Abstract
We have developed a simple, rapid, high-throughput cancer diagnosis system using functional nanoparticles (NPs) consisting of poly(catechin) capped-gold NPs (Au@PC NPs) and smaller nucleolin-binding aptamer (AS1411) conjugated gold NPs (AS1411-Au NPs). The AS1411-Au NPs/Au@PC NP is used as a targeting agent in laser desorption/ionization mass spectrometry (LDI-MS)-based tumor tissue imaging. Self-assembled core-shell Au@PC NPs are synthesized by a simple reaction of tetrachloroaurate(III) with catechin. Au@PC NPs with a well-defined and dense poly(catechin) shell (~40-60 nm) on the surface of each Au core (~60-80 nm) are obtained through careful control of the ratio of catechin to gold ions, as well as the pH of the reaction solution. Furthermore, we have shown that AS1411-conjugated Au NPs (13-nm) self-assembled on Au@PC NP can from a satellite-like gold nanocomposite. The high density of AS1411-Au NPs on the surface of Au@PC NP enhances multivalent binding with nucleolin molecules on tumor cell membranes. We have employed LDI-MS to detect AS1411-Au NPs/Au@PC NPs labeled nucleolin-overexpressing MCF-7 breast cancer cells through the monitoring of Au cluster ions ([Aun]+; 1 ≤ n ≤ 3). The ultrahigh signal amplification from Au NPs through the formation of a huge number of [Aun]+ ions results in a sensing platform with a limit of detection of 100 MCF-7 cells mL-1. Further, we have applied the satellite-like AS1411-Au NPs/Au@PC NP nanocomposite as a labeling agent for tumor tissue imaging by LDI-MS. Our nanocomposite-assisted LDI-MS imaging platform can be extended for simultaneous analysis of different tumor markers on cell membranes when using different ligand-modified metal nanoparticles.
Keywords: self-assembly, aptamers, gold nanocomposites, laser desorption/ionization mass spectrometry, tissue imaging
Introduction
Techniques for early cancer detection are important to reduce mortality rates of various cancers. Although traditional tissue section and cell morphological analysis can determine the degree of malignancy in clinical diagnosis, they are not sensitive enough for early cancer detection 1, 2. Biomedical imaging techniques, including X-ray computed tomography, magnetic resonance, ultrasound, endoscopy, and nuclear medicine functional imaging such as positron emission tomography and single-photon emission computed tomography are widely assisted with tissue analysis in clinical decision-making for cancer diagnosis 3-6. However, these imaging techniques for early diagnosis of cancer are limited due to low resolution, sensitivity, specificity and multiplexing ability. Thus, development of rapid, and highly specific imaging techniques with high resolution for early cancer detection remains a challenge.
Mass spectrometry imaging (MSI) has attracted much attention for analysis of biomolecules from tissue samples 7-11. Many MSI techniques, including matrix-assisted laser desorption/ionization (MALDI) 12-14, matrix-free laser desorption/ionization 15, 16, laser ablation electrospray ionization and desorption electrospray ionization 17-25, and secondary ion 26-29 MS are employed for imaging or mapping of biomolecules in tissue samples. MSI is able to identify multiple biomolecules simultaneously, but it requires a large database for molecular identification and its sensitivity and resolution for large proteins (molecular weight >80 kDa) are not great. The most convenient MALDI-MSI techniques are conducted in an organic matrix to assist desorption and ionization of biomolecules 12-14. However, high background signals from the organic matrix sometimes hinder identification of compounds with low molecular weights. The laser desorption/ionization MS (LDI-MS) techniques could be employed for the analysis the distribution of nanomaterials in the cell and tissue samples 30, 31. Recently, nanomaterials and nanostructured substrates prepared from metal, metal oxide, silicon and carbon materials have been widely used as matrices in MS analysis to achieve higher resolution and lower background noise 32-39. However, the LDI-MS techniques for tissue imaging can only detect highly abundant molecules, and therefore analysis of low-abundant proteins remains limited 40-43.
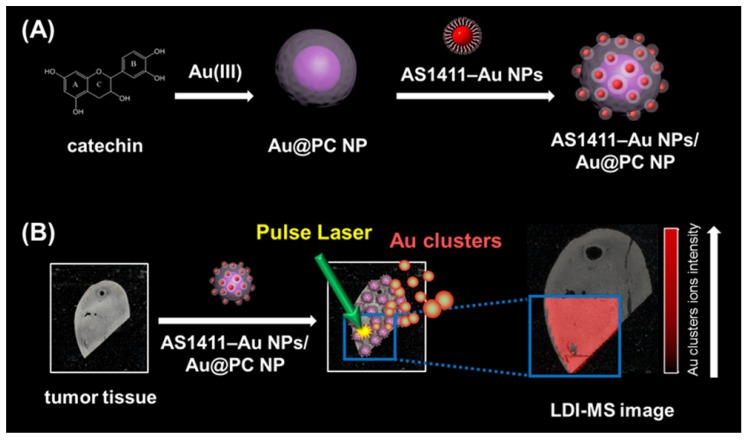
In this study, we have developed LDI-MS using functional nanocomposites for the detection of tumor cells and for tissue imaging (Scheme 1). Nucleolin binding aptamer (AS1411)-conjugated gold nanoparticles (AS1411-Au NPs) self-assembled on bigger core-shell gold-poly(catechin) nanoparticle (Au@PC NP) to prepare the functional nanocomposites (AS1411-Au NPs/Au@PC NP). Overexpression of nucleolin in some tumor cells was found to correlate with their longer half-life through binding to mRNA of Bcl-2 (an antiapoptotic protein) 44, 45. We demonstrated by LDI-MS analysis that nanosatellite-like AS1411-Au NPs/Au@PC NP nanocomposites bind selectively to tumor cells, mainly through a strong affinity of AS1411 aptamer toward nucleolin on tumor cell membranes [dissociation constant (Kd) is in pM to low nM range] 46. Gold cluster ions ([Aun]+) derived from AS1411-Au NPs/Au@PC NPs under LDI act as amplified signal reporters for the detection of tumor cells and for obtaining tumor tissue images. The AS1411-Au NPs/Au@PC NPs coupled with LDI-MS can detect down to 100 cells mL-1 of breast tumor MCF-7 cells. This simple and sensitive LDI-MS platform can discriminate normal from cancerous breast tissues.
Scheme 1.
Schematic representation of (A) the preparation of nucleolin-binding aptamer-modified gold nanoparticles (AS1411-Au NPs) and their conjugation to core-shell Au@poly(catechin) NP to form AS1411-Au NPs/Au@PC NP nanocomposite for (B) tumor tissue imaging through monitoring Au cluster ions by laser desorption/ionization mass spectrometry.
Experimental
Synthesis and Characterization of Core-shell Gold@poly(catechin) Nanoparticles
Poly(catechin) capped-gold nanoparticles (Au@PC NPs) were synthesized by catechin-mediated reduction of HAuCl4. Tris-acetate buffer solution (100 mM, pH 3.0-9.0, 1 mL) and HAuCl4 (5.0 mM, 1 mL) were added to a 20-mL vial containing deionized (DI) water (3 mL) with constant stirring. Then, freshly prepared catechin (0.1-4.0 mM, 5 mL) was added and the stirring continued for 3 h. The formation of Au NPs was indicated by the color change of the solution from yellow to brown/purple. The as-formed Au@PC NPs were purified via centrifugation [relative centrifugal force (RCF), 30,000 g] for 20 min, and the residue obtained was suspended in DI water. The purification process was repeated three times and the purified Au@PC NPs solutions were stored at 4 °C for 24 h before use. The particle sizes of Au@PC NPs were determined by transmission electron microscopy (TEM) (Tecnai 20 G2 S-Twin TEM, Philips/FEI, Hillsboro, OR, USA). The absorption spectra of the Au@PC NP solutions were recorded by a Synergy H1 multi-mode monochromatic microplate spectrophotometer (Biotek Instruments, Winooski, VT, USA). The X-ray diffraction (XRD) pattern of the Au@PC NPs deposited on Si substrates was recorded using a PANalytical X'Pert PRO diffractometer (PANalytical B.V., Almelo, Netherlands) and Cu-Kα radiation (λ = 0.15418 nm). The particle concentration of Au@PC NP was determined assuming them as ideal spherical particles and by using eq. 1.
| n = 3 m/4πr3s | (1) |
where, n is the number of Au particles per milliliter, m is the concentration of gold in the substance (g mL-1), r is the particle radius (cm), and s is the specific gravity of colloidal gold (19.3 g cm-3). Values of m and r were determined via inductively coupled plasma mass spectroscopy (ICP-MS; PerkinElmer ELAN 6000, Waltham, MA, USA) and TEM measurements, respectively. For the convenience of calculation, the obtained concentration in particles per mL was converted into molar concentration, by first converting it into number of particles per liter, followed by dividing it by Avogadro's number.
Preparation of Satellite-like AS1411-Au NPs/Au@PC NP Nanocomposites
Spherical Au NPs having average size of 13.3 nm were prepared by the reduction of HAuCl4 with trisodium citrate (please see details in the Supporting Information). The modified AS1411 aptamer used in this study has the sequence 5'-AAA AAA AAA AAA AAA AAA AAG GTG GTG GTG GTT GTG GTG GTG GTG G-3', which is comprised of two blocks, a 20-repeat polyadenine (A20) as an anchoring group to conjugate with Au NPs and a segment of the sequence providing AS1411 functionality. The functionalization of Au NPs with non-thiolated DNA was done by following previous reports with a slight modification 47-49. Briefly, aqueous citrate-stabilized Au NP solution was mixed with the AS1411 aptamer in 5 mM sodium phosphate buffer (pH 7.4) solution containing 50 mM NaCl in a 1.5 mL vial. The final concentrations of the Au NPs and the aptamer were 8 nM and 2.0 μM, respectively. After 2 h of incubation, the aptamer-modified Au NPs were salt-aged for 12 h in 500 mM NaCl and centrifuged at RCF of 30,000 g for 20 min to remove the unreacted aptamers. After three centrifuge/wash cycles, the colloids were resuspended in mimetic physiological buffer solution [PBS; 25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5.0 mM KCl, 1.0 mM MgCl2, and 1.0 mM CaCl2]. To determine the number of aptamer molecules on each Au NP, the amount of aptamer in the supernatant after centrifugation was measured using OliGreen dyes. The 2-folded diluted supernatant was mixed with OliGreen (5000-fold diluted) in PBS. The DNA-OliGreen complexes fluoresce ∼1000-fold more intensely than does the free and weakly fluorescent OliGreen. The concentration of AS1411 was calculated by measuring the fluorescence of the mixtures at 524 nm (excitation wavelength: 480 nm) with reference to a standard calibration curve. The nanosatellite AS1411-Au NPs/Au@PC NP was prepared by mixing 25 pM dispersion of Au@PC NPs and 1.0 nM of AS1411-Au NPs in PBS, followed by incubation for 12 h at room temperature to self-immobilize AS1411-Au NPs on Au@PC NP. The as-formed AS1411-Au NPs/Au@PC NP was stored at 4 °C.
Analysis of Tumor Cells by AS1411-Au NPs/Au@PC NP-LDI-MS
Cultured MCF-10A, MDA-MB-231 and MCF-7 cell (0-10,000 cells, 1.0-mL) samples were labeled separately with AS1411-Au NPs/Au@PC NP or rDNA-Au NPs/Au@PC NP (5 pM; rDNA: random DNA with the same length as AS1411) for 1 h in PBS. A portion of each suspended solution (1 mL) was passed through a track-etched polycarbonate membrane (PCM; pore size: 5 μm, diameter: 13 mm) by a syringe pump (1 mL min-1). The PCMs were then air-dried at room temperature for 10 min. The nanocomposite-bound cells remained on the PCMs, and the PCMs were attached to a stainless-steel LDI plate for MS experiments. MS experiments were conducted using an Autoflex III LDI time-of-flight (TOF) mass spectrometer (Bruker Daltonics, Bremen, Germany) in the reflectron positive-ion mode. SmartBeam laser from Nd:YAG (355 nm) at 100 Hz was used for sample desorption and ionization. A delayed extraction period of 30 ns was set to energetically stabilize the ions produced by laser irradiation, before accelerating through the TOF chamber, and an accelerating voltage of +20 to -20 kV was applied. Before MS analysis, the instruments were calibrated with Au clusters using their theoretical masses. Samples were irradiated with 2000 pulsed laser shots at 10 LDI target positions at a power density of 5.0 × 104 W cm-2.
Tissue Imaging
Human breast tumor and normal breast tissue microarray (T087b; 6 cases; diameter: 1.5 mm; thickness: 5 μm; US Biomax Inc., Derwood, MD, USA), human normal breast tissue section (BRE01; Pantomics Inc., Richmond, CA, USA), and human breast cancer section (BRE06; Pantomics Inc.) were used for tissue imaging. All the experiments involving tissue samples were conducted as per the protocols approved by the Animal Administration Committee of National Taiwan University. The tissue samples were deparaffinized and then labeled with AS1411-Au NPs/Au@PC NP, prepared from 0.2 nM AS1411-Au NPs and 5 pM Au@PC NP, in PBS for 1 h. The slide was washed three times with PBS before it was cast onto an MTP slide adapter II MALDI plate, and then air-dried at room temperature prior to LDI-MS measurements. LDI-MS imaging experiments were performed in the positive-ion mode using an Autoflex III LDI TOF mass spectrometer. The tissue slides were irradiated using a SmartBeam laser (355 nm Nd: YAG) operating at 100 Hz. A laser spot diameter of 30 μm and pixel step size of 150 μm were employed. The ion extraction period and accelerating voltage range were same as that applied for tumor cell analysis by LDI-MS. Prior to analysis, the instruments were calibrated with Au clusters using their theoretical masses. One spot was pulsed for a total of 1000 laser shots at a power density of 5.0 × 104 W cm-2.
See the Supporting information for the details on the materials, preparation and characterization of Au NPs (13-nm), cell cultures, and immunohistochemistry.
Results and discussion
Synthesis and Characterization of Au@PC NPs
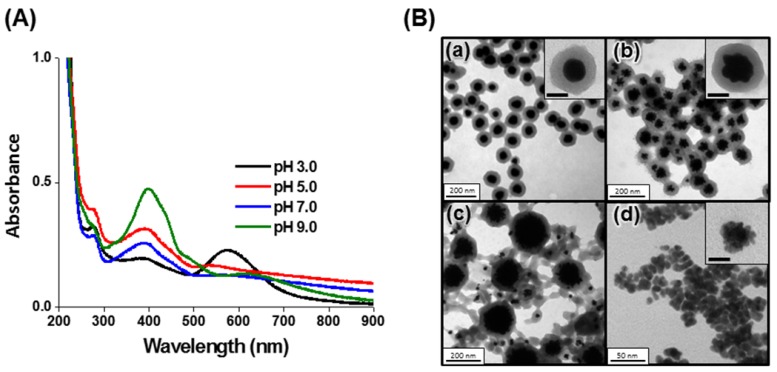
Core-shell Au@PC NPs were synthesized through a simple reaction of HAuCl4 with catechin, a polyphenolic flavonoid-type compound. The catechin acts as a reducing agent, while the as-formed poly(catechin) stabilize the nanocomposites. Oxidized catechin is suspected to self-polymerize to form poly(catechin), although the evidence for this reaction is indirect 50. A series of Au@PC NPs were prepared by varying the pH value (3.0-9.0) of reaction solution and the molar ratio of catechin to HAuCl4 (0.1-4.0). The core-shell Au@PC NPs, after 3 h of reaction, were formed only under acidic solution conditions (pH ≤ 5.0; Figure 1B, Supporting Information). The UV-vis absorption spectra of Au@PC NPs (synthesized at pH 3.0 and pH 5.0) both show two distinct absorption bands at ~575 nm and ~385 nm (Figure 1A, Supporting Information), which are attributed to the surface plasmon resonance (SPR) absorption of the Au NP core and oxidized poly(catechin) shell, respectively 51. It has been demonstrated elsewhere that the catechol B-ring of catechin (3′,4′-dihydroxyl moiety) possess stronger reducing ability than the resorcinol A-ring, and furthermore, that the reducing ability of catechin is pH-dependent 52, 53. Catechin has a stronger reducing ability upon increasing pH value because deprotonation of the catechol groups boosts its electron-donating potency 52, 53. Compared with Au@PC NP prepared at pH 3.0 or pH 5.0, irregular Au NPs were formed at pH 7.0 that have a wide size distribution (Figure 1B(c)), due to the higher growth rate of the gold core. As a result, poly(catechin) did not form a well dense shell on the surface of each Au core. Small-size, aggregated Au NPs (~8.8 nm) were formed at pH 9.0, probably due to the high nucleation rate of Au NPs and formation of less oxidized poly(catechin) to stabilize the Au NPs (Figure 1B(d)). Interestingly, we observed that the Au@PC NP prepared at pH 5.0 has a raspberry-shaped gold core, compared to the rounder shape of that formed at pH 3.0 (Figure 1B(a) and 1B(b)). A relatively fast reaction for growth of Au NPs at pH 5.0 might be responsible for the formation of anisotropic raspberry-shaped gold cores.
Figure 1.
(A) UV-vis absorption spectra and (B) TEM images of the Au@PC NPs synthesized from 0.5 mM HAuCl4 and 0.5 mM catechin in Tris-acetate buffer solution (10 mM) at (a) pH 3.0, (b) pH 5.0, (c) pH 7.0, and (d) pH 9.0. The insets to (B) show corresponding high-magnification TEM images. The scale bars in insets represent (a) 50 nm, (b) 50 nm and (d) 5 nm.
We further investigated the effect of catechin concentration on the synthesis of Au@PC NPs. A series of Au@PC NPs were synthesized in Tris-acetate solution (pH 3, 10 mM) with a constant concentration of HAuCl4 (0.5 mM) while varying the concentration of catechin (0.05-2.00 mM). Large-size Au NPs (~90 nm) without a poly(catechin) shell were observed when a low concentration of catechin was used (0.05 mM; Figure S1B(a), Supporting Information). A dense of poly(catechin) shell is formed on the Au NPs when the concentration catechin is 0.5 mM ([catechin]/[HAuCl4] = 1), and the as-prepared Au@PC NPs show a narrow SPR band at 575 nm, indicating that the Au@PC NPs were dispersed well in the solution (Figure S1(c)). Moreover, the shell of poly(catechin) becomes thinner, and a large degree of uncontrollable poly(catechin) sol appears at a higher concentration (2.0 mM) of catechin; as shown in Figure S1B(e). The formation of poly(catechin) sol mainly arises through the autoxidation of excess catechin. We further studied Au@PC NPs that were synthesized at a constant molar ratio ([catechin] = [HAuCl4]; [catechin]/[HAuCl4] = 0.1, 0.5, 1.0, or 5.0 mM) in Tris-acetate solution (pH 3.0, 10 mM). Under the optimized molar ratio of [catechin]/[HAuCl4] and pH value, all synthesized Au@PC NP have an ideal spherical Au NPs core and condensed poly(catechin) shell, and much less uncoated poly(catechin) (Figure S2, Supporting Information). The core sizes and shell thickness of all synthesized Au@PC NPs are listed in Table S1 (Supporting Information). The size of core NPs and shell thickness of poly(catechin) determined by TEM are expressed as mean ± standard deviations (100 counts). The representative histograms of the Au@PC NPs synthesized from 0.5 mM HAuCl4 and 0.5 mM catechin in Tris-acetate buffer solution (10 mM, pH 3.0-9.0) are shown in Figure S3 (Supporting Information). Overall, we can conclude that careful control of catechin concentration, the molar ratio of catechin to Au ions, and the solution pH results in the mediation of the nucleation and growth rates of Au NPs, and thereby varying the polymerization rate of oxidized catechin and the interaction of poly(catechin) with Au NP. Consequently, a well-defined poly(catechin) shell with a narrow-size distribution of Au NPs can be formed.
We further characterized the optimized Au@PC NP (core size ~70 nm, shell thickness ~50 nm), which were synthesized through the reaction of HAuCl4 (0.5 mM) with catechin (0.5 mM) in 10 mM Tris-acetate (pH 3.0). The optimized Au@PC NP has a zeta potential of ca. -30 mV. The high zeta potential and a dense shell of poly(catechin) endow the Au@PC NP with stability in biological mimetic solution [25 mM Tris-HCl (pH 7.4), 150 m M NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 100 μM BSA; Figure S4, Supporting Information]. The X-ray diffraction (XRD) pattern of Au@PC NPs reveal that the Au core dominates the (111) crystal plane of fcc Au (Figure S5, Supporting Information). The poly(catechin) in Au@PC NPs is about 22 wt% as obtained from thermal gravimetric analysis (TGA) data (Figure S6, Supporting Information). Fourier transform infrared spectroscopy (FTIR) of Au@PC NP reveals a large number of phenolic -OH (3357 cm-1) and aromatic -C=C (1610 cm-1) groups (Figure S7, Supporting Information), suggesting that the interaction Au@PC NP with AS1411-Au NPs is mainly through hydrogen bonding and π-π stacking interactions.
Preparation and Characterization of AS1411-Au NPs/Au@PC NP
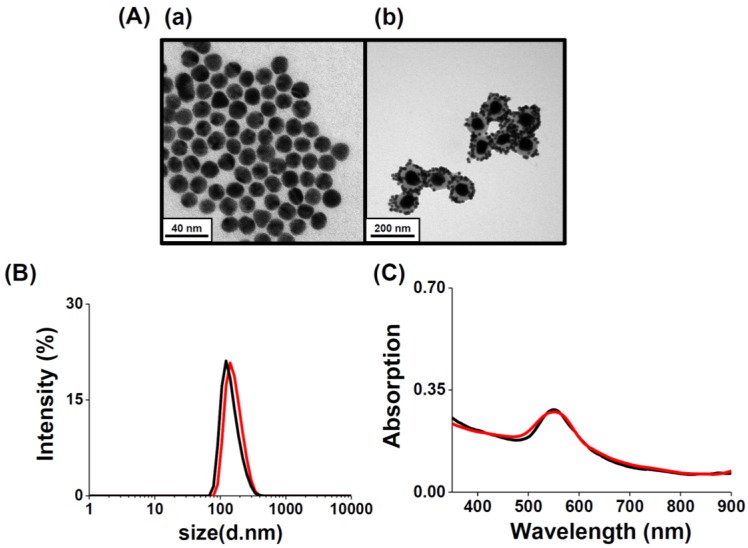
Aptamer-assembled Au NPs (AS1411-Au NPs) were obtained by mixing Au NPs (13-nm) with aptamer solutions, followed by salt-aging and purification. The number of AS1411 molecules on each NP was estimated by measuring the fluorescence of the supernatant after staining with OliGreen fluorescent dye, and found to be 30. Similar to Au-S bonding, the polyA sequence at the 5'-terminus of the aptamer can preferentially bind with the Au NPs with high affinity 47-49. Non-thiolated, poly(adenine)-terminal AS1411 aptamers are significantly cheaper than thiolated ones, and possess high flexibility after modifying the Au NPs 47, 48. Moreover, the as-prepared AS1411-Au NPs remain stable in 2-fold diluted human plasma after 2 h incubation; the AS1411-Au NPs are not aggregated (Figure S8, Supporting Information), and AS1411 released from the Au NPs is less than 5%. The steric effect of the G-quadruplex structure of AS1411 blocks the aminothiol and protein molecules in plasma from accessing and interacting with the Au NP surfaces. Satellite-like AS1411-Au NPs/Au@PC NP was prepared by simply mixing 1.0 nM of AS1411-Au NPs and 25 pM of Au@PC NP [prepared from 0.5 mM HAuCl4 solution and 0.5 mM catechin solution in 10 mM Tris-acetate (pH 3.0)] in PBS. AS1411-Au NPs were self-assembled on Au@PC NP, mainly through multivalent π-π stacking and hydrogen bonding between nucleobase-poly(catechin), although hydrophobic interactions cannot not be excluded. We note that the AS1411-Au NPs (>95%) were bound on the Au@PC NP demonstrated by the agarose gel electrophoresis separation (data not shown), indicating ca. 40 AS1411-Au NPs on per Au@PC NP. The AS1411 molecules on the Au NPs that are opposite to the interaction side of the AS1411-Au NPs and Au@PC NP retain their targeting ability to nucleolin proteins on cell membranes. As evident from TEM images of AS1411-Au NPs/Au@PC NP, the AS1411-Au NPs are uniformly distributed on the Au@PC NP (Figure 2A(b)). Absorption spectra studies and dynamic light scattering (DLS) measurements further reveal that self-assembled AS1411-Au NPs/Au@PC NP nanocomposites are very stable in biological mimetic solution (Figure 2B and 2C). Furthermore, in vitro cytotoxicity assays have revealed superior biocompatibility of AS1411-Au NPs/Au@PC NP (Figure S9, Supporting Information).
Figure 2.
(A) TEM images of as-prepared (a) AS1411-Au NPs and (b) AS1411-Au NPs/Au@PC NP nanocomposites. (B) DLS spectra and (C) UV-vis absorption spectra of as-synthesized AS1411-Au NPs/Au@PC NP (2-fold diluted) in (black) DI water and (red) biological mimetic solution.
LDI-MS of AS1411-Au NPs/Au@PC NP
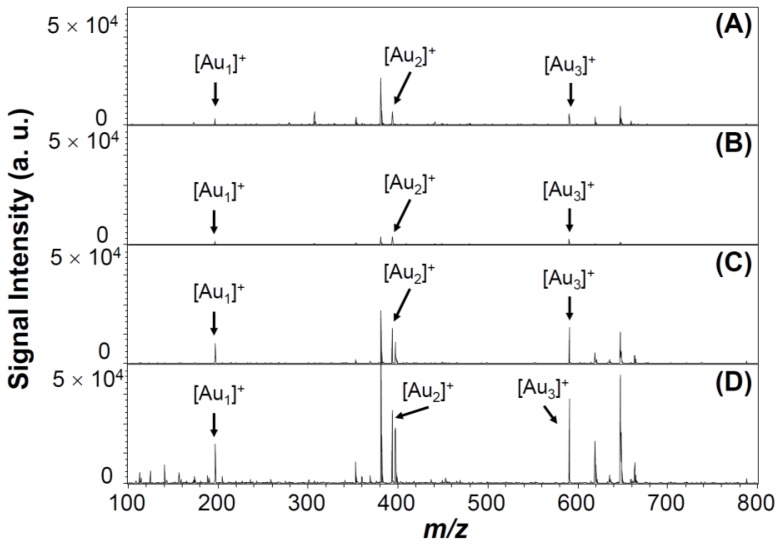
Under pulsed laser irradiation of Au NPs, photoabsorption occurs that results in desorption, fragmentation, and ionization of surface Au atoms and/or whole Au NPs, through photothermal evaporation and Coulomb explosion processes 54-56. The photothermal evaporation happens as a result of transfer of thermal energy to the lattice system of metallic nanoparticles, and results in the formation of neutral and charged metallic clusters upon reaching the lattice temperature (Tℓ) 54-56. In contrast, Coulomb explosion is the ejection of a large number of electrons from metallic nanoparticles, followed by generation of positively charged metallic cluster ions 54-56. LDI-induced formation of Au cluster ions by fragmentation and ionization from Au NPs depends on nanoparticle properties (composition, crystallization, size, and surface ligand), environmental temperature and pressure, as well as laser parameters (wavelength, intensity, and pulse width) 57-59. The Au NPs may transfer their adsorption laser energy to surface AS1411 ligands, and the dense “mantle” of aptamer may strongly inhibit the Au NPs' ejection of electrons, thereby resulting in large decreases in the Tℓ and electron temperature (Te). As a result, the formation of Au cluster ions [Aun]+ (1 ≤ n ≤ 3) from the Au NPs is reduced to 60 % when they were capped with AS1411 (Figure 3A and 3B). However, the intensity of Au cluster ions formed from AS1411-Au NPs is enhanced after anchoring on the Au@PC NP (Figure 3D). The AS1411-Au NPs/Au@PC NP exhibits much stronger Au cluster ions intensity than that of AS1411-Au NPs and Au@PC NP. This phenomenon arises mainly because of the high UV-region absorption coefficients of poly(catechin) in the nanocomposites. The poly(catechin) probably absorbs the laser energy and then transfers it to the AS1411-Au NPs, thereby facilitating formation of [Aun]+ cluster ions. The dense poly(catechin) layer, rich with stacked phenyl rings, may assist electron ejection from the interface of AS1411 and Au NPs, resulting in efficient formation of [Aun]+ cluster ions 54-56.
Figure 3.
LDI-MS spectra of (A) Au NPs (1.0 nM), (B) AS1411-Au NPs (1.0 nM), (C) Au@PC NP (25 pM), and (D) AS1411-Au NPs/Au@PC NP ([AS1411-Au NPs] = 1.0 nM; [Au@PC NP] = 25 pM). Signals at m/z 196.97, 393.93, and 590.90 are assigned to [Au1]+, [Au2]+, and [Au3]+ ions, respectively. 500 pulsed laser shots were applied at five LDI-targeted positions at a laser power density of 5.0 × 104 W cm-2. Signal intensities are plotted in arbitrary units (a. u.).
Specific Detection of Tumor Cells
We applied the AS1411-Au NPs/Au@PC NP-LDI-MS platform to the analysis of nucleolin-overexpressing MCF-7 cells. The multivalent AS1411-Au NPs on Au@PC NP have superior cooperative affinity toward nucleolin on cell membranes 60-63, and an innately higher efficiency to produce Au cluster ions than that of AS1411-Au NPs. As a result, use of AS1411-Au NPs/Au@PC NP for the LDI-MS based detection of MCF-7 cells results in ~4 folded stronger [Aun]+ signals compared to that using AS1411-Au NPs (Figure 4a and 4b). The AS1411-Au NPs/Au@PC NP probe allows detection of MCF-7 cells as low as ~100 cells mL-1 through monitoring the [Au1]+ signal by LDI-MS (Figure S10, Supporting Information). As a control, random DNA (rDNA)-conjugated Au NPs/Au@PC NP for the analysis of MCF-7 cells show negligible [Aun]+ signals compared with that of AS1411-Au NPs/Au@PC NP (Figure 4c), revealing the high specificity provided by AS1411. The self-assembled rDNA-Au NPs/Au@PC NP show very similar structure and UV-vis absorption to AS1411-Au NPs/Au@PC NP (Figure 2 and Figure S11, Supporting Information). The signal from random DNA (rDNA)-conjugated Au NPs/Au@PC NP (rDNA-Au NPs/Au@PC NP) for the detection of MCF-7 cells is probably due to the nonspecific interaction of the nanocomposites with the cells or plate substrate.
Figure 4.
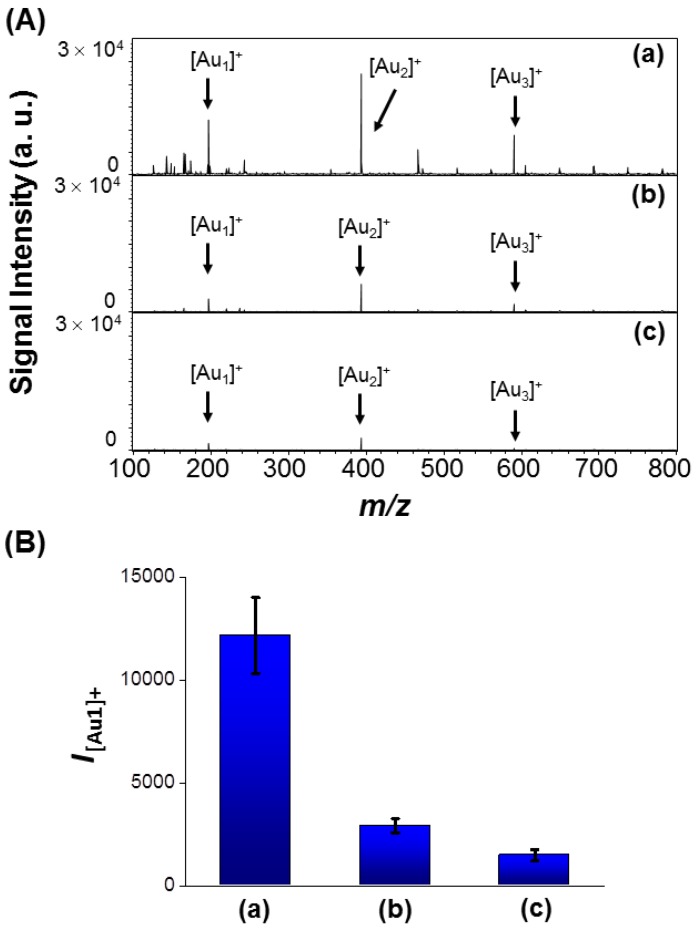
(A) LDI-MS spectra recorded using (a) AS1411-Au NPs/Au@PC NP ([AS1411-Au NPs] = 0.2 nM; [Au@PC NP] = 5 pM), (b) AS1411-Au NPs (0.2 nM), and (c) rDNA-Au NPs/Au@PC NP ([random DNA-Au NPs] = 0.2 nM; [Au@PC NP] = 5 pM) as probes for the detection of 104 MCF-7 cells. (B) Peak intensities of [Au1]+ (I[Au1]+) obtained from (a) AS1411-Au NPs/Au@PC NP, (b) AS1411-Au NPs, and (c) rDNA-Au NPs/Au@PC NP after labeling of MCF-7 cells.
To further show the specificity of the assay, the AS1411-Au NPs/Au@PC NP-LDI-MS platform was applied for analysis of two other cell lines, MCF-10A (normal mammary epithelial cells) and MDA-MB-231 (breast adenocarcinoma cells). [Au1]+ signal intensity obtained for MCF-7 cell line is about 7.3 fold, and MDA-MB-231 cell line is about 15.5-fold higher than that obtained for normal-like MCF-10A cell line (Figure S12, Supporting Information). This result is consistent with that obtained from western blotting analysis (Figure S13, Supporting Information). Moreover, the intensity of the [Au1]+ signal decreases with the increase in the ratio of cell numbers of MCF-10A to MCF-7 in the co-culture samples (Figure S14, Supporting Information). Most human breast cancer cells overexpress nucleolin; however, MCF-7 cells show lower overexpression of nucleolin than that of MDA-MB-231 cells 45, 64. Reports show that, nucleolin located on the breast cell membrane acts as a receptor for several oncogenic ligands, and transformation in the expression and localization of nucleolin can induce many oncogenic effects, such as stabilization of AKT, Bcl-2, Bcl-XL, and IL-2 mRNAs 64, 65. Analysis of tumor cells by other mass spectrometry techniques require sample pretreatment, labeling and purification, and are time consuming, and difficult for specific targeting of tumor cells for quantitation. In contrast, our newly developed platform is more sensitive, specific, and simple, moreover it shows great potential for quantification of tumor cells. Cell surface nucleolin is involved in the proliferation and angiogenesis of tumor cells 66; and accordingly, nucleolin have become an attractive tumor marker and a target for anti-tumor immunotherapies 67. Therefore, our sensing platform can be applied for the determination of nucleolin levels in tumor cells for analysis of the degree of malignancy and metastasis.
Tumor Tissue Imaging
LDI-MS-based tissue imaging techniques, using a suitable organic matrix or nanoparticle substrates, have been widely used for the analysis of small molecules such as lipid components and drugs 12-14, 32-39. However, reports on the analysis of proteins on cell membranes by LDI-MS is rare. Here, we applied the AS1411-Au NPs/Au@PC NP-LDI-MS platform for analysis of nucleolin expression in a typical tissue microarray by monitoring the [Au1]+ signal. The AS1411-Au NPs/Au@PC NPs labeled breast carcinoma or normal tissues were analyzed by LDI-MS. LDI-MS images in Figure S15, Supporting Information, show the total intensity of [Au1]+ signal obtained for breast carcinoma microarray tissue section was over 20-fold higher than that of normal breast microarray tissue section. Our results demonstrate that our AS1411-Au NPs/Au@PC NP-LDI-MS system can be used to analyze nucleolin expression in tissue samples. The resolution of our LDI-MS imaging system ~150 μm that does not match up to fluorescence imaging, however, the strong binding of the nanocomposite with high specificity to the targeted membrane proteins and high signal amplification by Au NPs enable our platform to achieve high targeting ability and sensitivity.
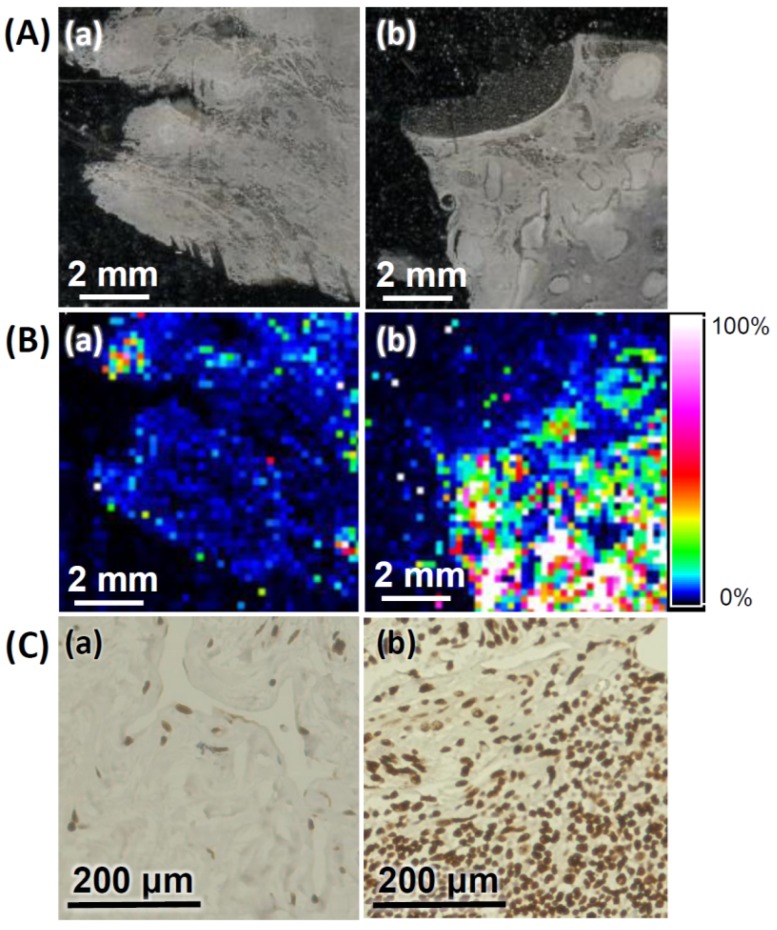
We further applied the AS1411-Au NPs/Au@PC NP-LDI-MS imaging system to analyze the tissue sections of human breast invasive ductal carcinoma; from a 47 years old female, and normal human breast tissue sections from a 33 years old female. Representative MS images generated through monitoring of [Au1]+ signal intensity clearly indicate the tumor tissues relative to normal tissues have much stronger signal intensities (Figure 5B), signifying that our newly developed MSI platform can be employed for the diagnosis of malignancies. Further analysis of nucleolin expression and distribution in breast tissue sections by immunohistochemistry (IHC) show results consistent with those obtained through LDI-MS. The representative IHC image of breast tumor tissue clearly shows the developed brown color being deeper (Figure 5C). Traditional IHC analysis with chromogenic detection requires tedious sandwich binding and enzyme-catalytic color development (amplification) 68. Moreover, IHC analysis is difficult for identification of co-localized multi-proteins, and its dynamic range is narrow. Our simple LDI-MS detection platform has shown a wide dynamic range (>3 orders) for quantitation of tumor cells (Figure S10). Moreover, our AS1411-Au NPs/Au@PC NP-LDI-MS probe shows significant potential for the LDI-MS analysis of co-localized multi-proteins in cell membranes by functionalizing different metal (e.g. gold, silver, platinum) nanoparticles with highly recognized ligands.
Figure 5.
(A) Optical images of (a) normal breast and (b) breast tumor tissues. (B) LDI-MS images of the [Au1]+ intensity obtained for (a) normal breast and (b) breast tumor tissues after labeling with AS1411-Au NPs/Au@PC NPs. (C) Immunostaining of nucleolin in (a) normal breast and (b) tumor breast tissues. The dark brown and blue colors represent the staining of nucleolin and nuclei, respectively. Parameters of LDI-MS imaging: 60 × 60 pixel, laser spot size: 30 μm, and step size: 150 μm. Other conditions were the same as those described in Figure 4.
Conclusions
We synthesized Au@PC NPs, each with an Au NP core stabilized with a dense poly(catechin) shell. The molar ratio of catechin to Au ions and the pH value highly influence the size and morphology of the Au-core, and the density and thickness of the poly(catechin) shell in the synthesis of Au@PC NPs. The poly(catechin) shell on the Au NP is an excellent matrix for immobilization of AS1411 aptamer-modified Au NPs, and acts an efficient pulse light energy transducer. The self-assembled nanosatellite AS1411-Au NPs/Au@PC NP possess high specificity toward the target nucleolin overexpressed tumor cells. Compared to our previous aptamer-Au NPs modified graphene oxide nanocomposite for protein targeting 69, the preparation of self-assembled and label-free AS1411-Au NPs/Au@PC NP is relatively straightforward. Moreover, the AS1411-Au NPs/Au@PC NP provide a huge signal of [Aun]+ cluster ions in the LDI-MS analysis. We also demonstrated the AS1411-Au NPs/Au@PC NP-LDI-MS as a sensitive platform for detection of tumor cells and tumor tissue imaging. In the future, we will extend our aptamer-Au NPs/Au@PC NP nanocomposite for theragnosis applications by loading anti-tumor drugs on the poly(catechin) shell.
Supplementary Material
Supplementary experimental section, table and figures.
Acknowledgments
We are grateful to the Ministry of Science and Technology (MOST) of Taiwan for providing financial support for this study under contracts 104-2923-M-002-006-MY3, 104-2113-M-002-008-MY3 and 104-2628-M-019-001-MY3. The assistance of Ms. Ya-Yun Yang and Ms. Ching-Yen Lin from the Instrument Center of National Taiwan University (NTU) for TEM measurement is appreciated. We would like to thank the Proteomics MS Core Facility at Department of Chemistry at National Taiwan University for assistance with the LDI-MS measurements.
Biographies
 Yu-Ting Tseng is a Ph.D. candidate at the Department of Chemistry, National Taiwan University, Taiwan under the supervision of Dr. Huan-Tsung Chang. She received her Bachelor of Science Degree in Food Science (2011) and Master of Science Degree in Institute for Bioscience and Biotechnology (2013) at National Taiwan Ocean University (NTOU). Her research interest is synthesis of functional nanocomposites for biosensor and nanodrug applications.
Yu-Ting Tseng is a Ph.D. candidate at the Department of Chemistry, National Taiwan University, Taiwan under the supervision of Dr. Huan-Tsung Chang. She received her Bachelor of Science Degree in Food Science (2011) and Master of Science Degree in Institute for Bioscience and Biotechnology (2013) at National Taiwan Ocean University (NTOU). Her research interest is synthesis of functional nanocomposites for biosensor and nanodrug applications.
 Scott G. Harroun is a Ph.D. in Chemistry student at Université de Montréal, Montréal, QC, Canada. He previously worked as a Research Assistant in the Department of Chemistry, National Taiwan University, Taipei, Taiwan. Before that, he earned his M.Sc. in Applied Science and B.Sc. (Honours) in Chemistry degrees at Saint Mary's University, Halifax, NS, Canada. He has co-authored more than 15 publications, and has won several conference awards and scholarships, including an Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) for his doctoral studies. His research interests include surface-enhanced Raman spectroscopy (SERS), DNA nanotechnology, and bioanalytical applications of nanomaterials.
Scott G. Harroun is a Ph.D. in Chemistry student at Université de Montréal, Montréal, QC, Canada. He previously worked as a Research Assistant in the Department of Chemistry, National Taiwan University, Taipei, Taiwan. Before that, he earned his M.Sc. in Applied Science and B.Sc. (Honours) in Chemistry degrees at Saint Mary's University, Halifax, NS, Canada. He has co-authored more than 15 publications, and has won several conference awards and scholarships, including an Alexander Graham Bell Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) for his doctoral studies. His research interests include surface-enhanced Raman spectroscopy (SERS), DNA nanotechnology, and bioanalytical applications of nanomaterials.
 Chien-Wei Wu is currently doing his Doctoral studies at the Department of Chemistry, National Taiwan University, Taiwan under the supervision of Dr. Huan-Tsung Chang. He received his M. S. in the department of Bioscience and biotechnology at National Taiwan Ocean University (NTOU) in 2015. His research interest is synthesis of functional nanomateria for energy storage, conversion and biosensor applications.
Chien-Wei Wu is currently doing his Doctoral studies at the Department of Chemistry, National Taiwan University, Taiwan under the supervision of Dr. Huan-Tsung Chang. He received his M. S. in the department of Bioscience and biotechnology at National Taiwan Ocean University (NTOU) in 2015. His research interest is synthesis of functional nanomateria for energy storage, conversion and biosensor applications.
 Ju-Yi Mao obtained his Master of Science Degree from the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan (2016). His master thesis is on “Tuning the Orientation of Proteins on the Surface of Nanoparticles Through Genetic Engineering for Detection of HPV Virus” under the co-supervision of Dr. Han-Jia Lin and Dr. Chih-Ching Huang. Currently, He is a Ph. D. student enrolled in the Doctoral Degree Program in Marine Biotechnology in National Taiwan Ocean University under the supervision of Dr. Chih-Ching Huang. He is currently committed in development of protein engineering nanomaterials for DNA detection systems.
Ju-Yi Mao obtained his Master of Science Degree from the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan (2016). His master thesis is on “Tuning the Orientation of Proteins on the Surface of Nanoparticles Through Genetic Engineering for Detection of HPV Virus” under the co-supervision of Dr. Han-Jia Lin and Dr. Chih-Ching Huang. Currently, He is a Ph. D. student enrolled in the Doctoral Degree Program in Marine Biotechnology in National Taiwan Ocean University under the supervision of Dr. Chih-Ching Huang. He is currently committed in development of protein engineering nanomaterials for DNA detection systems.
 Huan-Tsung Chang is currently a Professor of the Department of Chemistry, National Taiwan University. He obtained his PhD from the Department of Chemistry, Iowa State University, in 1994 with Dr Edward S. Yeung. His current research interests include nanotechnology, green chemistry, biosensors, and mass spectrometry.
Huan-Tsung Chang is currently a Professor of the Department of Chemistry, National Taiwan University. He obtained his PhD from the Department of Chemistry, Iowa State University, in 1994 with Dr Edward S. Yeung. His current research interests include nanotechnology, green chemistry, biosensors, and mass spectrometry.
 Chih-Ching Huang is a Professor at the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan. He received his PhD degree in Analytical Chemistry from the Department of Chemistry under the supervision of Dr. Huan-Tsung Chang, National Taiwan University, Taiwan (2004). He worked as Postdoctoral fellow at the same laboratory (2006-2008). In 2008, he joined the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan. His research work is mainly focused on development of nanosensors and nanodrugs.
Chih-Ching Huang is a Professor at the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan. He received his PhD degree in Analytical Chemistry from the Department of Chemistry under the supervision of Dr. Huan-Tsung Chang, National Taiwan University, Taiwan (2004). He worked as Postdoctoral fellow at the same laboratory (2006-2008). In 2008, he joined the Department of Bioscience and Biotechnology, National Taiwan Ocean University, Taiwan. His research work is mainly focused on development of nanosensors and nanodrugs.
References
- 1.Ong SH, Jin XC, Jayasooriah, Sinniah R. Image analysis of tissue sections. Comput Biol Med. 1996;26:269–70. doi: 10.1016/0010-4825(96)00004-2. [DOI] [PubMed] [Google Scholar]
- 2.He L, Long LR, Antani S. et al. Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed. 2012;107:538–56. doi: 10.1016/j.cmpb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fass L. Imaging and cancer: a review. Mol Oncol. 2008;2:115–52. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vakoc BJ, Fukumura D, Jain RK. et al. Cancer imaging by optical coherence tomography: preclinical progress and clinical potential. Nat Rev Cancer. 2012;12:363–8. doi: 10.1038/nrc3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway JRW, Carragher NO, Timpson P. Developments in preclinical cancer imaging: innovating the discovery of therapeutics. Nat Rev Cancer. 2014;14:314–28. doi: 10.1038/nrc3724. [DOI] [PubMed] [Google Scholar]
- 6.Weissleder R, Nahrendorf M. Advancing biomedical imaging. Proc Natl Acad Sci U S A. 2015;112:14424–8. doi: 10.1073/pnas.1508524112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237–77. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gessel MM, Norris JL, Caprioli RM. MALDI imaging mass spectrometry: spatial molecular analysis to enable a new age of discovery. J Proteomics. 2014;107:71–82. doi: 10.1016/j.jprot.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole LM, Clench MR. Mass spectrometry imaging tools in oncology. Biomark Med. 2015;9:863–8. doi: 10.2217/bmm.15.61. [DOI] [PubMed] [Google Scholar]
- 10.Laskin J, Lanekoff I. Ambient mass spectrometry imaging using direct liquid extraction techniques. Anal Chem. 2016;88:52–73. doi: 10.1021/acs.analchem.5b04188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spengler B. Mass spectrometry imaging of biomolecular information. Anal Chem. 2015;87:64–82. doi: 10.1021/ac504543v. [DOI] [PubMed] [Google Scholar]
- 12.Fernández JA, Ochoa B, Fresnedo O. et al. Matrix-assisted laser desorption ionization imaging mass spectrometry in lipidomics. Anal Bioanal Chem. 2011;401:29–51. doi: 10.1007/s00216-011-4696-x. [DOI] [PubMed] [Google Scholar]
- 13.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309–42. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cillero-Pastor B, Heeren RM. Matrix-assisted laser desorption ionization mass spectrometry imaging for peptide and protein analyses: a critical review of on-tissue digestion. J Proteome Res. 2014;13:325–35. doi: 10.1021/pr400743a. [DOI] [PubMed] [Google Scholar]
- 15.Hölscher D, Shroff R, Knop K. et al. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 2009;60:907–18. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 16.Yan B, Kim ST, Kim CS. et al. Multiplexed imaging of nanoparticles in tissues using laser desorption/ionization mass spectrometry. J Am Chem Soc. 2013;135:12564–7. doi: 10.1021/ja406553f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098–106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 18.Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2010;82:982–8. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Dill AL, Eberlin LS. et al. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32:218–43. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiseman JM, Ifa DR, Zhu Y. et al. Desorption electrospray ionization mass spectrometry: imaging drugs and metabolites in tissues. Proc Natl Acad Sci U S A. 2008;105:18120–5. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiseman JM, Puolitaival SM, Takáts Z. et al. Mass spectrometric profiling of intact biological tissue by using desorption electrospray ionization. Angew Chem Int Ed. 2005;44:7094–7. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 22.Laskin J, Heath BS, Roach PJ. et al. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84:141–8. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanekoff I, Thomas M, Carson JP. et al. Imaging nicotine in rat brain tissue by use of nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2013;85:882–9. doi: 10.1021/ac302308p. [DOI] [PubMed] [Google Scholar]
- 24.Calligaris D, Caragacianu D, Liu X. et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci U S A. 2014;111:15184–9. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu C-C, Chou P-T, Zare RN. Imaging of proteins in tissue samples using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2015;87:11171–5. doi: 10.1021/acs.analchem.5b03389. [DOI] [PubMed] [Google Scholar]
- 26.Boxer SG, Kraft ML, Weber PK. Advances in imaging secondary ion mass spectrometry for biological samples. Annu Rev Biophys. 2009;38:53–74. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 27.Passarelli MK, Winograd N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS) Biochim Biophys Acta Mol Cell Biol Lipids. 2011;1811:976–90. doi: 10.1016/j.bbalip.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passarelli MK, Ewing AG, Winograd N. Single-cell lipidomics: characterizing and imaging lipids on the surface of individual Aplysia californica neurons with cluster secondary ion mass spectrometry. Anal Chem. 2013;85:2231–8. doi: 10.1021/ac303038j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraft ML, Klitzing HA. Imaging lipids with secondary ion mass spectrometry. Biochim Biophys Acta Mol Cell Biol Lipids. 2014;1841:1108–19. doi: 10.1016/j.bbalip.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Hou S, Sikora KN, Tang R. et al. Quantitative differentiation of cell surface-bound and internalized cationic gold nanoparticles using mass spectrometry. ACS Nano. 2016;10:6731–6736. doi: 10.1021/acsnano.6b02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Xiong C, Liu H. et al. Mass spectrometry imaging reveals the sub-organ distribution of carbon nanomaterials. Nat Nanotechnol. 2015;10:176–82. doi: 10.1038/nnano.2014.282. [DOI] [PubMed] [Google Scholar]
- 32.Arakawa R, Kawasaki H. Functionalized nanoparticles and nanostructured surfaces for surface-assisted laser desorption/ionization mass spectrometry. Anal Sci. 2010;26:1229–40. doi: 10.2116/analsci.26.1229. [DOI] [PubMed] [Google Scholar]
- 33.Chiang C-K, Chen W-T, Chang H-T. Nanoparticle-based mass spectrometry for the analysis of biomolecules. Chem Soc Rev. 2011;40:1269–81. doi: 10.1039/c0cs00050g. [DOI] [PubMed] [Google Scholar]
- 34.Pilolli R, Palmisano F, Cioffi N. Gold nanomaterials as a new tool for bioanalytical applications of laser desorption ionization mass spectrometry. Anal Bioanal Chem. 2012;402:601–23. doi: 10.1007/s00216-011-5120-2. [DOI] [PubMed] [Google Scholar]
- 35.Sekuła J, Nizioł J, Rode W. et al. Silver nanostructures in laser desorption/ionization mass spectrometry and mass spectrometry imaging. Analyst. 2015;140:6195–209. doi: 10.1039/c5an00943j. [DOI] [PubMed] [Google Scholar]
- 36.Tsao C-W, Yang Z-J. High sensitivity and high detection specificity of gold-nanoparticle-grafted nanostructured silicon mass spectrometry for glucose analysis. ACS Appl Mater Interfaces. 2015;7:22630–7. doi: 10.1021/acsami.5b07395. [DOI] [PubMed] [Google Scholar]
- 37.Xu G, Liu S, Peng J. et al. Facile synthesis of gold@graphitized mesoporous silica nanocomposite and its surface-assisted laser desorption/ionization for time-of-flight mass spectroscopy. ACS Appl Mater Interfaces. 2015;7:2032–8. doi: 10.1021/am507894y. [DOI] [PubMed] [Google Scholar]
- 38.Abdelhamid HN, Wu H-F. Gold nanoparticles assisted laser desorption/ionization mass spectrometry and applications: from simple molecules to intact cells. Anal Bioanal Chem. 2016;408:4485–502. doi: 10.1007/s00216-016-9374-6. [DOI] [PubMed] [Google Scholar]
- 39.Hinman SS, Chen C-Y, Duan J. et al. Calcinated gold nanoparticle arrays for on-chip, multiplexed and matrix-free mass spectrometric analysis of peptides and small molecules. Nanoscale. 2016;8:1665–75. doi: 10.1039/c5nr06635b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shrivas K, Hayasaka T, Sugiura Y. et al. Method for simultaneous imaging of endogenous low molecular weight metabolites in mouse brain using TiO2 nanoparticles in nanoparticle-assisted laser desorption/ionization-imaging mass spectrometry. Anal Chem. 2011;83:7283–9. doi: 10.1021/ac201602s. [DOI] [PubMed] [Google Scholar]
- 41.Morosi L, Spinelli P, Zucchetti M. et al. Determination of paclitaxel distribution in solid tumors by nano-particle assisted laser desorption ionization mass spectrometry imaging. PLoS One. 2013;8:e72532. doi: 10.1371/journal.pone.0072532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekula J, Niziol J, Rode W. et al. Gold nanoparticle-enhanced target (AuNPET) as universal solution for laser desorption/ionization mass spectrometry analysis and imaging of low molecular weight compounds. Anal Chim Acta. 2015;875:61–72. doi: 10.1016/j.aca.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 43.Nizioł J, Ossoliński K, Ossoliński T. et al. Surface-transfer mass spectrometry imaging of renal tissue on gold nanoparticle enhanced target. Anal Chem. 2016;88:7365–71. doi: 10.1021/acs.analchem.6b01859. [DOI] [PubMed] [Google Scholar]
- 44.Otake Y, Soundararajan S, Sengupta TK. et al. Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood. 2007;109:3069–75. doi: 10.1182/blood-2006-08-043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soundararajan S, Chen W, Spicer EK. et al. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–65. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 46.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J Biol Chem. 1999;274:15908–12. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 47.Huang S-S, Wei S-C, Chang H-T. et al. Gold nanoparticles modified with self-assembled hybrid monolayer of triblock aptamers as a photoreversible anticoagulant. J Control Release. 2016;221:9–17. doi: 10.1016/j.jconrel.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 48.Pei H, Li F, Wan Y. et al. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J Am Chem Soc. 2012;134:11876–9. doi: 10.1021/ja304118z. [DOI] [PubMed] [Google Scholar]
- 49.Wang W, Ding X, He M. et al. Kinetic adsorption profile and conformation evolution at the DNA-gold nanoparticle interface probed by dynamic light scattering. Anal Chem. 2014;86:10186–92. doi: 10.1021/ac502440h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.García-Viguera C, Bridle P, Bakker J. The effect of pH on the formation of colored compounds in model solutions containing anthocyanins, catechin, and acetaldehyde. Vitis. 1994;33:37–40. [Google Scholar]
- 51.Raula M, Maity D, Rashid MH. et al. In situ formation of chiral core-shell nanostructures with raspberry-like gold cores and dense organic shells using catechin and their catalytic application. J Mater Chem. 2012;22:18335–44. [Google Scholar]
- 52.Kumamoto M, Sonda T, Nagayama K. et al. Effects of pH and metal ions on antioxidative activities of catechins. Biosci Biotechnol Biochem. 2001;65:126–32. doi: 10.1271/bbb.65.126. [DOI] [PubMed] [Google Scholar]
- 53.Janeiro P, Oliveira Brett AM. Catechin electrochemical oxidation mechanisms. Anal Chim Acta. 2004;518:109–15. [Google Scholar]
- 54.Werner D, Hashimoto S. Improved working model for interpreting the excitation wavelength- and fluence-dependent response in pulsed laser-induced size reduction of aqueous gold nanoparticles. J Phys Chem C. 2011;115:5063–72. [Google Scholar]
- 55.Hashimoto S, Werner D, Uwada T. Studies on the interaction of pulsed lasers with plasmonic gold nanoparticles toward light manipulation, heat management, and nanofabrication. J Photochem Photobiol C Photochem Rev. 2012;13:28–54. [Google Scholar]
- 56.McLean JA, Stumpo KA, Russell DH. Size-selected (2-10 nm) gold nanoparticles for matrix assisted laser desorption ionization of peptides. J Am Chem Soc. 2005;127:5304–5. doi: 10.1021/ja043907w. [DOI] [PubMed] [Google Scholar]
- 57.Pustovalov VK, Smetannikov AS, Zharov VP. Photothermal and accompanied phenomena of selective nanophotothermolysis with gold nanoparticles and laser pulses. Laser Phys Lett. 2008;5:775–92. [Google Scholar]
- 58.Liu Y-C, Li Y-J, Huang C-C. Information derived from cluster ions from DNA-modified gold nanoparticles under laser desorption/ionization: analysis of coverage, structure, and single-nucleotide polymorphism. Anal Chem. 2013;85:1021–8. doi: 10.1021/ac302847n. [DOI] [PubMed] [Google Scholar]
- 59.Li Y-J, Chiu W-J, Unnikrishnan B. et al. Monitoring thrombin generation and screening anticoagulants through pulse laser-induced fragmentation of biofunctional nanogold on cellulose membranes. ACS Appl Mater Interfaces. 2014;6:15253–61. doi: 10.1021/am503615c. [DOI] [PubMed] [Google Scholar]
- 60.Xiong X, Lv Y, Chen T. et al. Nucleic acid aptamers for living cell analysis. Annu Rev Anal Chem. 2014;7:405–26. doi: 10.1146/annurev-anchem-071213-015944. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, Zhao H, He L. et al. Aptamer density dependent cellular uptake of lipid-capped polymer nanoparticles for polyvalent targeted delivery of vinorelbine to cancer cells. RSC Adv. 2015;5:16931–9. [Google Scholar]
- 62.Ocsoy I, Gulbakan B, Shukoor MI. et al. Aptamer-conjugated multifunctional nanoflowers as a platform for targeting, capture, and detection in laser desorption ionization mass spectrometry. ACS Nano. 2013;7:417–27. doi: 10.1021/nn304458m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hasegawa H, Savory N, Abe K, Ikebukuro K. Methods for improving aptamer binding affinity. Molecules. 2016;21:421. doi: 10.3390/molecules21040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C-Y, Gherzi R, Andersen JS. et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–48. [PMC free article] [PubMed] [Google Scholar]
- 65.Palmieri D, Richmond T, Piovan C. et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc Natl Acad Sci U S A. 2015;112:9418–23. doi: 10.1073/pnas.1507087112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Destouches D, El Khoury D, Hamma-Kourbali Y. et al. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS One. 2008;3:e2518. doi: 10.1371/journal.pone.0002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koutsioumpa M, Papadimitriou E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat Anticancer Drug Discov. 2014;9:137–52. doi: 10.2174/1574892808666131119095953. [DOI] [PubMed] [Google Scholar]
- 68.Miettinen M. Immunohistochemistry of soft tissue tumours - review with emphasis on 10 markers. Histopathology. 2014;64:101–18. doi: 10.1111/his.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang R-C, Chiu W-J, Po-Jung Lai I. et al. Multivalent aptamer/gold nanoparticle-modified graphene oxide for mass spectrometry-based tumor tissue imaging. Sci Rep. 2015;5:10292. doi: 10.1038/srep10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary experimental section, table and figures.