Abstract
Purpose of review
To describe the current knowledge on the cross-talk between connexins and microRNAs (miRs) in bone cells.
Recent findings
Connexins play a crucial role on bone development and maintenance, and disruptions in their abundance or localization can affect how bone perceives and responds to mechanical, hormonal, and pharmacological stimuli. Connexin expression can be modified by miRs, which modulate connexin mRNA and protein levels. Recently, different manners by which miRs and connexins can interact in bone have been identified, including mechanisms that mediate miR exchange between cells in direct contact through gap junctions, or between distant cells via extracellular vesicles (EVs).
Summary
We bring to light the relationship between miRs and connexins in bone tissue, with special focus on regulatory effects of miRs and connexins on gene expression, as well as the mechanisms that mediate miR exchange between cells in direct contact through gap junctions, or between distant cells via EVs.
Keywords: microRNA, gap junction, connexin, musculoskeletal tissue, bone, extracellular vesicles
Introduction
The evolution from unicellular to multicellular organisms was an enormous step in the advancement of organism development, requiring a system to coordinate the exchange of molecules among cells and the generation of a vast array of membrane specializations. In particular, skeletal development and homeostasis depends on the tight control of bone cell proliferation, differentiation and activity, orchestrated through regulated cell-to-cell communication [1–3]. This coordinated intercellular communication via the exchange of signaling molecules and genetic material can occur between adjacent cells in direct contact through gap junction channels formed by connexins [4,5] or between distant cells via extracellular vesicle- (EV) mediated signaling [6]. Intercellular microRNA (miR) transfer has recently been shown to play a critical role in regulating a variety of key cellular processes involved in the maintenance of skeletal tissue [7]. Herein, we will discuss the complex regulatory signaling mechanisms involved in controlling bone cells through intracellular bidirectional miR and connexin post-translational regulation, as well as intercellular signaling via diverse mechanisms of miR transfer.
Gap junctions: structure and function
Gap junctions, a type of membrane specialization, are clusters of membrane channels that enable electrometabolic coupling and control the passage of ions, such as Ca2+, H+, Na+, K+, Cl−, sugars, amino acids, nucleotides, and vitamins up to 1.5 nm in diameter through conduit-like structures named connexons [8]. In addition, recent findings have shown that larger biomolecules, such as nucleic acids, can also go through these channels [4].
Gap junction proteins or connexins contain four transmembrane domains, two extracellular loops, cytoplasmic N- and C-terminal domains, and a single cytoplasmic loop. Connexins are named based on their expected molecular weight; for example, connexin (Cx) 43 and Cx37 are approximately 43 and 37 kDa in size, respectively [8,9]. Six connexin molecules assemble in the Golgi apparatus to form a connexon (or hemichannel) that is then transported to the plasma membrane, where it is coupled to another connexon of an adjacent cell, forming a continuous aqueous channel between the cells [10]. In addition to mediating the exchange of molecules between cells, connexins can also participate in intracellular signaling by triggering biochemical signals through their structural domains [11].
While connexins are expressed in nearly every cell type, with the exception of red blood cells, spermatozoids, and differentiated skeletal muscle cells of adult vertebrates, the composition and cellular membrane quantity of gap junction channels varies greatly between cell types [8]. Connexin expression and channel activity is controlled by numerous regulatory factors. To ensure proper activity, the control of opening and closure of the channels is regulated by voltage, ion concentration, cytoplasmic pH, amino sulfonates, phosphorylation, lipophiles, cyclic nucleotides, and others [9], although the precise mechanisms are not completely known. In addition, the half-life of the intercellular gap junctions is only a couple of hours, undergoing degradation in the endo-lysosomal network about 10 times faster than other membrane proteins [8]. The rapid turnover allows for the dynamic and efficient regulation of connexin abundance and localization, which facilitates the rapid exchange of molecules and downstream signaling upon cell requirement. Further, gap junction protein expression and activity are also controlled by several post-translational mechanisms, including phosphorylation, oxidation/reduction, protein-protein interaction, and regulation of mRNA/protein levels by small noncoding RNA, particularly miRs.
The essential role that connexins play in the development and maintenance of bone cells has been demonstrated in numerous studies [12]. Cx43 was the first connexin to be identified and it is the most highly studied and expressed connexin in bone tissue. Mutations of the Cx43 gene are associated with occulodentodigital dysplasia (ODDD), a disorder characterized by abnormalities that include weak enamel, small or missing teeth, early tooth loss, and broad long bones [13]. Although Cx45 and Cx46 are also expressed in bone [14,15], their role is still unknown. Global Cx40-deficient newborn mice show defective axial and appendicular bone, with abnormal rib development, lower limb malformations and delayed ossification in anklebones [16]. This phenotype is due to abnormal endochondral ossification, and the role of Cx40 in adult bone is not known. More recently, Cx37 deletion was associated with reduced osteoclastogenesis and modifications in bone geometry and Wnt/β-catenin signaling [17,18].
Cx43 expression in osteoblasts and osteocytes is important for proper bone development and health. Thus, analysis of embryos with global Cx43 deletion shows delayed bone formation and defective osteoblast differentiation, which affects both endochondral and intramembranous bone formation [19]. Milder skeletal phenotypes are observed in mice lacking Cx43 in osteochondroprogenitors, osteoblast precursors, and mature osteoblasts, which is progressively less evident as the connexin is deleted in more mature cells [20]. Further, Cx43 targeted deletion in osteocytes does not alter bone mineral density, but results in osteocyte apoptosis and modifications in the composition of the bone matrix and bone mechanical properties. In addition, osteoclast differentiation is impaired in mice lacking Cx43 in osteoclast precursors, resulting in reduced bone resorption [21]. Thus, all these pieces of evidence point out that the fine-tuned control of gap junction/connexin expression is crucial for bone health.
microRNA biogenesis, structure, and function
miRs are a class of small noncoding RNA gene products of approximately 22 nucleotides in length that function in the post-transcriptional regulation of gene expression in diverse organisms, including plants and animals [22]. Since their first description in 1993, thousands of new miRs have been discovered [23]. miRs are named using the “miR” prefix along with an exclusive identification number (for example, miR-21, miR-218, miR-5, etc.), which follows the order in which that miR was described. However, there are a few exceptions such as the names for let-7 and lin-4, which are maintained for historical reasons. The gene name encoding the miR receives the same three-letter prefix [24].
miR biogenesis is a complex biological process that involves multiple steps and requires numerous transcription factors, binding-proteins, and regulatory enzymes (Fig. 1). miRs are encoded in long endogenous transcripts that are transcribed as long primary transcripts (primiRs), which form a long single-molecule hairpin structure [22]. Pri-miRs then undergo a specific enzymatic cleavage driven by the microprocessor complex composed of Drosha and DGCR8, which generates a shorter hairpin structure precursor miR (pre-miRs). Following cleavage, the pre-miR is exported by exportin-5 from the nucleus to the cytoplasm, where it undergoes additional cleavage by Dicer into a mature miR duplex [25]. Upon loading onto an argonaute (AGO) protein, one strand is lost whereas the other complementary miR strand is incorporated into the RNA-induced silencing complex (RISC) where the miR and the corresponding messenger RNA interact. Specifically, miRs bind to the 3′- or 5′- untranslated regions (UTR) of mRNAs inducing repression of gene expression in a complementarity-dependent level. One single miR might target several genes, whereas several miRs can target a single gene [22].
Fig. 1. Model for the proposed mechanisms of intercellular miRNA cell-to-cell transfer.
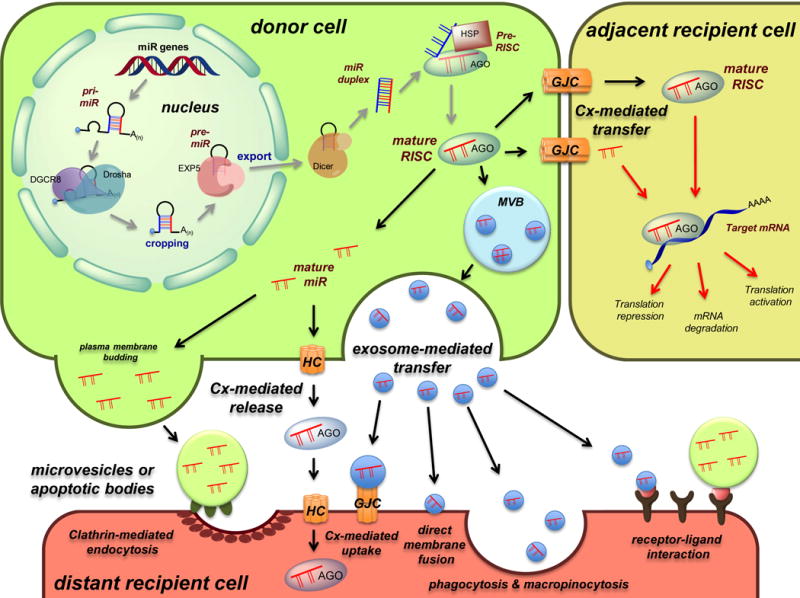
Schematic representation of miR biogenesis (grey arrows), see text for more details. Upon completion of the biogenesis process, the mature RISC complex can regulate mRNA gene expression in the donor cell or can be transferred to adjacent or distant recipient cells to modulate mRNA expression by different manners (red arrows). Cell-to-cell transfer mechanisms include connexin-mediated transfer (via direct gap junctions or through hemichannels) and exosome-mediated transfer (via direct membrane fusion, clathrin-mediated endocytosis, phagocytosis and micropinocytosis, receptor-ligand interaction, or connexin-mediated uptake). HC: hemichannel, GJC: gap junction channel, MVB: multivesicular body.
Extensive research across numerous fields has demonstrated the important role of miRs in regulating many essential biological processes including tissue development, and cell differentiation, activity, and survival, as well as in the onset and progression of numerous diseases in various tissues [26]. In particular, miRs have been shown to play an important role in the process of skeletal development and bone remodeling under physiological conditions and in metabolic bone diseases, including osteoporosis [27–29]. The role of miRs in skeletal development was first demonstrated by in vivo studies in which Dicer was deleted in bone cells, which resulted in an overall reduction in miR expression. These decreases in miR expression led to significant growth and skeletal defects, as a consequence of reductions in proliferation and differentiation of chondrocytes and osteoblasts [29]. Moreover, a variety of miRs have been shown to play a role in bone remodeling by regulating osteoblast and osteoclast differentiation, activity and survival [29,30]. Although the critical role of miRs in regulating bone development and homeostasis has been demonstrated by numerous studies, the mechanisms of their action and their roles in bone cell signaling still remain unclear.
Recent studies have demonstrated that miRs generated in one cell can be transferred and regulate mRNA expression in neighboring or distant cells through different mechanisms, which include connexin channel-mediated transfer, as well as EV-directed transport, illustrated in Fig. 1 [6,31,32].
Extracellular vesicles
Extracellular vesicles (EVs) made up of organelle-free cytosol surrounded by a lipid bilayer membrane are released from numerous cell types, including osteoblasts, osteocytes, and osteoclasts [33]; and have been recently shown to play an important role in mediating cell-to-cell signaling [34–36]. EVs, which include exosomes (30-100 nm), microvesicles (50-2000 nm), and apoptotic bodies (500-5000 nm) are classified depending on their size and are formed and released through different mechanisms [35] (Fig. 1). Exosomes, the smallest type of EVs, are formed by a process involving the inward budding of the endosomal membrane leading to the formation of multivesicular bodies (MVB). Following fusion of the outer MVB membrane with the plasma membrane, the MVBs are then released into the extracellular space as exosomes [32]. Microvesicles and apoptotic bodies are formed through the outward budding of the plasma membrane [33]. The EV membrane provides a protective environment, maintaining the stability of the molecular cargo contained within the EV in the extracellular environment.
Upon extracellular release, EVs can mediate intercellular signaling through the transfer of their cargo, including proteins, lipids, mRNAs, and miRs, to neighboring or distant cells [37]. The recipient cell can take up EVs through a variety of different mechanisms such as direct membrane fusion, clathrin-mediated endocytosis, phagocytosis and micropinocytosis, or receptor-ligand interactions [32] (Fig. 1). Further, recent work by Soares et al. found that Cx43 hemichannels are present on the exosomal surface and that these connexin channels can interact with connexin channels present on the plasma membranes of recipient cells facilitating the transfer of exosomal cargo to recipient cells [38]. The physiological relevance of this EV-mediated transfer in skeletal tissues has been demonstrated by regulation of bone remodeling by altering osteoblast and osteoclast differentiation and function following EV release and uptake between bone cells [7].
Cross-regulation of connexins and miRs in bone cells
Computational methods predict several RNA- and DNA-binding motifs present in the sequences of connexins [39]. Further, studies in recent years have shown that connexin expression is functionally regulated by miRs in bone cells. In particular, miR-206, initially considered as muscle-specific, is expressed in osteoblastic cells and is involved in the regulation of Cx43 expression [40]. Thus, Cx43 expression is repressed by osteoblastic-targeted miR-206 overexpression in mice, resulting in low bone formation and reduced bone mass. Another study showed that miR-206 derived from cells of the hematopoietic and osteoblastic lineages is upregulated, whereas Cx43 expression is reduced in a rabbit model of glucocorticoid-induced osteonecrosis of the femoral head [41], further supporting the role of miR-206 as a bone miR that targets Cx43. miR-23a is also expressed in osteoblastic cells, and its overexpression in human osteosarcoma cells leads to downregulation of Cx43 expression and inhibition of the increase in Cx43 expression during osteoblast differentiation [42]. Overexpression of miR-23a also is accompanied by delayed osteoblast differentiation, suggesting that the miR can inhibit osteoblastogenesis by repressing Cx43 expression. On the other hand, miR-211 expression is increased during osteogenic differentiation in human induced pluripotent stem cells, and transfection of a miR-211 inhibitor abrogated osteoblast differentiation and mineralization in vitro [43]. This study showed that the effect of miR-211 on osteoblast differentiation is mediated by an increase in autophagy, downstream of upregulation of Atg14 expression. Although Cx43 expression has been shown to regulate autophagy in osteoblastic cells and to associate with Atg14 in the absence of serum [44], it is not known whether the effect of miR-211/Atg14 in osteoblast differentiation is mediated by Cx43.
We have recently shown that bones from old mice exhibit reduced Cx43 expression, and that miR-21 levels are decreased in bones from both old and Cx43-deficient mice [45]. Deletion of Cx43 from osteocytic cells results in reduced miR-21 and increased miR-218 expression levels; both changes are associated with increased apoptosis in other cell types [26]. In addition, deletion of miR21 in bone ex vivo is sufficient to increase osteocyte apoptosis. These findings indicate the cell autonomous requirement of Cx43 for the maintenance of miR-21/miR-218 levels in osteocytic cells and identify a new type of interaction not yet explored in osteocytic cells, by which connexins regulate miR expression.
Cell-to-cell signaling via intercellular miR transfer
In addition to the well-established regulation of mRNA expression by miRs within the cell, recent studies have begun to demonstrate the role of miRs in mediating intercellular signaling through various mechanisms, involving both connexin channel-mediated transfer and EV-directed transport [5,31,32,39,47] (Fig. 1). Upon transfer, these miRs can alter mRNA expression and mediate downstream cellular effects in the recipient cell [31]. It was also recently discovered that miRs can function as ligands and directly interact with toll-like receptor (TLRs) [31], demonstrating yet another complexity associated with extracellular-mediated miR signaling.
Connexin mediated miR transfer
Besides the interactions between connexin and miRs in the reciprocal regulation of their expression, gap junctions have been shown to transfer mature miRs between adjacent cells [5,31,47,48]. This has been demonstrated in a variety of cell types under both physiological and pathological conditions, as highlighted by Lim et al. [48] and in the review by Lemcke et al. [5]. Gap junctions have also been shown to transfer small interference (si)RNA, which are similar in size to miRs (20-30 nucleotides) [4]. Interestingly, gap junction-mediated transfer of siRNA appears to be dependent upon the connexin subtype, as evidenced by the findings that Cx43 gap junctions, but not Cx32 or Cx26 allowed cell-to-cell movement of siRNA. More recently, it was demonstrated that Cx43 with intact channel activity is required for miR intercellular transfer, with recipient cell having up to 30% of miR level of the donor cell [47]. This evidence suggests that connexin channels might also have specificity for transferring particular miRs. Additionally, AGO-bound miRs have been shown to pass between cells through gap junctions, where they can post-transcriptionally regulate target mRNAs in the recipient cells. It has also been hypothesized that these highly stable, AGO-bound miRs can pass through hemichannels into the extracellular space, and later be transferred to a distant cells, in which miRs can regulate gene expression [31]. Although this gap junction mediated miR transfer has not been shown specifically in bone cells, these studies highlight the possible role of connexin channel-mediated miR transfer in the regulation of skeletal development and bone remodeling.
Extracellular vesicles modulate miR transfer
In addition to gap junction-mediated miR transfer, EVs have been shown to transport miRs that mediate intercellular communication [6,49]. EVs are released from numerous cell types [33], including many bone-associated cells such as mesenchymal stem cells (MSCs), osteoblast precursors, mature osteoblasts, osteocytes, and osteoclasts [7]. In recent years, EV-mediated miR transfer has gained the attention of numerous research fields; and an increasing number of studies have demonstrated the involvement of EV-derived miRs in regulating a vast array of cellular processes including proliferation, differentiation, activity, and survival [50]. During the process of plasma membrane budding miRs are incorporated into microvesicles and apoptotic bodies. Further, miRs are selectively packaged into MVBs, which are then released to the extracellular space as exosomes. Following extracellular release, these EV-contained miRs can then be taken up by adjacent or distant recipient cells through various uptake mechanisms, where they can regulate a variety of cellular processes [32]. Several studies have shown the physiological importance of EV-mediated miR transfer, demonstrating miR-mediated regulation of gene expression in recipient cells and the downstream cellular effects associated with mRNA regulation [31].
Notably, an increasing number of studies have demonstrated the role of EV-derived miRs in controlling bone cells during skeletal development and homeostasis [7]. These miRs have been shown to regulate various aspects of bone remodeling from controlling bone cell differentiation to regulating cellular activity and survival. miRs have been shown to play a critical role in regulating key skeletal cell intracellular signaling including Wnt, insulin, TGF-β, and calcium signaling pathways [51]. Current work in the field is beginning to identify the effects of EVs derived from bone cells and determine the role of specific EV-contained miRs in mediated these cellular effects. Specifically, bone cells including osteoblast precursors, mature osteoblasts, and mature osteoclasts have been shown to release miR-containing EVs that regulate bone remodeling through either stimulating or inhibiting bone cell differentiation and activity (recently reviewed by Xie and colleagues [7]).
In particular, osteoblast precursor-derived EVs have been shown to contain miRs involved in enhancing osteoblast differentiation (miR-199b [52], miR-218 [53], miR-181a [54], miR-196a [55,56], let-7 [57,58] family of miRs), as well as miRs that inhibit osteoblast differentiation (miR-135b [59,60], miR-204 [61], miR-855 [62]). Bone regulatory miRs have also been identified in EVs released by mature osteoblast including miRs involved in promoting osteoblast differentiation (let 7 [57,58], miR-335 [63], miR-378 [64], and miR-677 [51]) and miRs that inhibit osteoblast differentiation (miR-30d [65], miR-133b [66], miR-140 [67]). Further, EVs derived from both osteoblast precursors and mature osteoblasts have been shown to contain miRs that regulate osteoclast activity. Specifically, MSC-derived EVs containing miRs stimulate osteoclast differentiation (miR-148a [68]), and mature osteoblast-derived EV containing miRs (miR-503 [69]) inhibit osteoclast differentiation. Mature osteoclasts have also been shown to release EVs containing miR-214, which was found to both inhibit osteoblast differentiation and also stimulate osteoclast activity [70,71]. Thus, miRs can either increase or decrease the differentiation and function of osteoblasts and osteoclasts, depending on the particular miR and the stage of cell differentiation. Taken together, these findings illustrate the diverse roles of bone-derived EV-contained miRs in controlling bone cell activity, demonstrating the importance intercellular miR transfer in mediating bone cell-cell signaling.
While promising, these findings demonstrate the need for future studies to identify the contribution of extracellular miR transfer in regulating skeletal development and bone remodeling. Thus, highlighting the potential involvement of both connexin channel-mediated and EV directed miR transport as a means of modulating bone cell signaling under both normal and disease conditions.
Clinical applications/therapeutic potential
Due to the diverse regulatory capabilities of miRs, there is extensive therapeutic potential in understanding and modulating intercellular miR transfer. Extracellular miRs can serve as biomarkers for different disease conditions, allowing for less invasive and more specific examination of different pathologies [27]. In particular for bone, a recent meta-analysis study showed that miR levels in tissue from individuals with osteosarcoma can predict the overall survival of the patients [72]. Further, cell-directed miR delivery appears to be a promising potential therapeutic treatment method for a vast array of diseases, through the use of both cell-based and cell-free delivery mechanisms [73,74].
The number of clinical trials using MSCs to treat various musculoskeletal conditions has drastically increased in recent years, including their beneficial use in treating osteoarthritis and other bone defects [75]. MSCs possess extensive potential to be used for cell-directed therapeutic treatment methods due to their phenotypic plasticity and the expression of key molecules involved in regulating osteogenesis, including miRs, which can be released in response to external stimuli [73]. In addition, MSCs can be used as vehicles for drug delivery because they can be easily manipulated and loaded particularly with miRs [76]. Moreover, MSCs express connexins that form channels able to mediate cell-to-cell transfer of miRs [77,78]. These findings highlight the potential therapeutic applications of using MSC cell based treatment methods and the importance of understanding and exploiting gap junction-mediated miR transfer.
In addition to direct cell-to-cell communication, recent studies have demonstrated the therapeutic potential of MSC-derived exosomes as a cell-free method to treat musculoskeletal conditions [74,75]. MSC-derived exosomes, as well as exosomes from other sources, selectively package miRs to regulate cellular processes and, in particular, osteogenic differentiation by potentially modifying RNA degradation, mRNA surveillance, RNA transport and Wnt signaling [62,74]. In addition, umbilical cord-derived MSCs were found to release factors that stimulated cartilage and bone repair in a calvaria critical defect rat model [79] and, similarly, exosomes released by human-induced pluripotent stem cell derived from MSCs were able to repair critical-sized bone defects in osteoporotic rats by stimulating both osteogenesis and angiogenesis [80]. Further, like MSCs, exosomes and other MSC-derived EVs have been used as vehicles to deliver drugs or bioactive molecules and genetic material including miRs [81]. Although there are still numerous unknowns with regards to exosomes as means of therapeutic treatment, the use of EVs is promising due to their highly stable nature and ability to transport a variety of important regulatory molecules, combined with the promising initial findings in studies testing their regenerative capabilities [82]. Exosomes, especially those derived from MSCs, appear to possess extensive therapeutic potential and merit further investigation, thus highlighting the importance of studying exosomal miR based delivery methods as a means to treat and prevent musculoskeletal diseases.
Conclusions
Recent findings have demonstrated that both connexins and miRs modulate the differentiation and function of bone cells, via complementary and opposing effects. Further, new light has been shed on the interaction between connexins and miRs, a very complex process that involves bidirectional miR- and connexin-mediated regulation of gene/protein expression. In addition, gap junction- and hemichannel-mediated miR transport to both adjacent and distant cells plays a role in cell-to-cell signaling in bone. Transport of miRs and/or Cx43 via exosomes is another sophisticated way of interaction, which enhances the successful delivery of the molecules and also regulates molecular signaling in the recipient cell. In addition, the presence of Cx43 on the surface of exosomes facilitates the docking with the plasma membrane of a recipient cell and the delivery of the exosome cargo (in particular, of miRs).
In this review we have highlighted the relationship between miRs and connexins with emphasis on their known contribution to the regulation of skeletal development and bone remodeling. Numerous studies have demonstrated the critical roles of both miRs and connexins in maintaining proper bone cell signaling and skeletal homeostasis; however, there are still numerous unanswered questions regarding this area of research. These unknowns include the specific mechanisms that link and control the post-translational regulation of miRs and connexins and those that control intercellular miR transfer to both adjacent and distant cells.
Human and Animal Rights and Informed Consent.
The protocols involving mice were approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine. This article does not contain studies with human subjects performed by any of the authors.
Acknowledgments
This research was supported by the National Institutes of Health (R01-AR067210 and R01-AR053643 to LIP). HMD is supported by an NIH T32-AR065971 grant. RPC received a scholarship from Coordination of Improvement of Higher Level Personnel (CAPES), Brazil (PDE# 232636/2014-1).
Footnotes
Lilian I. Plotkin: ORCID: orcid.org/0000-0002-9537-4544
Rafael Pacheco-Costa: ORCID: orcid.org/0000-0001-7259-079X
Hannah M. Davis: ORCID: orcid.org/0000-0002-8398-1259
Compliance with Ethical Standards
Conflict of Interest
Lilian I. Plotkin, Rafael Pacheco-Costa, and Hannah M. Davis declare that no potential conflicts of interest.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Plotkin LI, Stains JP. Connexins and pannexins in the skeleton: gap junctions, hemichannels and more. Cell Mol Life Sci. 2015;72:2853–2867. doi: 10.1007/s00018-015-1963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batra N, Kar R, Jiang JX. Gap junctions and hemichannels in signal transmission, function and development of bone. Biochim Biophys Acta. 2012;1818:1909–1918. doi: 10.1016/j.bbamem.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saez JC, Martinez AD, Branes MC, et al. Regulation of gap junctions by protein phosphorylation. Braz J Med Biol Res. 1998;31:593–600. doi: 10.1590/s0100-879x1998000500001. [DOI] [PubMed] [Google Scholar]
- 4.Valiunas V, Polosina YY, Miller H, et al. Connexin-Specific Cell to Cell Transfer of Short Interfering RNA by Gap Junctions. J Physiol. 2005;568:459–468. doi: 10.1113/jphysiol.2005.090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemcke H, Steinhoff G, David R. Gap junctional shuttling of miRNA—A novel pathway of intercellular gene regulation and its prospects in clinical application. Cell Signal. 2015;27:2506–2514. doi: 10.1016/j.cellsig.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Chen Y, Zhang L, et al. The roles of bone-derived exosomes and exosomal microRNAs in regulating bone remodelling. J Cell Mol Med. 2016:1–9. doi: 10.1111/jcmm.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans WH. Cell communication across gap junctions: a historical perspective and current developments. Biochem Soc Trans. 2015;43:450–459. doi: 10.1042/BST20150056. [DOI] [PubMed] [Google Scholar]
- 9.Oshima A. Structure and closure of connexin gap junction channels. FEBS Lett. 2014;588:1230–1237. doi: 10.1016/j.febslet.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 10.Martin PE, Evans WH. Incorporation of connexins into plasma membranes and gap junctions. Cardiovasc Res. 2004;62:378–387. doi: 10.1016/j.cardiores.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Plotkin LI, Bellido T. Beyond gap junctions: Connexin43 and bone cell signaling. Bone. 2013;52:157–166. doi: 10.1016/j.bone.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plotkin LI. Connexin 43 hemichannels and intracellular signaling in bone cells. Front Physiol. 2014;5:131. doi: 10.3389/fphys.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paznekas WA, Boyadjiev SA, Shapiro RE, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koval M, Harley JE, Hick E, et al. Connexin46 is retained as monomers in a trans-Golgi compartment of osteoblastic cells. J Cell Biol. 1997;137:847–857. doi: 10.1083/jcb.137.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minkoff R, Rundus VR, Parker SB, et al. Gap junction proteins exhibit early and specific expression during intramembranous bone formation in the developing chick mandible. Anat Embryol (Berl) 1994;190:231–241. doi: 10.1007/BF00234301. [DOI] [PubMed] [Google Scholar]
- 16.Pizard A, Burgon PG, Paul DL, et al. Connexin 40, a target of transcription factor Tbx5, patterns wrist, digits, and sternum. Mol Cell Biol. 2005;25:5073–5083. doi: 10.1128/MCB.25.12.5073-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacheco-Costa R, Hassan I, Reginato RD, et al. High Bone Mass in Mice Lacking Cx37 Due to Defective Osteoclast Differentiation. J Biol Chem. 2014;289:8508–8520. doi: 10.1074/jbc.M113.529735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacheco-Costa R, Kadakia JR, Atkinson EG, et al. Connexin37 deficiency alters organic bone matrix, cortical bone geometry, and increases Wnt/beta-catenin signaling. Bone. 2017;97:105–113. doi: 10.1016/j.bone.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Lecanda F, Warlow PM, Sheikh S, et al. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol. 2000;151:931–944. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plotkin LI, Laird DW, Amedee J. Role of connexins and pannexins during ontogeny, regeneration, and pathologies of bone. BMC Cell Biology. 2016;17:29–38. doi: 10.1186/s12860-016-0088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternlieb M, Paul E, Donahue HJ, et al. Ablation of connexin 43 in osteoclasts leads to decreased in vivo osteoclastogenesis. J Bone Miner Res. 2012;27:S53. [Google Scholar]
- 22.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 23.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 24.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tetreault N, De G V. miRNAs: their discovery, biogenesis and mechanism of action. Clin Biochem. 2013;46:842–845. doi: 10.1016/j.clinbiochem.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 27.Sun M, Zhou X, Chen L, et al. The Regulatory Roles of MicroRNAs in Bone Remodeling and Perspectives as Biomarkers in Osteoporosis. Biomed Res Int. 2016;2016:1652417. doi: 10.1155/2016/1652417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ell B, Kang Y. MicroRNAs as regulators of bone homeostasis and bone metastasis. Bonekey Rep. 2014;3:549. doi: 10.1038/bonekey.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014;71:4747–4761. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Eerden C. MicroRNAs in the skeleton: cell-restricted or potent intercellular communicators? Arch Biochem Biophys. 2014;561:46–55. doi: 10.1016/j.abb.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: A Collision of Two Paradigms. Trends Biochem Sci. 2016;41:883–892. doi: 10.1016/j.tibs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014:3. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo CA, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Schwab A, Meyering SS, Lepene B, et al. Extracellular vesicles from infected cells: potential for direct pathogenesis. Front Microbiol. 2015;6:1132. doi: 10.3389/fmicb.2015.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buzas EI, Gyorgy B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10:356–364. doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 37.Ciardiello C, Cavallini L, Spinelli C, et al. Focus on Extracellular Vesicles: New Frontiers of Cell-to-Cell Communication in Cancer. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38 •.Soares AR, Martins-Marques T, Ribeiro-Rodrigues T, et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci Rep. 2015;5:13243. doi: 10.1038/srep13243. This article shows the presence of Cx43 hemichannels in exosomes and its important participation on the delivery of nucleic acid to recipient cells and their trasnference between exosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela-Eirin M, Varela-Vazquez A, Mateos MR, et al. Recruitment of RNA molecules by connexin RNA-binding motifs: implication in RNA and DNA transport through microvesicles and exosomes. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbamcr.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Inose H, Ochi H, Kimura A, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci U S A. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G, Luo G, Bo Z, et al. Impaired osteogenic differentiation associated with connexin43/microRNA-206 in steroid-induced avascular necrosis of the femoral head. Exp Mol Pathol. 2016;101:89–99. doi: 10.1016/j.yexmp.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 42 •.Gindin Y, Jiang Y, Francis P, et al. miR-23a impairs bone differentiation in osteosarcoma via down-regulation of GJA1. Front Genet. 2015;6:233. doi: 10.3389/fgene.2015.00233. This article evidence the Cx43 regulation by miR in a osteoblastic cell lineage. This article shows Cx43 regulation by miRs in a cell of osteoblastic lineage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozeki N, Hase N, Hiyama T, et al. MicroRNA-211 and autophagy-related gene 14 signaling regulate osteoblast-like cell differentiation of human induced pluripotent stem cells. Exp Cell Res. 2017 doi: 10.1016/j.yexcr.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 44.Bejarano E, Yuste A, Patel B, et al. Connexins modulate autophagosome biogenesis. Nat Cell Biol. 2014 doi: 10.1038/ncb2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45 ••.Davis HM, Pacheco-Costa R, Atkinson EG, et al. Disruption of the Cx43/miR 21 pathway leads to osteocyte apoptosis and increased osteoclastogenesis with aging. Aging Cell. 2017 doi: 10.1111/acel.12586. This article shows the requirement of Cx43 and miR21 to maintain osteocyte survival and identified RANKL and HMGB1 as two molecules inved with elevated osteoclastogenesis. In addition, provides the first description of miR regulation via connexin in osteocytic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu CH, Sui BD, Du FY, et al. miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep. 2017;7:43191. doi: 10.1038/srep43191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zong L, Zhu Y, Liang R, et al. Gap junction mediated miRNA intercellular transfer and gene regulation: A novel mechanism for intercellular genetic communication. Sci Rep. 2016;6:19884. doi: 10.1038/srep19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim PK, Bliss SA, Patel SA, et al. Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 2011;71:1550–1560. doi: 10.1158/0008-5472.CAN-10-2372. [DOI] [PubMed] [Google Scholar]
- 49.Vojtech L, Woo S, Hughes S, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng L, Wang Y, Peng Y, et al. Osteoblast-derived microvesicles: A novel mechanism for communication between osteoblasts and osteoclasts. Bone. 2015;79:37–42. doi: 10.1016/j.bone.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Cui Y, Luan J, Li H, et al. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016;590:185–192. doi: 10.1002/1873-3468.12024. [DOI] [PubMed] [Google Scholar]
- 52.Lauvrak SU, Munthe E, Kresse SH, et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J Cancer. 2013;109:2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassan MQ, Maeda Y, Taipaleenmaki H, et al. miR-218 Directs a Wnt Signaling Circuit to Promote Differentiation of Osteoblasts and Osteomimicry of Metastatic Cancer Cells. J Biol Chem. 2012;287:42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhushan R, Grunhagen J, Becker J, et al. miR-181a promotes osteoblastic differentiation through repression of TGF-beta signaling molecules. Int J Biochem Cell Biol. 2013;45:696–705. doi: 10.1016/j.biocel.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 55.Kim YJ, Bae SW, Yu SS, et al. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 56.Qin Y, Wang L, Gao Z, et al. Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Sci Rep. 2016;6:21961. doi: 10.1038/srep21961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Egea V, Zahler S, Rieth N, et al. Tissue inhibitor of metalloproteinase-1 (TIMP-1) regulates mesenchymal stem cells through let-7f microRNA and Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2012;109:E309–E316. doi: 10.1073/pnas.1115083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei J, Li H, Wang S, et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014;23:1452–1463. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, et al. MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev. 2010;19:877–885. doi: 10.1089/scd.2009.0112. [DOI] [PubMed] [Google Scholar]
- 60.Xu S, Cecilia SG, De VK, et al. Upregulation of miR-135b is involved in the impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients. PLoS ONE. 2013;8:e79752. doi: 10.1371/journal.pone.0079752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, Zhao L, Xing L, et al. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu JF, Yang GH, Pan XH, et al. Altered microRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. PLoS ONE. 2014;9:e114627. doi: 10.1371/journal.pone.0114627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Tu Q, Bonewald LF, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26:1953–1963. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You L, Gu W, Chen L, et al. MiR-378 overexpression attenuates high glucose-suppressed osteogenic differentiation through targeting CASP3 and activating I3K/Akt signaling pathway. Int J Clin Exp Pathol. 2014;7:7249–7261. [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hwang S, Park SK, Lee HY, et al. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014;588:2957–2963. doi: 10.1016/j.febslet.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 68.Cheng P, Chen C, He HB, et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. 2013;28:1180–1190. doi: 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 69.Chen C, Cheng P, Xie H, et al. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res. 2014;29:338–347. doi: 10.1002/jbmr.2032. [DOI] [PubMed] [Google Scholar]
- 70.Zhao C, Sun W, Zhang P, et al. miR-214 promotes osteoclastogenesis by targeting Pten/I3k/Akt pathway. RNA Biol. 2015;12:343–353. doi: 10.1080/15476286.2015.1017205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71 ••.Li D, Liu J, Guo B, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7:10872. doi: 10.1038/ncomms10872. This article shows the osteoclast-derived exosomes released from ovariectomized mice and elderly fractured women contain a miR able to target osteoblasts and then inhibit bone formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim YH, Goh TS, Lee CS, et al. Prognostic value of microRNAs in osteosarcoma: A meta-analysis. Oncotarget. 2017 doi: 10.18632/oncotarget.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng S, Gao D, Gao C, et al. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review) Mol Med Rep. 2016 doi: 10.3892/mmr.2016.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burke J, Kolhe R, Hunter M, et al. Stem Cell-Derived Exosomes: A Potential Alternative Therapeutic Agent in Orthopaedics. Stem Cells Int. 2016;2016:5802529. doi: 10.1155/2016/5802529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burke J, Hunter M, Kolhe R, et al. Therapeutic potential of mesenchymal stem cell based therapy for osteoarthritis. Clin Transl Med. 2016;5:27. doi: 10.1186/s40169-016-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sasso L, Hosamuddin H, Emanueli C. Extracellular vesicles at the cross-line between basic science and clinical needs. Microcirculation. 2017:24. doi: 10.1111/micc.12333. [DOI] [PubMed] [Google Scholar]
- 77.Valiunas V, Doronin S, Valiuniene L, et al. Human mesenchymal stem cells make cardiac connexins and form functional gap junctions. J Physiol. 2004;555:617–626. doi: 10.1113/jphysiol.2003.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hahn JY, Cho HJ, Kang HJ, et al. Pre-treatment of mesenchymal stem cells with a combination of growth factors enhances gap junction formation, cytoprotective effect on cardiomyocytes, and therapeutic efficacy for myocardial infarction. J Am Coll Cardiol. 2008;51:933–943. doi: 10.1016/j.jacc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 79.Wang KX, Xu LL, Rui YF, et al. The effects of secretion factors from umbilical cord derived mesenchymal stem cells on osteogenic differentiation of mesenchymal stem cells. PLoS ONE. 2015;10:e0120593. doi: 10.1371/journal.pone.0120593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qi X, Zhang J, Yuan H, et al. Exosomes Secreted by Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells Repair Critical-Sized Bone Defects through Enhanced Angiogenesis and Osteogenesis in Osteoporotic Rats. Int J Biol Sci. 2016;12:836–849. doi: 10.7150/ijbs.14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular Vesicles: Novel Mediators of Cell Communication In Metabolic Disease. Trends Endocrinol Metab. 2017;28:3–18. doi: 10.1016/j.tem.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Heldring N, Mager I, Wood MJ, et al. Therapeutic Potential of Multipotent Mesenchymal Stromal Cells and Their Extracellular Vesicles. Hum Gene Ther. 2015;26:506–517. doi: 10.1089/hum.2015.072. [DOI] [PubMed] [Google Scholar]


