nm DNA threads decorated with discrete 11-nm nucleosome particles (12, 13). The hierarchical model (Fig. 1A) proposes that primary DNA-nucleosome polymers fold into secondary 30-nm fibers. Electron microscopy (EM) and x-ray crystallography studies of up to 2 kb of DNA reconstituted with nucleosomes in vitro support two different structural models of the 30-nm fiber, referred to as the solenoid and zigzag fiber models (7–9). The solenoid fiber structure has a diameter of 33 nm with six nucleosomes every 11 nm along the fiber axis (7). The two-start zigzag fiber has a diameter of 27.2 to 29.9 nm with five to six nucleosomes every 11 nm (8, 9). The 30-nm fiber is thought to assemble into helically folded 120-nm chromonema, 300- and 700-nm chromatids, and mitotic chromosomes (Fig. 1A) (14–18). The chromonema structures (measured between 100 and 130 nm) are based on EM studies of permeabilized nuclei from which other components had been extracted with detergents and high salt to visualize chromatin (10, 11).
However, there have been cryo-EM (19, 20), x-ray scattering (21), and electron spectroscopy imaging (ESI) studies (22, 23) o f the nucleus do not support the hierarchical chromatin-folding model. However, the 3D sampling volume of ESI is limited, and other cellular components have to be extracted to visualize the weak phosphorous signals of DNA. In cryo-EM tomography, details arise from the phase contrast between the atoms of the molecules and those of the vitreous ice. In recent cryo-EM studies of thin lamellae of nuclear membrane regions of cultured cells prepared with cryo-focused ion-beam milling (24), microtubules, ribosomes, and nuclear pore structures could be visualized. However, the contrast of DNA in vitreous ice is very poor (25), and chromatin cannot be identified unambiguously or have its ultrastructure and 3D organization reconstructed through large nuclear volumes.
Super-resolution light microscopy and fluorescent labels can provide an estimate of relative DNA and chromatin compaction at specific genomic loci (26–31). However, EM is required to directly visualize chromatin ultrastructure. To visualize chromatin in situ requires heavy-metal stains that selectively enhance the contrast of DNA under the electron microscope. Conventional EM stains, such as osmium tetroxide (OsO4), uranium acetate, and lead salts, preferentially bind to lipids, proteins, and RNA, respectively, and either do not react with DNA or do not stain it selectively (32–34). Osmium ammine binds to DNA but requires harsh acid treatments that destroy native chromatin structure (35, 36). To overcome these limitations, we have developed a DNA-labeling method, ChromEM, which, together with advances in multitilt EMT, enables the chromatin ultrastructure and 3D organization of megabases of DNA to be visualized in the nucleus of resting and mitotic human cells in situ.
A cell-based screen identifies a fluorescent DNA-binding dye that photo-oxidizes diaminobenzidine (DAB)
Upon excitation, there are certain fluorophores that not only emit a photon to return to the ground state but also undergo intersystem crossing (37) (Fig. 1B). In cells, this leads to the local generation of reactive oxygen species that can be harnessed in situ to catalyze the polymerization of DAB on the surface of fluorescently labeled macromolecules (38–40), enabling visualization by EM (39, 41). For example, miniSOG (mini singlet oxygen generator), a green fluorescent protein that photo-oxidizes DAB, has been used as a genetic tag to visualize proteins with correlated light and EM (42, 43). We reasoned that a fluorescent DNA-binding dye that photo-oxidizes DAB would be a powerful probe to visualize DNA and chromatin ultrastructure in the nucleus. To identify such a probe, we developed a cell-based assay and screened fluorescent DNA dyes for their ability to photo-oxidize DAB (Fig. 1C). Wavelengths below 400 nm induce DAB autopolymerization, resulting in brown precipitates and nonspecific staining (39, 44, 45). Therefore, we focused on DNA-binding dyes that are excited at longer wavelengths. Human osteosarcoma U2OS cells were fixed with glutaraldehyde, stained with fluorescent DNA dyes, and then excited that in the presence of DAB. The photo-oxidation of DAB was identified by the appearance of dark precipitates in the nucleus. An example of the data for some of the far-red dyes that we screened is shown (fig. S1A). Of the DNA dyes we screened, only DRAQ5 (deep-red fluorescing anthraquinone Nr. 5) excitation photo-oxidized DAB (Fig. 1D and Movie 1).
Movie 1. Time-lapse imaging of DAB photo-oxidation upon excitation of DRAQ5-labeled DNA.
U2OS cells were fixed with glutaraldehyde, DNA-stained with DRAQ5, bathed in DAB, and excited by continuous illumination for 6 min (light flash) under the microscope. The movie shows the live transmitted-light images of DRAQ5-induced DAB photo-oxidation on chromatin in the nucleus. DAB polymerization can be identified by the appearance of dark precipitates in cell nuclei.
DRAQ5 is a membrane-permeable anthraquinone dye [excitation wavelength/maximum emission wavelength of 646 nm/697 nm] that binds to double-stranded DNA and is used routinely in live-cell imaging studies (fig. S1B) (46, 47). There are up to 14 minor groove-binding sites for DRAQ5 in the 147 bp of DNA that wrap around each nucleosome core particle (fig. S1C). DRAQ5 does not have an A-T–base pair binding preference and labels chromatin in the nucleus with a pattern similar to H2B fused to green fluorescent protein (48). We show that regardless of whether DRAQ5 is used to stain DNA in live cells before fixation or after either paraformaldehyde or glutaraldehyde fixation, DRAQ5 excitation catalyzes DAB polymerization on chromatin in the nucleus (fig. S1, D and E). In live cells, DRAQ5 binding to DNA could potentially displace RNA polymerase II–transcription factor complexes (49) and histone H1 (50). Therefore, we fixed cells and DNA-chromatin complexes with glutaraldehyde before staining DNA with DRAQ5. Glutaraldehyde is a protein cross-linker that preserves cellular ultra-structure and minimizes the diffusion of DAB precipitates (39).
ChromEM staining enables DNA in the nucleus to be visualized by EM
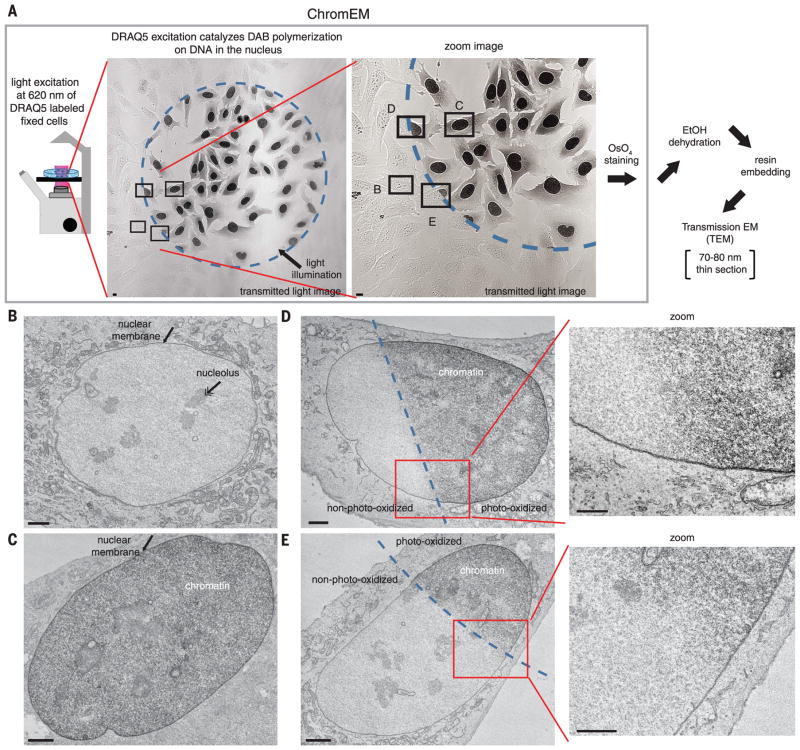
In EM, OsO4 is generally used to fix and stain cell membranes (51). However, OsO4 also binds to DAB polymers with a high affinity (41). Therefore, we determined if OsO4 stains DAB precipitates on the surface of DRAQ5-labeled DNA, enabling chromatin to be visualized by EM. U2OS cells were fixed, labeled with DRAQ5, and excited in the presence of DAB. The transmitted-light image shows that dark DAB precipitates only form in the nucleus of cells within the excitation field (Fig. 2A, dashed circle). The entire plate of cells was stained with OsO4, and thin sections (70 to 80 nm) were prepared for analysis by transmission EM (TEM) (Fig. 2, B to E). In the nucleus of non–photo-oxidized cells, OsO4 stains the nuclear membrane and the nucleolus but not chromatin (Fig. 2B), consistent with previous EM studies (52). However, in DAB photo-oxidized cells, DNA and chromatin in the nucleus are stained by OsO4 and visible in EM micrographs (Fig. 2C). The TEM images of cells bisected by the excitation field are particularly convincing (Fig. 2, A, D, and E). DNA and chromatin are only stained in the half of the cell nucleus that was in the excitation field, whereas cytoplasmic structures and membranes are stained evenly throughout the cell (Fig. 2, D and E). We conclude that DRAQ5 photo-oxidation catalyzes the deposition of osmiophilic DAB polymers on DNA, enabling it to be visualized in the nucleus by EM. We refer to DRAQ5 DNA labeling, DAB photo-oxidation, and OsO4 staining of chromatin as ChromEM.
Fig. 2. ChromEM: DRAQ5 excitation photo-oxidizes DAB on DNA in the nucleus and enables chromatin to be visualized by osmium staining in EM.
(A) U2OS cells were fixed with glutaraldehyde, stained with DRAQ5, and DAB photo-oxidized for 7 min. Cells were then stained with OsO4 and thin sections prepared for TEM. EtOH, ethanol. Scale bar, 10 μm. (B to E) Magnified views of the cells in the labeled rectangles in panel (A). TEM images of a cell outside the photo-oxidation field (B), inside the photo-oxidation field (C), and bisected by the excitation field (D) and (E). Zoom-in (red box) is shown to the right. Nuclear membrane (arrow) and nucleolus (double arrow) are shown. Scale bar, 2 μm.
ChromEM and multitilt EMT enable chromatin ultrastructure to be resolved in situ
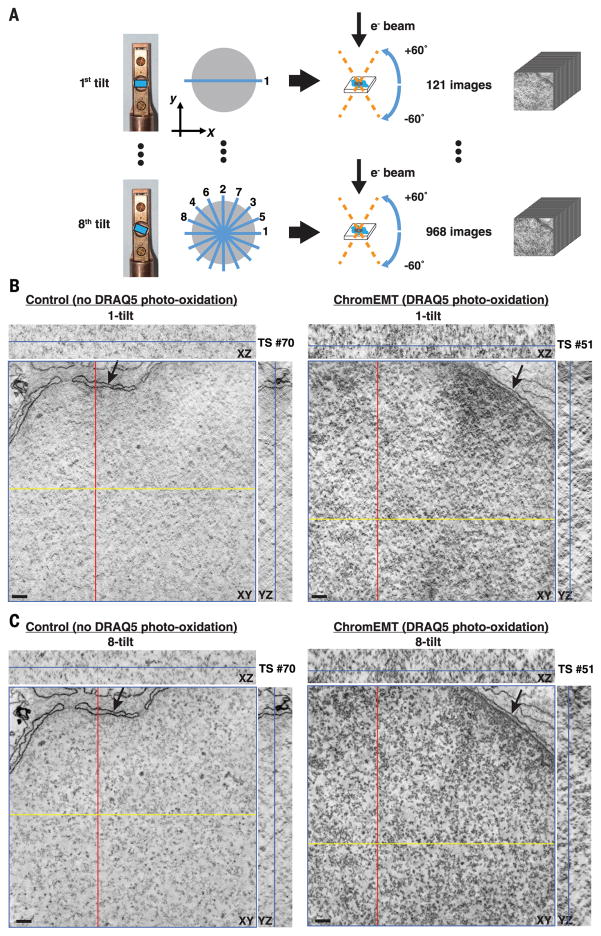
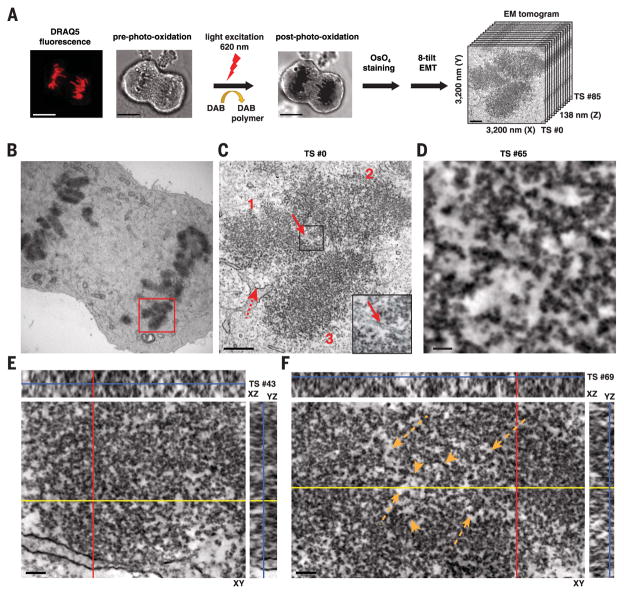
TEM micrographs of ChromEM-stained DNA are 2D projections of chromatin in 70-nm-thick sections. To visualize individual chromatin polymers and reconstruct their 3D ultrastructure through large nuclear volumes requires EMT. Human small-airway epithelial cells (SAECs) were labeled with DRAQ5, incubated with DAB, and either excited to catalyze DAB photo-oxidation or left unexcited (control). Cells were then stained with OsO4. Typically, a single-tilt EMT series (Fig. 3A) is used. However, the reconstructed cube volume of densities has a “missing wedge” of data along the axis of the tilting plane (53). To improve on this and increase axial resolutions (54), we applied an eight-tilt collection scheme (Fig. 3A) (55) and reconstructed the images using an extended TxBR software package (56, 57).
Fig. 3. ChromEMT and eight-tilt EMT enable chromatin to be visualized with high-contrast and spatial resolutions through large 3D volumes.
SAECs were fixed with glutaraldehyde and stained with DRAQ5. Cells were either left untreated (control) or excited for 6 min to photo-oxidize DAB. Samples were stained with OsO4, cut into 250-nm sections, and imaged by EMT. (A) Single-(121 images) and eight-tilt EMT data sets (968 images) were collected by rotating the sample block from –60° to +60° at the depicted orientations (blue lines). (B) Collected data set was reconstructed into an EM tomogram of 1666 nm (x) by 1619 nm (y) by 231 nm (z). Tomographic slices (TSs) from single-tilt data sets of control (left) and photo-oxidized nuclei (right). The xy image and the respective xz and yz cross-sectional views are shown.The blue line in the xz and yz cross sections shows the z position of the xy image. Scale bar, 100 nm. (C) Same as (B) for an eight-tilt data set.
Single- and eight-tilt–series EMTs were collected for both control and photo-oxidized cells. The EMT data sets comprise 141 individual 1.64-nm-thick tomographic slices (TSs) that go from the top (TS #0) to the bottom (TS #140) of the volume. OsO4 reacts with lipids in the membranes of the nuclear envelope and provides a useful normalization reference for the enhanced contrast of chromatin, specifically in photo-oxidized samples (Fig. 3, B and C). In control cells, there are some isolated particles and amorphous threads that are weakly stained by OsO4 (Fig. 3B, left panel) and visible with multitilt averaging (Fig. 3C, left panel), but chromatin is not visible or stained by OsO4. However, in DAB photo-oxidized cells, chromatin chains can be visualized with high contrast and definition (Fig. 3, B and C, right panels). Chromatin is the darkly stained particulate polymer that percolates and twists through the nuclear volume. The advantage of eight-tilt versus single-tilt EMT is also apparent. The tomographic averaging of 968 images results in more refined structures and improved axial resolutions that allow the trajectory of chromatin chains to be visualized through the 3D volume.
Chromatin is a disordered 5- to 24-nm-diameter granular chain that is packed together at different concentration densities in the nucleus
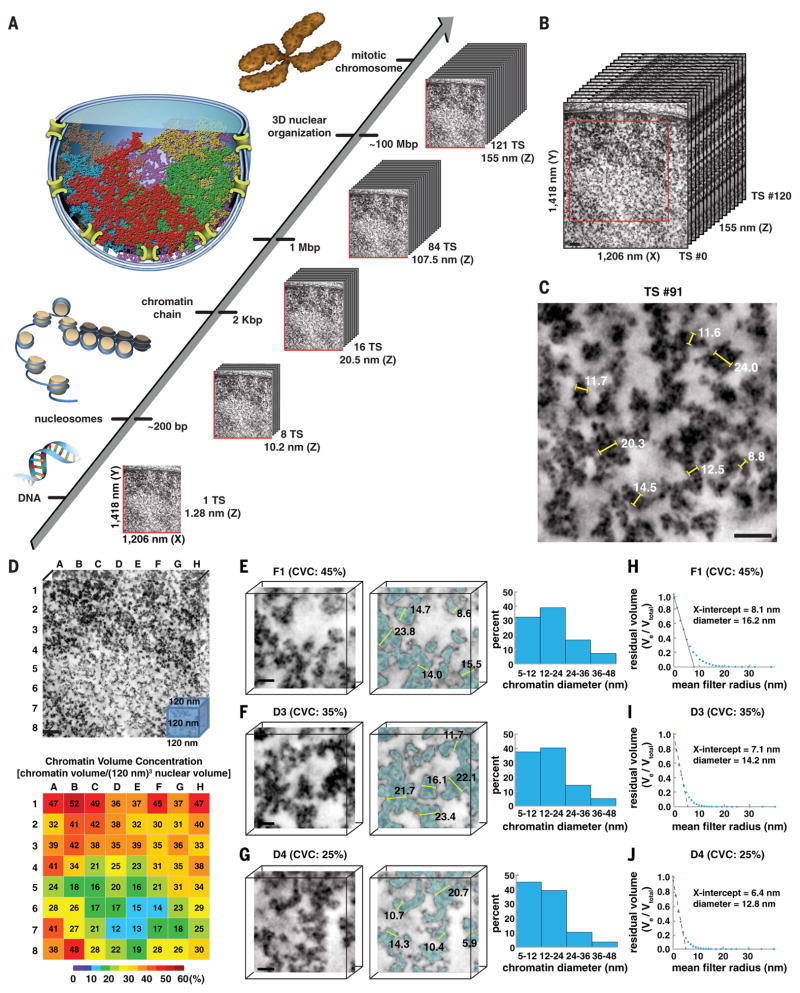
ChromEM and multitilt EMT (which we refer to as ChromEMT) enable the direct visualization of chromatin across a critical range of structural and biologically relevant scales in the nucleus, from individual chromatin chains to heterochromatin domains and mitotic chromosomes in serial TSs (Fig. 4A). To visualize chromatin ultrastructure and packing in interphase cells, we collected an eight-tilt EMT data set of ChromEM-stained SAECs (fig. S2). The EMT volume (1206-nm–by–1418-nm–by–155-nm volume) comprises 121 serial TSs (Fig. 4B). Each TS is 1.28 nm thick, which enables individual chromatin chains to be resolved. By compiling serial TSs into a “movie,” chromatin chains can be visualized as a continuum from the top to the bottom of the entire nuclear volume (Movie 2). Stepping through serial TSs, gaps appear in the nuclear membrane that correspond to the insertion sites of nuclear pore complexes (NPCs). There are a total of five nuclear pore insertion sites in the EMT volume. The NPC protein basket is not visible, underscoring the selectivity of ChromEM staining for DNA and chromatin. At the nuclear membrane, chromatin is packed together at higher concentrations, which makes the trajectory of individual chains difficult to follow. Chromatin chains weave back and forth and interact with each other and the lamina at multiple points and shortly interspersed intervals, resulting in a dense 3D mesh and heterochromatin domain.
Fig. 4. Chromatin is a disordered chain that has diameters between 5 and 24 nm and is packed together at different concentration densities in interphase nuclei.
(A) ChromEMT enables the ultrastructure of individual chromatin chains, megabase domains, and mitotic chromosomes to be resolved and visualized as a continuum in serial slices through large 3D volumes. (B) Reconstructed eight-tilt EMT data set (SAEC #1) of ChromEM-stained SAECs comprising 121 TSs (each 1.28 nm thick). Scale bar, 100 nm. To visualize chromatin and 3D organization as a continuum through the entire EMT data set, we compiled serial slices into a movie (Movie 2). (C) Manual measurements of chromatin diameters in a single TS. Scale bar, 50 nm. (D) The central EMT volume [red box in (B), 963 nm by 963 nm by 120 nm] was divided into an 8-by-8 grid comprising 64 subvolumes of 120-nm cubes. Chromatin volume concentrations (CVCs) are shown in the heat map. Scale bar, 100 nm. (E to G) The surface-thickness function was used to determine chromatin diameters in subvolumes with high (45%), medium (35%), and low (25%) CVCs. Irrespective of CVC, there are two major bin peak distributions for chromatin diameter: 5 to 12 nm and 12 to 24 nm. Scale bar, 20 nm. (H to J) Continuous erosion analysis to determine average chromatin diameter. The residual chromatin volume (Ve/Vtotal) is plotted against the spherical mean filter radius. The average radius of chromatin in each subvolume is the x-axis intercept of a linear fit of the first five erosion factor sizes.
Movie 2. The chromatin ultrastructure and 3D organization of the human genome in the nucleus.
ChromEM staining and multitilt EMT enable the ultrastructure of individual chromatin chains to be resolved and their 3D organization to be visualized as a continuum in the context of the nuclear architecture. Resting human SAECs were fixed, stained with ChromEM, and an eight-tilt EM tomogram (29,000×) was collected of a 250-nm-thick section (SAEC #1 from Fig. 4B). The reconstructed EMT volume is 1206 nm (x) by 1418 nm (y) by 155 nm (z) and comprises 121 TSs (each 1.28 nm thick). We compiled serial TS slices (TS #0 to TS #120) into a movie that enables chromatin to be visualized as a continuum from the top to the bottom of the nuclear volume. Chromatin is a disordered particulate chain that is packed together at different concentrations in the nucleus, with higher densities at the nuclear lamina. The gaps in the nuclear membrane correspond to the insertion sites of nuclear pores.
The hierarchical folding model predicts that 30- and 120-nm fibers are the predominant structural forms of chromatin in the nucleus (Fig. 1A). However, manual measurements in a single TS image indicate that chromatin diameters vary between ~8 and 24 nm (Fig. 4C). A caveat is that we are only analyzing chromatin in a small region and fraction of the entire nuclear volume. The concentration and staining density of chromatin appears to be different in certain regions of the nucleus, for example, heterochromatin domains at the nuclear membrane versus central regions (Fig. 4B and Movie 2). This leads to two interrelated questions: What is the 3D concentration of chromatin in interphase nuclei and are higher packing densities associated with the assembly of chromatin into higher-order 30- to 120-nm fibers (Fig. 1A)?
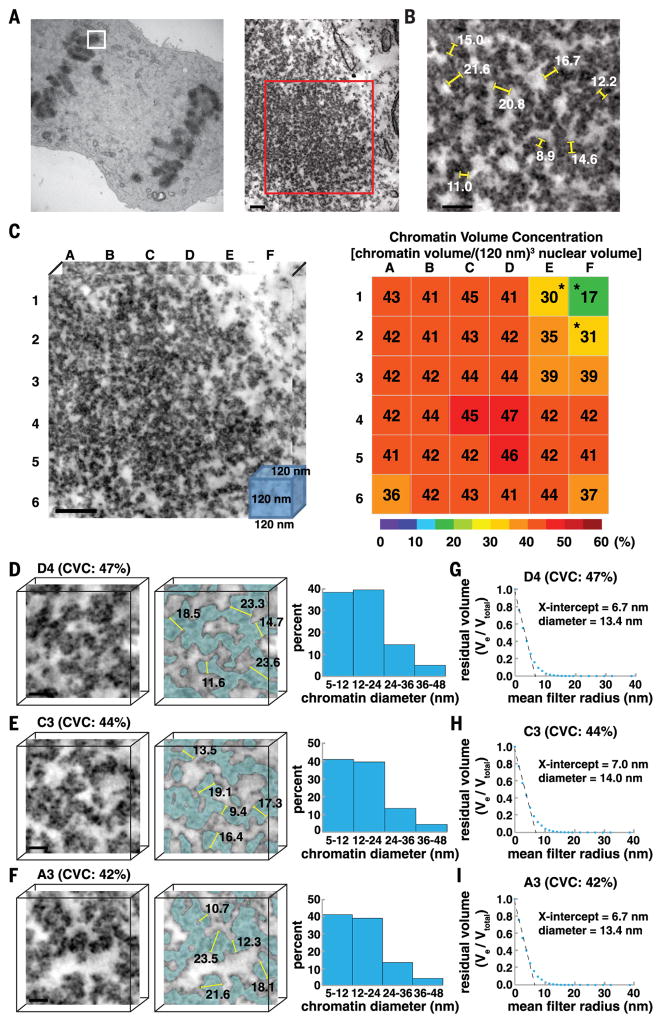
To address these questions, we developed a spatially aware analytical pipeline to systematically quantify chromatin diameters and 3D packing in large nuclear volumes. The convoluted trajectories of chromatin (Movie 2) and large volumes make manual segmentation impractical. However, the high contrast of ChromEM-stained chromatin facilitates automated segmentation. We explored several different algorithms and workflows, for example, local contrast-limited adaptive histogram equalization (CLAHE) (58) followed by global thresholding using either the Li (59) or Otsu (60) methods (fig. S3A). CLAHE with a block size of 100 nm followed by global Li thresholding had the highest accuracy (88 to 94%) and precision (59 to 92%) when compared with manual segmentation of chromatin (ground truth) in the same EMT subvolumes (fig. S3B and table S1).
To determine the 3D concentration of chromatin in the nucleus, we subdivided the EMT nuclear volume (Fig. 4B, red box) into an 8-by-8 gridded reference map comprising 64 subtomogram volumes (Fig. 4D). Each subtomogram is a cube that has x, y, and z dimensions of 120 nm. We chose a 120-nm3 volume as it is large enough to capture higher-order 30- and 120-nm chromatin fibers. We define the chromatin volume concentration (CVC) as the percentage of chromatin volume in each 120-nm3 nuclear volume (chromatin volume/120-nm3 volume). The results are displayed in a heat map: low CVC (0 to 20%, blue), medium CVC (20 to 35%, green to yellow), and high CVC (35 to 60%, orange to red) (Fig. 4D). In interphase SAECs, CVCs range from 12% (D7) to 52% (B1), with a mean value of 30 ± 10%. Heterochromatin domains at the nuclear envelope have higher CVCs (37 to 52%) compared to subvolumes toward the center of the nucleus (12 to 21%) (Fig. 4D). We conclude that chromatin chains are packed together at different concentration densities and spatial distributions in the nucleus of resting human cells.
Next, we determined if high CVCs are associated with the assembly of discrete higher-order chromatin fibers. To this end, we first rendered the 3D surface of chromatin as a polygonal mesh of interconnecting triangles and then measured chromatin diameter using the Amira surface-thickness function (61). Chromatin diameter is calculated by measuring the shortest normal distance between the vertices of triangles on each opposing surface. To illustrate how this works, we applied the surface-thickness function to two 21.7-nm cylinders (fig. S4, A to C). The results are shown as a histogram with major and minor bin peak distributions of 20 to 25 nm and 35 to 40 nm, respectively (fig. S4D). The bin distribution is due to imperfect polygonal meshing of 3D objects (fig. S4C). The major bin distribution peak contains the cylinder diameter, and the minor peak contains the length of each cylinder.
We applied the surface-thickness algorithm to determine the chromatin diameters in sub-volumes that have high to low CVCs: F1 (45%), D3 (35%), and D4 (25%) (Fig. 4, E to G). There are two major bin distribution peaks for chromatin diameters, 5 to 12 nm and 12 to 24 nm (Fig. 4, E to G). The two bins reflect the structural heterogeneity of chromatin chains that have different diameters along their lengths. In nuclear volumes that have CVCs ≥35%, the 12- to 24-nm–chromatin diameter bin is more frequent (fig. S5).
We also developed an alternative method to estimate the average chromatin diameter that does not require polygonal meshing. In continuous erosion analysis (fig. S6), chromatin voxels are assigned a value of 1 and interchromatin space a value of 0. Spherical mean filters of increasing radii are then used to erode the respective volumes. The average chromatin radius is the x-axis intercept of a plot of residual chromatin volume (Ve/Vtotal) versus spherical mean filter erosion radii, where Ve is the eroded volume. The average diameter of chromatin in subtomograms with different CVCs is 16.2 nm (F1, 45%), 14.2 nm (D3, 35%), and 12.8 nm (D4, 25%) (Fig. 4, H to J). These data are consistent with surface-thickness estimates of chromatin-diameter distributions.
In addition, we analyzed two additional SAEC EMT data sets (figs. S7 and S8). CVC distributions (SAEC #2, 16 to 52%; SAEC #3, 14 to 50%) are similar to SAEC #1. Furthermore, there are two major bin distributions for chromatin diameters, 5 to 12 nm and 12 to 24 nm (figs. S7, C and D, and S8, C and D).
ChromEM stains higher-order 30-nm fibers induced in in vitro purified chicken erythrocyte nuclei treated with magnesium chloride (MgCl2)
We fail to detect 30- and 120-nm higher-order chromatin fibers in SAECs even in nuclear regions with high CVCs (Fig. 4, E to J, and figs. S5, S7, C and D, and S8, C and D). X-ray scattering and cryo-EM studies from Langmore et al. (62) and Scheffer et al. (63) observed 40- and 30-nm fibers, respectively, in nuclei purified from hypotonically lysed chicken erythrocytes and treated with MgCl2 to induce chromatin compaction. Therefore, we repeated these experiments to determine if ChromEM can stain and detect the induction of higher-order chromatin fibers in these conditions. After hypotonic swelling and lysis, we purified nuclei from chicken erythrocytes and induced chromatin compaction with 2 mM MgCl2 (fig. S9A). Nuclei were then plated on poly-lysine coated plates, fixed with glutaraldehyde, and processed for ChromEM staining and eight-tilt EMT (fig. S9, A and B).
Multitilt tomography series requires plastic-embedded samples that can tolerate higher levels of irradiation than the vitreous-ice embedding of cryo-EM samples. A caveat is that plastic embedding and beam-induced mass loss can decrease a structure’s volume (53). However, similar to previous studies of MgCl2-treated chicken erythrocytes (62, 63), we observe large chromatin clusters that span 100 to 200 nm (fig. S9C). The high resolutions of multitilt EMT and ChromEM reveal that these large clusters are made up of individual chromatin fibers. At the edges of clusters, it is easier to visualize individual chromatin fibers (fig. S9D), which have diameters between 30 and 40 nm and are made up of closely interacting particles arranged in semiordered arrays along their axes.
In addition to manual measurements, we also determined if our automated segmentation and surface-thickness workflows detect higher-order chromatin fibers. The lack of space between chromatin fibers in the densely packed clusters make measurements with the surface-thickness algorithm more fraught than in the SAEC data sets. Therefore, we applied the algorithm to sub-volumes at the edges of clusters. The two major bin distributions for chromatin diameter are 12 to 24 nm and 24 to 36 nm. There is also a high frequency of diameters greater than 48 nm, which likely reflects the merged diameters of closely interacting chromatin fibers within the clusters that cannot be distinguished by the algorithm (fig. S9E). These data demonstrate that our automated work-flows and algorithms detect higher-order chromatin fibers. We conclude that ChromEM stains higher-order chromatin clusters and fibers and enables their ultrastructure and 3D volumes to be reconstructed in EMT data sets.
DNA assembles disordered chromatin chains that have different particle arrangements, conformations, and compact 3D motifs
In contrast to higher-order fibers induced in MgCl2-treated chicken erythrocyte nuclei, in nonlysed human SAECs, chromatin is a disordered chain with diameters ranging from 5 to 24 nm (Fig. 4, E to J; figs. S5, S7, C and D, and S8, C and D; and Movie 2). In nuclear sub-volumes that have high CVCs (≥38%) the peak chromatin diameter distribution is 12 to 24 nm (fig. S5). However, in subvolumes that have low CVCs (≤20%), the 5- to 12-nm-diameter bin distribution dominates (Fig. 5A and fig. S5). Consistent with this, we observe chromatin chains that have thin threads decorated with discrete single particles in these subvolumes (Fig. 5, B and C). These chromatin chains resemble beads-on-a-string structures of nucleosomes and DNA reconstituted in vitro in the absence of linker histones (12). The DNA–nucleosome core particle has dimensions of 11 nm (face) by 5.5 nm (side) by 11 nm (height) (fig. S1C) (8). Our resolution is not sufficient for detailed docking, but manual placements without regard to orientation show that the EMT particle densities match the general dimensions of nucleosomes (movies S1 and S2).
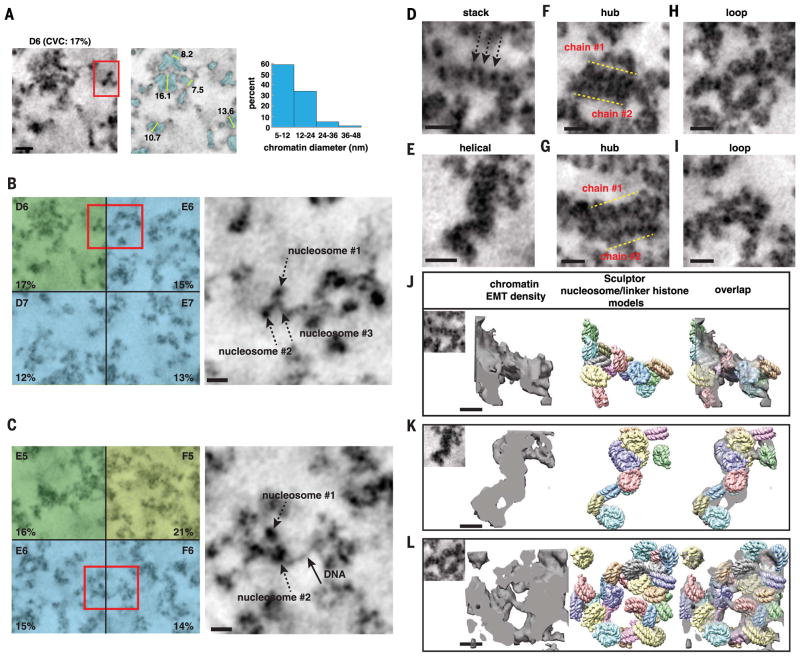
Fig. 5. DNA and nucleosomes form disordered chromatin chains that have different particle arrangements, conformations, densities, and 3D motifs.
(A) In interphase subvolumes that have low CVCs (<20%), chromatin chains have predominantly 5- to 12-nm diameter bin distributions (see fig. S5). Left, single TS (D6, SAEC #1); middle, segmented chromatin; right, chromatin-diameter range. Scale bar, 20 nm. (B to C) Magnified images of chromatin in volumes with low CVCs (red boxes). Chromatin chains have thin threads decorated with discrete particles (right panels: single TS, scale bar 20 nm). See movies S1 and S2. (D to I) Gallery of different structures and motifs: (D) short linear nucleosome stack, (E) helical twist, (F) and (G) two chromatin chains interact in parallel to form a hub, and (H) and (I) loops between and within chromatin chains. Scale bar, 20 nm. (J to L) Sculptor models (middle) of nucleosome–linker histone H5 (PDB 4QLC) in EMT densities of chromatin stack, helical twist, and loop (left panels). Overlap of the Sculptor models and EMT densities (right). One Sculptor solution is shown. Cross-correlation values: 82% (J), 92% (K), and 86% (L). Additional Sculptor solutions in fig. S11. Scale bar, 11 nm.
DNA in chromatin chains with discrete particles and in nuclear subvolumes with low CVCs would be more accessible for transcription, and these structural features may be hallmarks of active euchromatin. The majority of chromatin chains have continuous granular structures and many different particle arrangements (Fig. 5, D to I, and fig. S10). For example, we observe chromatin chains with short linear segments of what appear to be stacked nucleosomes (Fig. 5D, arrows). There are chromatin chains with distinct helical twists (Fig. 5E). There are also numerous instances of chromatin chains that converge and interact closely for short sections. In Fig. 5, F and G, two chromatin chains interact in parallel to form a 3D “hub” and have a combined diameter of ~22 to 24 nm at their intersection. Another recurring motif is the formation of loops of all different sizes between and within chromatin chains (Fig. 5, H and I). There are many variations on each of these motifs and a catalog of other conformations.
DRAQ5 excitation catalyzes the deposition of osmiophilic DAB polymers on the surface of DNA and any intimately associated DNA- and chromatin-interacting proteins in the nucleus. Thus, modeling disordered chromatin-chain structures is challenging because of their variability as well as the large number of unknowns in situ compared to in vitro structures of highly purified and uniform DNA-nucleosome units. The simultaneous multidomain fitting function (64) in Sculptor (65) is a genetic algorithm–based computational method for simultaneously fitting multiple atomic structures into an EM density map at resolutions as low as 40 Å. A genetic algorithm is a method for solving both constrained and unconstrained optimization problems by using a selection process that mimics evolution by repeatedly modifying a population of individual solutions (66). We tested the ability of Sculptor to model nucleosome–linker histone units into the cryo-EM density of the 30-nm chromatin fiber [EM Data Bank (EMDB) 2601] (fig. S11, A and B) (9). We simulated an EM density for the atomic structure of chicken linker histone H5 bound to linker DNAs at the dyad of a DNA–core nucleo-some particle [Protein Data Bank (PDB) 4QLC] (67) (fig. S11A) and then used Sculptor to simultaneously fit 12 nucleosome–linker histone units into the 30-nm fiber density map. The Sculptor model closely matches the experimentally determined known nucleosome organization of the 30-nm chromatin fiber (fig. S11B, middle panel), validating our approach.
We then applied a similar approach to model nucleosome–linker histone H5 units in chromatin-chain EMT densities (Fig. 5, D, E, and H). The EMT volumes of chromatin chains were used as a starting point to estimate the initial number of nucleosome–linker histone units to model. Sculptor alters the position of each nucleosome–linker histone H5 unit for 500 generations (64). We ran five independent Sculptor evolutions for each chromatin-chain EMT density (fig. S11, C to E). All of the Sculptor nucleosome–linker histone models have cross-correlation values between 80 and 92% (Fig. 5, J to L, and fig. S11, C to E). Not surprisingly, there are differences between each of the solutions as well as unaccounted for densities. The unmatched densities may reflect off-dyad H1 binding (67), histone-tail structures, or proteins other than nucleosomes that interact with DNA and chromatin in the nucleus, such as high-mobility groups (HMGs) (68). In addition, DAB polymers and OsO4 staining could add density and increase thickness by an estimated 1 to 2 nm. Notwithstanding, our data indicate that chromatin chains could comprise different arrangements of nucleosome–linker histone units. Sculptor models suggest that disordered chromatin chains have three to four nucleosomes per 11 nm, more than primary beads-on-a-string DNA-nucleosome polymers but less than 30-nm fibers (six nucleosomes) (9).
ChromEMT enables the ultrastructure and 3D organization of chromatin to be visualized in situ in human mitotic chromosomes
The hierarchical folding of 30-nm fibers into 120-nm chromonema to 300-nm chromatid and 700-nm helical loops is thought to be required for chromatin compaction into mitotic chromosomes (Fig. 1A) (17, 69, 70). Mitotic chromosomes have a 2:1 mass ratio of protein:DNA (71). EM stains such as uranyl acetate bind to both proteins and DNA in mitotic chromosomes, resulting in a black, densely stained, and opaque structure. To determine the ultrastructure and organization of chromatin in mitotic chromosomes, we used ChromEMT. U2OS cells are a human osteosarcoma cell line with a high mitotic index compared to SAECs, which facilitates the identification of mitotic cells by microscopy in the absence of synchronization. U2OS cells were fixed with glutaraldehyde and then stained with DRAQ5. A mitotic cell that appears to be in anaphase stage was identified by its characteristic morphology in light and fluorescence microscopy. DRAQ5 fluorescence exclusively labels DNA in mitotic chromosomes (Fig. 6A), demonstrating the specificity of DRAQ5 staining. Furthermore, we show that DRAQ5 photo-oxidation catalyzes the deposition of DAB polymers on DNA in mitotic chromosomes and can be visualized by OsO4 staining in TEM images of 70- and 250-nm serial sections (Fig. 6B and fig. S12A).
Fig. 6. ChromEMT enables chromatin ultrastructure and 3D organization to be visualized in situ in human mitotic chromosomes.
(A) Mitotic U2OS cells were fixed with glutaraldehyde, stained with DRAQ5, and photo-oxidized for 3 min. Cells were stained with OsO4 and prepared for EM. Scale bar, 10 μm. (B) TEM image (250-nm section). (C) An eight-tilt EMT data set (3200 nm by 3200 nm by 138 nm) comprising 86 TSs was collected of three chromosomes [red box in (B)]. To visualize chromatin ultrastructure and 3D organization as a continuum through mitotic chromosomes, see Movie 3. TS (1.6 nm thick) image of three chromosomes (1, 2, and 3). Inset shows the microtubule structure. Microtubule (solid arrow), membrane fragments (dashed arrow). Scale bar, 100 nm. (D) A magnified image of chromatin in chromosome 2. Scale bar, 40 nm. (E) Chromatin ultrastructure and organization in chromosome 1. The xz and yz cross sections are shown. Scale bar, 100 nm. (F) TS #69 showing chromatin ultrastructure and organization in chromosome 2. The unstained space in between chromatin has a reticular pattern that permeates the 3D volume (dashed arrows). Straight chromatin chains are observed at the borders of unstained venous structures (arrowheads). Scale bar, 100 nm.
To visualize individual chromatin chains and 3D packing in mitotic chromosomes, we collected an eight-tilt EMT data set (Fig. 6B, red box). The reconstructed tomogram (3200-nm–by–3200-nm–by–138-nm volume) comprises 86 TSs, each 1.6 nm thick, and captures sections of three different chromosomes, including the tip of one chromosome and two additional chromosome sections (Fig. 6, A and C). A movie of compiled serial TSs enables chromatin to be visualized as a continuum through a 3D volume of mitotic chromosomes (Movie 3).
Movie 3. ChromEM and multitilt EMT enable chromatin structure and organization to be visualized in mitotic chromosomes in situ.
Mitotic U2OS cells (anaphase) were fixed, stained with ChromEM (Fig. 6C), and an eight-tilt EM tomogram (11,000×) was collected of three chromosomes (1, 2, and 3). The reconstructed EMT volume is 3200 nm (x) by 3200 nm (y) by 138 nm (z) comprising 86 TSs (each 1.6 nm thick). We compiled serial TS slices (TS #0 to TS #85) into a movie, enabling chromatin ultrastructure and organization to be visualized as a continuum through large 3D volumes of mitotic chromosomes in situ. Disordered chromatin chains (darkly stained polymers) are packed together at high uniform densities in mitotic chromosome scaffolds. OsO4-stained membrane fragments and microtubule surfaces (parallel lines) can also be seen.
Chromatin ultrastructure can be visualized with high definition and contrast (Fig. 6D). Furthermore, individual chromatin chains can be resolved from one another in xz and yz in large 3D volumes of mitotic chromosome scaffolds (Fig. 6, E and F, and fig. S12B). Similar to interphase cells, chromatin chains are structurally heterogeneous with many different conformations. In mitotic chromosomes, the unstained regions have a distinct reticular pattern that percolates through the entire 3D volume (Fig. 6F, dashed arrows). Furthermore, at the border of these unstained venous structures, chromatin chains are improbably straight (Fig. 6F, arrowheads). Together, these features suggest that unstained scaffolding factors constrain and compact flexible chromatin chains into the mitotic chromosome architecture.
The summed density advantage of multitilt EMT enables very weak OsO4 staining at the surface of microtubules to be detected. Microtubule surfaces are visible as parallel threads (Fig. 6C, inset, and fig. S12C, arrow). The trajectory of an individual microtubule can be followed in serial TSs as it travels toward the center of a chromosome (fig. S12C). The in situ microtubule diameter (20 to 24 nm) is consistent with its known diameter in vitro (72) and provides an internal control and reference with which to compare chromatin diameters in mitotic chromosomes. It is immediately apparent that chromatin-chain diameters are less than that of microtubules, and we do not observe higher-order 120- to 700-nm chromatin fibers in human mitotic chromosomes in situ.
Chromatin is a disordered 5- to 24-nm-diameter chain that is packed together at high concentration densities in mitotic chromosomes
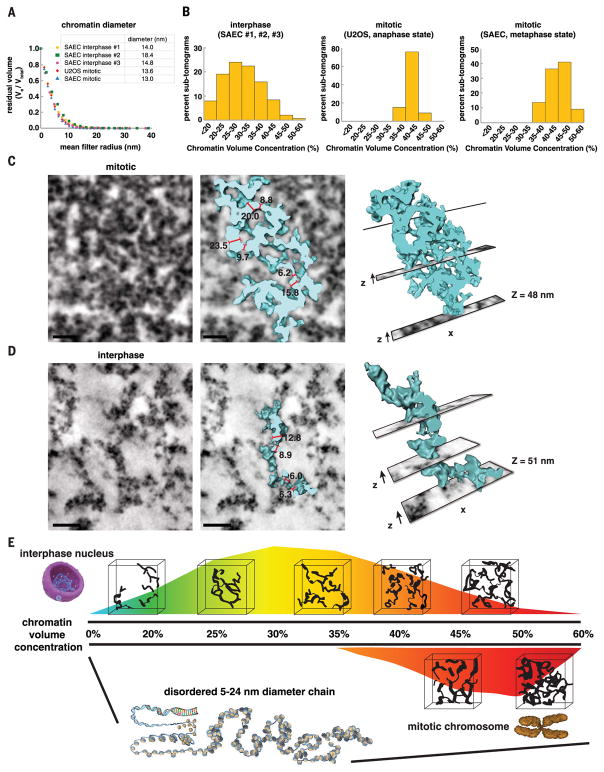
To analyze and compare chromatin in mitotic chromosomes, we collected an eight-tilt EMT data set of a mitotic chromosome at the same magnification (29,000×) as interphase data sets (Fig. 7A and Movie 4). Manual measurements indicate that chromatin diameter ranges are ~8 to 21 nm (Fig. 7B). We applied the automated segmentation protocol and analytical pipeline we developed for interphase chromatin to mitotic chromosomes (fig. S3C and table S2). In mitotic chromosomes, CVCs range from 35% (E2) to 47% (D4), with a mean of 42 ± 2.5% (Fig. 7C). These data demonstrate that the concentration of chromatin for the same unit nuclear volume (120-nm cube) has a higher and narrower distribution range in mitotic chromosomes than in interphase nuclei. However, chromatin has a similar disordered structure and diameter in interphase nuclei and mitotic chromosomes. Chromatin-diameter bin distributions are 5 to 12 nm and 12 to 24 nm (Fig. 7, D to F), and the average diameter of chromatin in D4, C3, and A3 is 13.4, 14.0, and 13.4 nm, respectively (Fig. 7, G to I).
Fig. 7. In mitotic chromosomes, disordered 5- to 24-nm diameter chromatin chains are packed together at high 3D concentration densities.
(A) An eight-tilt EMTdata set (1024 nm by 1280 nm by 180 nm; 141 total TSs, each 1.28 nm thick) of a mitotic chromosome (white box, left panel) at 29,000× (red box, right panel). Scale bar, 100 nm.To visualize chromatin ultrastructure and 3D organization as a continuum, see Movie 4. (B) Manual measurements of chromatin diameter in a single TS. Scale bar, 50 nm. (C) The mitotic chromosome [red box in (A), 722 nm by 722 nm by 120 nm] was divided into 36 subvolumes of 120-nm cubes. CVCs are shown in a heat map. Asterisks indicate cytoplasmic fractions. Scale bar, 100 nm. (D to F) Surface-thickness estimates of chromatin diameters.There are two major bin distributions: 5 to 12 nm and 12 to 24 nm (right panel). Scale bar, 20 nm. (G to I) Continuous erosion analysis to estimate average chromatin diameter (x-axis intercept).
Movie 4. The chromatin ultrastructure and 3D organization of mitotic chromosomes.
A 29,000× reconstructed eight-tilt EMT data set of ChromEM-stained chromatin in a human mitotic chromosome (1024 nm by 1280 nm by 180 nm; 141 total TSs, each 1.28 nm thick) from Fig. 7A. We compiled serial TS slices (TS #0 to TS #140) into a movie, enabling chromatin ultrastructure and organization to be visualized as a continuum.
To further test and extend our conclusions regarding chromatin structure and packing, we generated ChromEMT data sets of primary SAECs at the metaphase stage of mitosis (fig. S13, A and B). CVCs range from 35% (A4) to 55% (A7), with a mean of 45 ± 4.1% (fig. S13C). There are two bin distributions for chromatin diameter, 5 to 12 nm and 12 to 24 nm (fig. S13D). Furthermore, continuous erosion analysis shows that the average diameter of chromatin in A7, D6, and F3 sub-tomograms is 16.4, 13.6, and 12.4 nm, respectively (fig. S13E). These data are similar to measurements in mitotic U2OS data sets (Fig. 7, D to I). We conclude that in human cells, chromatin is a disordered primary polymer chain that is packed together at different concentrations in interphase and mitotic chromosomes.
Disordered chromatin chains bend and flex and are packed together at different densities in interphase cells and mitotic chromosomes
By applying continuous erosion analysis to the entire EMT volumes, we show that the global average diameter of chromatin is ~14 nm in both interphase cells and mitotic chromosomes (Fig. 8A). These data indicate that the general primary polymer structure of chromatin is similar in interphase and mitotic chromosomes. In mitotic chromosomes, chromatin is packed together at high CVCs with a narrow distribution range from 40 to 55% (Fig. 8B). However, in interphase nuclei (cumulative frequency), there is a broad range of different CVCs that exhibit a normal distribution from 12 to 52% (Fig. 8B).
Fig. 8. Disordered 5- to 24-nm-diameter chromatin chains are flexible and can be packed together at different concentration densities in interphase nuclei and mitotic chromosomes.
(A) Global average of chromatin diameters in interphase and mitotic EMT data sets using continuous erosion analysis. (B) Histogram of subvolume CVC frequencies in interphase and mitotic cells. (C) In mitotic chromosomes, an individual chromatin chain is traced, which bends back on itself at short intervals along its length, forming a compact 3D structure. Left, single TS (1.6 nm thick); right, rendered chromatin surface; middle, overlay. Scale bar, 40 nm. (D) In interphase cells, an individual chromatin chain is traced, which has a more extended curvilinear structure than in mitotic chromosomes. Left, single TS (1.28 nm thick); right, rendered chromatin surface; middle, overlay. Scale bar, 40 nm. (E) Higher-disorder 3D chromatin packing. Chromatin is a flexible disordered 5- to 24-nm-diameter granular chain that is packed together at different 3D volume concentration density distributions in interphase nuclei and mitotic chromosomes.
We conclude that chromatin is a flexible 5- to 24-nm-diameter granular chain that is packed together at different concentration densities in interphase and mitotic chromosomes. In mitotic chromosomes, chromatin chains bend back on themselves at shortly interspersed intervals, and there are more dense interactions between chains (Fig. 8C). In interphase cells, chromatin chains have more extended curvilinear structures and less frequent contacts between and within chains (Fig. 8D). Thus, instead of higher-order folding, we propose that higher disorder enables chromatin chains to be packed together at different densities to achieve different levels of compaction in interphase and mitotic cells. This would explain the surprising elasticity of mitotic chromosomes in atomic force microscopy measurements (73, 74), which has been difficult to reconcile with the hierarchical folding model.
Discussion
In summary, ChromEMT enables the ultrastructure and 3D organization of individual chromatin polymers, heterochromatin domains, and mitotic chromosomes to be visualized in situ in single cells (Fig. 4A). ChromEM staining does not require genetically modified cells, overexpression of tagged histone fusions, or incorporation of nucleotide analogs (75), all of which could perturb DNA structure and function. As such, ChromEMT provides a facile and universal method to compare the structure of genomic DNA in different kingdoms of life. To offset the possibility that DRAQ5 binding could disrupt DNA interactions (49, 50), we fixed cells with glutaraldehyde before labeling. However, glutaraldehyde cross-linking could induce higher-order structures that are not present in living cells. Samples prepared in vitreous ice are thought to preserve native structure. A recent study showed that there are no morphological differences in chromatin structure between cryofrozen and glutaraldehyde-fixed chromocenters (23). Moreover, we observe the exact opposite of higher-order fibers even in densely packed metaphase chromosomes (fig. S13).
In previous EM studies where higher-order fibers were observed (14–18), other cellular components had to be extracted with detergents and salts to visualize chromatin. We show that ChromEM stains and detects the induction of higher-order 30-nm chromatin fibers in MgCl2-treated hypotonically lysed chicken erythrocyte nuclei. However, we do not observe higher-order fibers in human interphase and mitotic cells in situ (Figs. 4, 6, and 7 and figs. S7, S8, and S13). It is possible that 30-nm fibers and 120-nm chromonema are exceedingly rare and/or peculiar to specialized cell types and states. Alternatively, 30- and 120-nm fibers may be induced in in vitro conditions and extracted nuclei (7–11). The formation of 30-nm chromatin structures requires the selective intrafiber binding of neighboring nucleosomes, which is favored in dilute solutions (76) but perhaps not the crowded milieu of the nucleus. Cryo-EM (19, 20), x-ray scattering (21), ESI (22, 23), and superresolution studies (30) also failed to detect higher-order fibers with a 30-nm periodicity, suggesting that chromatin may have an 11-nm spacing. Consistent with these data, the global average diameter of chromatin in ChromEMT data sets is ~14 nm (Fig. 8A). However, we show that the average diameter reflects the structural heterogeneity of chromatin chains as opposed to in vitro beads-on-a-string structures that have uniform 11-nm diameters (12). Instead, we show that chromatin is a disordered granular chain with varying diameters between 5 and 24 nm and many different nucleosome particle arrangements, unknown densities, and structural conformations. There are many more chromatin structures in situ than have been observed in vitro. In human cells, there are a possible 15 million different DNA polymer–nucleosome units comprising distinct combinations of H1, H2A, H2B, and H3 isoforms (10 H1s, 7 H2As, 4 H2Bs, 8 H3s), his-tone posttranslational modifications, DNA-linker variations (77–79), and HMG proteins (68). Notably, the chromatin loops, hubs, and bends in situ (Fig. 5, J to L, and fig. S11) resemble in silico structural models where DNA-linker lengths and H1 binding modes are varied (80, 81). The diverse array of chromatin conformations in situ is exciting and provides a basis for how different DNA sequences, linker lengths, histone variants, modifications, and nuclear protein interactions could be integrated to exquisitely fine-tune the structure, activity, and accessibility of genomic DNA.
We conclude that it is not necessary for DNA to fold into discrete higher-order chromatin fibers (Fig. 1A) to compact the human genome. In contrast to higher-order fibers that have longer and fixed persistence lengths (82–84), disordered chromatin chains with varying diameters are flexible and can bend at different lengths to achieve a range of packing densities. Chromatin chains have more extended curvilinear structures in interphase cells (Fig. 8D) and collapse into arrays of small compact loops in mitotic chromosome scaffolds (Fig. 8C). These data help to explain the rapid dynamics of chromatin condensation at mitosis (76) and how epigenetic interactions and structures could be inherited through cell division (85). We propose a model in which the assembly of 3D domains with CVCs that exceed a certain threshold determines DNA accessibility and compaction in the nucleus (Fig. 8E). Interphase subvolumes have CVCs ranging from 12 to 52% with distinct spatial distribution patterns in the nucleus (Fig. 4D and figs. S7A and S8A), whereas mitotic chromosomes have CVCs >40% (Fig. 7C and fig. S13C). Mitotic chromosomes and heterochromatin domains at the nuclear membrane are transcriptionally silent and have CVCs >40% (Figs. 4D and 7C and figs. S7A, S8A, and S13C) (86, 87). CVCs >40% may exceed the critical overlap concentration at which semiflexible chromatin chains become topologically entangled (88), resulting in phase separated “gels” that limit the diffusion and access of large macromolecular assemblies, such as RNA polymerase. Thus, it will be exciting to explore if the 3D concentration of chromatin in the nucleus is a simple and universal self-organizing principle that determines the functional activity and accessibility of genomic DNA.
Supplementary Material
Acknowledgments
We thank members of the O’Shea and Ellisman laboratories for their insights and comments. We are grateful to the Waitt Advanced Biophotonics Center for computational support and S. Navlakha for advice on image analysis. This work was supported by grants from the W. M. Keck Foundation and the NIH (grant 5U01EB021247). The research of C.C.O. is supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute. C.C.O. is also supported by the William Scandling Trust, the Price Family Foundation, and The Leona M. and Harry B. Helmsley Charitable Trust grant. Salk core services were supported by grant P30CA014195 from the National Cancer Institute. University of California, San Diego (UCSD), National Center for Microscopy and Imaging Research (NCMIR) technologies and instrumentation are supported by grant GM103412 from the National Institute of General Medical Sciences. The EM tomograms are deposited in the Cell Image Library: www.cellimagelibrary.org/groups/49801.
Footnotes
REFERENCES AND NOTES
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 4.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickmore WA, van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: Evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 9.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 10.Belmont AS, Braunfeld MB, Sedat JW, Agard DA. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- 11.Belmont AS, Bruce K. Visualization of G1 chromosomes: A folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olins AL, Senior MB, Olins DE. Ultrastructural features of chromatin nu bodies. J Cell Biol. 1976;68:787–793. doi: 10.1083/jcb.68.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olins AL, Carlson RD, Olins DE. Visualization of chromatin substructure: Upsilon bodies. J Cell Biol. 1975;64:528–537. doi: 10.1083/jcb.64.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedat J, Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42:331–350. doi: 10.1101/SQB.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Rattner JB, Lin CC. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42:291–296. doi: 10.1016/S0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 16.Belmont AS, Sedat JW, Agard DA. A three-dimensional approach to mitotic chromosome structure: Evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS. Visualization of early chromosome condensation. J Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dehghani H, Dellaire G, Bazett-Jones DP. Organization of chromatin in the interphase mammalian cell. Micron. 2005;36:95–108. doi: 10.1016/j.micron.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDowall AW, Smith JM, Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986;5:1395–1402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmed K, et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLOS ONE. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fussner E, et al. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012;13:992–996. doi: 10.1038/embor.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahamid J, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 25.Bouchet-Marquis C, Dubochet J, Fakan S. Cryoelectron microscopy of vitrified sections: A new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol. 2006;125:43–51. doi: 10.1007/s00418-005-0093-x. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda A, et al. Condensed mitotic chromosome structure nanometer resolution using PALM and EGFP- histones. PLOS ONE. 2010;5:e12768. doi: 10.1371/journal.pone.0012768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smeets D, et al. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin. 2014;7:8. doi: 10.1186/1756-8935-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zessin PJ, Finan K, Heilemann M. Super-resolution fluorescence imaging of chromosomal DNA. J Struct Biol. 2012;177:344–348. doi: 10.1016/j.jsb.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 30.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 31.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanaichi T, et al. A stable lead by modification of Sato’s method. J Electron Microsc (Tokyo) 1986;35:304–306. [PubMed] [Google Scholar]
- 33.Watson ML. Staining of tissue sections for electron microscopy with heavy metals. J Biophys Biochem Cytol. 1958;4:727–730. doi: 10.1083/jcb.4.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huxley HE, Zubay G. Preferential staining of nucleic acid-containing structures for electron microscopy. J Biophys Biochem Cytol. 1961;11:273–296. doi: 10.1083/jcb.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogliati R, Gautier A. Mise en évidence de l’ADN et des polysaccharides à l’aide d’un nouveau réactif “de type Schiff”. C R Acad Sci Hebd Seances Acad Sci D. 1973;276:3041–3044. [PubMed] [Google Scholar]
- 36.Olins AL, Moyer BA, Kim SH, Allison DP. Synthesis of a more stable osmium ammine electron-dense DNA stain. J Histochem Cytochem. 1989;37:395–398. doi: 10.1177/37.3.2465337. [DOI] [PubMed] [Google Scholar]
- 37.Cló E, Snyder JW, Ogilby PR, Gothelf KV. Control and selectivity of photosensitized singlet oxygen production: Challenges in complex biological systems. ChemBioChem. 2007;8:475–481. doi: 10.1002/cbic.200600454. [DOI] [PubMed] [Google Scholar]
- 38.Bentivoglio M, Su HS. Photoconversion of fluorescent retrograde tracers. Neurosci Lett. 1990;113:127–133. doi: 10.1016/0304-3940(90)90291-G. [DOI] [PubMed] [Google Scholar]
- 39.Deerinck TJ, et al. Fluorescence photooxidation with eosin: A method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou HD, Deerinck TJ, Bushong E, Ellisman MH, O’Shea CC. Visualizing viral protein structures in cells using genetic probes for correlated light and electron microscopy. Methods. 2015;90:39–48. doi: 10.1016/j.ymeth.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham RC, Jr, Karnovsky MJ. The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: Ultrastructural cytochemistry by a new technique. J Histochem Cytochem. 1966;14:291–302. doi: 10.1177/14.4.291. [DOI] [PubMed] [Google Scholar]
- 42.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLOS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ou HD, et al. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garton HJ, Schoenwolf GC. Improving the efficacy of fluorescent labeling for histological tracking of cells in early mammalian and avian embryos. Anat Rec. 1996;244:112–117. doi: 10.1002/(SICI)1097-0185(199601)244:1<112::AID-AR11>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 45.Balercia G, Chen S, Bentivoglio M. Electron microscopic analysis of fluorescent neuronal labeling after photoconversion. J Neurosci Methods. 1992;45:87–98. doi: 10.1016/at0165-0270(92)90046-G. [DOI] [PubMed] [Google Scholar]
- 46.Smith PJ, Wiltshire M, Errington RJ. DRAQ5 labeling of nuclear DNA in live and fixed cells. Current Protoc Cytom. 2004;28:7.25.1–7.21.11. doi: 10.1002/0471142956.cy0725s28. [DOI] [PubMed] [Google Scholar]
- 47.Njoh KL, et al. Spectral analysis of the DNA targeting bisalkylaminoanthraquinone DRAQ5 in intact living cells. Cytometry A. 2006;69A:805–814. doi: 10.1002/cyto.a.20308. [DOI] [PubMed] [Google Scholar]
- 48.Martin RM, Leonhardt H, Cardoso MC. DNA labeling in living cells. Cytometry A. 2005;67A:45–52. doi: 10.1002/cyto.a.20172. [DOI] [PubMed] [Google Scholar]
- 49.Richard E, et al. Short exposure to the DNA intercalator DRAQ5 dislocates the transcription machinery and induces cell death. Photochem Photobiol. 2011;87:256–261. doi: 10.1111/j.1751-1097.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 50.Wojcik K, Dobrucki JW. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells—influence on chromatin organization and histone-DNA interactions. Cytometry A. 2008;73A:555–562. doi: 10.1002/cyto.a.20573. [DOI] [PubMed] [Google Scholar]
- 51.Bozzola JJ, Russell LD. Electron Microscopy Principles and Techniques for Biologists. Johnes and Bartlett Publishers; Boston: 1992. [Google Scholar]
- 52.Stockert JC, Colman OD. Observations on nucleolar staining with osmium tetroxide. Experientia. 1974;30:751–752. doi: 10.1007/BF01924164. [DOI] [PubMed] [Google Scholar]
- 53.Frank J, editor. Electron Tomography: Methods for Three-Dimensional Visualization of Structures in the Cell. 2. Springer; 2010. [Google Scholar]
- 54.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 55.Phan S, et al. 3D reconstruction of biological structures: Automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Adv Struct Chem Imaging. 2017;2:8. doi: 10.1186/s40679-016-0021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phan S, et al. TxBR montage reconstruction for large field electron tomography. J Struct Biol. 2012;180:154–164. doi: 10.1016/j.jsb.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Lawrence A, Bouwer JC, Perkins G, Ellisman MH. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J Struct Biol. 2006;154:144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Zuiderveld K. In: Graphics Gems IV. Paul SH, editor. Academic Press Professional; San Diego, CA: 1994. pp. 474–485. [Google Scholar]
- 59.Li CH, Tam PKS. An iterative algorithm for minimum cross entropy thresholding. Pattern Recognit Lett. 1998;19:771–776. doi: 10.1016/S0167-8655(98)00057-9. [DOI] [Google Scholar]
- 60.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 61.Stalling D, Westerhoff M, Hege H-C. In: The Visualization Handbook. Hansen CD, Johnson CR, editors. Butterworth-Heinemann; Burlington, MA: 2005. pp. 749–767. [Google Scholar]
- 62.Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–622. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 63.Scheffer MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci USA. 2011;108:16992–16997. doi: 10.1073/pnas.1108268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rusu M, Birmanns S. Evolutionary tabu search strategies for the simultaneous registration of multiple atomic structures in cryo-EM reconstructions. J Struct Biol. 2010;170:164–171. doi: 10.1016/j.jsb.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birmanns S, Rusu M, Wriggers W. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J Struct Biol. 2011;173:428–435. doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis LD, Mitchell M. Handbook of Genetic Algorithms. Van Nostrand Reinhold; New York: 1991. [Google Scholar]
- 67.Zhou BR, et al. Structural mechanisms of nucleosome recognition by linker histones. Mol Cell. 2015;59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst) 2015;36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 69.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-X. [DOI] [PubMed] [Google Scholar]
- 70.König P, Braunfeld MB, Sedat JW, Agard DA. The three-dimensional structure of in vitro reconstituted Xenopus laevis chromosomes by EM tomography. Chromosoma. 2007;116:349–372. doi: 10.1007/s00412-007-0101-0. [DOI] [PubMed] [Google Scholar]
- 71.Salzman NP, Moore DE, Mendelsohn J. Isolation and characterization of human metaphase chromosomes. Proc Natl Acad Sci USA. 1966;56:1449–1456. doi: 10.1073/pnas.56.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandelkow EM, Mandelkow E. Unstained microtubules studied by cryo-electron microscopy: Substructure, supertwist and disassembly. J Mol Biol. 1985;181:123–135. doi: 10.1016/0022-2836(85)90330-4. [DOI] [PubMed] [Google Scholar]
- 73.Gállego I, Oncins G, Sisquella X, Fernàndez-Busquets X, Daban JR. Nanotribology results show that DNA forms a mechanically resistant 2D network in metaphase chromatin plates. Biophys J. 2010;99:3951–3958. doi: 10.1016/j.bpj.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gállego I, Castro-Hartmann P, Caravaca JM, Caño S, Daban JR. Dense chromatin plates in metaphase chromosomes. Eur Biophys J. 2009;38:503–522. doi: 10.1007/s00249-008-0401-1. [DOI] [PubMed] [Google Scholar]
- 75.Ngo JT, et al. Click-EM for imaging metabolically tagged nonprotein biomolecules. Nat Chem Biol. 2016;12:459–465. doi: 10.1038/nchembio.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014;123:225–237. doi: 10.1007/s00412-014-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 80.Collepardo-Guevara R, Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc Natl Acad Sci USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlick T, Hayes J, Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J Biol Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langowski J. Polymer chain models of DNA and chromatin. Eur Phys J E Soft Matter. 2006;19:241–249. doi: 10.1140/epje/i2005-10067-9. [DOI] [PubMed] [Google Scholar]
- 83.Tark-Dame M, van Driel R, Heermann DW. Chromatin folding—from biology to polymer models and back. J Cell Sci. 2011;124:839–845. doi: 10.1242/jcs.077628. [DOI] [PubMed] [Google Scholar]
- 84.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci USA. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 86.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 87.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 88.Raghavan SR, Douglas JF. The conundrum of gel formation by molecular nanofibers, wormlike micelles, and filamentous proteins: Gelation without cross-links? Soft Matter. 2012;8:8539–8546. doi: 10.1039/c2sm25107h. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.