Abstract
Outbreaks of foodborne diseases related to fresh produce have been increasing in North America and Europe. Viral foodborne pathogens are poorly understood, suffering from insufficient awareness and surveillance due to the limits on knowledge, availability, and costs of related technologies and devices. Current foodborne viruses are emphasized and newly emerging foodborne viruses are beginning to attract interest. To face current challenges regarding foodborne pathogens, a point-of-care (POC) concept has been introduced to food testing technology and device. POC device development involves technologies such as microfluidics, nanomaterials, biosensors and other advanced techniques. These advanced technologies, together with the challenges in developing foodborne virus detection assays and devices, are described and analysed in this critical review. Advanced technologies provide a path forward for foodborne virus detection, but more research and development will be needed to provide the level of manufacturing capacity required.
Keywords: Foodborne virus, microfluidics, nanomaterials, biosensor, point-of-care.
Overview of Foodborne Viruses and Detection
Food safety and its related issues are attracting interest worldwide because they are closely related to human lives and health conditions. Pathogens from the environment may contaminate food and food products, thus foodborne pathogens and their detection are directly related to human life and safety. Foodborne viruses, among all other pathogens, are relatively new, gaining more attention due to their emerging contamination events and the small scale of outbreaks.
Global public food safety issues have been increasing in recent years. Foodborne disease outbreaks related to fresh produce have been increasing in North America and European Union 1. In the United States, norovirus is the main pathogen, responsible for 59%, followed by Salmonella, which is responsible for 18% of foodborne diseases related to fresh produce. In European Union, norovirus is responsible for 53%, followed by Salmonella, which is responsible for 20% of foodborne diseases related to fresh produce. In Canada, Salmonella is the main pathogen, responsible for 50%, and hepatitis A virus is responsible for 0.1% of foodborne diseases from fresh produce 2. Although viruses are not the major pathogen in Canadian fresh produce, they are prevalent in farm-level infection such as hepatitis E virus (34%), porcine enteric calicivirus (20%), and rotavirus (7%) in finisher pigs 3. These viruses are hypothesised to infect humans zoonotically through swine and pork exposure.
Food Safety and Its Assessment
Food safety issues are of significant importance in food manufacture and transportation to different regions domestically and internationally 4. Food contamination from pathogens like viruses and bacteria; and chemicals like metals, pesticides, and other adulterants may cause severe and wide-reaching life burdens and economic losses. The damage of food hazards to human health may range from acute to chronic periods of time, and include organ damage, health issues, and diseases including cancer.
Food-derived health hazards may cause outbreaks locally or be spread out in both developing and developed countries, making them unpredictable 5. Hence the understanding of foodborne pathogens in proper handling and preparation of food is crucial in ensuring food quality and the preservation of societal health. Understanding foodborne pathogens is particularly important for food manufacturing, packaging, and transportation processes.
Most importantly, proper monitoring and pathogen detection systems, and their application and development to match modern technology, will ensure food quality and safety 28. Effective food safety monitoring can be used to monitor and control food quality at a large scale, at multiple locations, and across production points from manufacture to food consumption. Ideally these control points should include all steps or procedures involved in food handling from farm to fork, including food production, storage, transportation, delivery, and consumption. Unfortunately current food safety and its monitoring and assessment practices do not meet expectations, mostly due to the high cost of traditional laboratory tests and time consuming operations requiring expensive equipment and specialised trained professionals.
Foodborne Pathogens
Current disease burdens of foodborne pathogen remain unknown with an ongoing debate over predictions of foodborne diseases and deaths increasing or decreasing 6. Even though foodborne infectious diseases are reported rarely from a few industrialised countries with few pathogens, evidence shows that food safety remains a dynamic situation. With the ongoing establishment and development of food industry and healthcare systems in developing countries, food safety standards and pathogen detection tools will reveal more details in the dynamic situation and its related factors.
Foodborne pathogens are broadly understood to fit into three categories: bacteria, viruses, and parasites 7. Bacterial foodborne pathogens, such as Listeria monocytogenes, Salmonella, Campylobacter, and Escherichia coli, are well-studied and well-recognised by researchers and policy makers, and thus will not be the target of this review. Parasitic foodborne pathogens rarely cause extensive food issues and infection outbreaks due to better food safety education and manufacturing technology. This review focuses on viral foodborne pathogens since they are seemingly increasing in pace and constantly emerging in food safety issues.
Viral foodborne pathogens are poorly understood, with insufficient awareness and surveillance, probably because of the limits of knowledge, availability, and costs of related technology and device 72. Current foodborne virus efforts are focused on norovirus, rotavirus, and hepatitis viruses. Foodborne viruses emerging currently, such as SARS coronavirus, avian influenza virus, and Nipah virus are causing considerable illness and mortality in humans 8, 9. Current situations in foodborne viruses and their monitoring and detecting require the instigation of an ongoing collaboration and dialogue between food safety experts, public health, and veterinary experts to enhance multidisciplinary skills and technologies.
Foodborne Virus Types and Categories
The most common foodborne viruses are norovirus, rotavirus, and hepatitis viruses. These viruses can either be single or double stranded, and either RNA or DNA viruses (Table 1). The only common point shared between them is that they can contaminate water or food and thus categorised as foodborne viruses.
Table 1.
Pathogenic viruses responsible for foodborne illness.
| Viruses | Associated Foods | Infected doses | Disease Names | References |
|---|---|---|---|---|
| Norovirus | Vegetables, shellfish, oysters, water & ice | 100 copies/mL | Viral gastroenteritis, stomach flu | 11,12,13,42 |
| Rotavirus | Meats, water | Unknown, presumed to be low | Viral gastroenteritis in children | 15,17,19 |
| Hepatitis A | Sandwiches, fruits, vegetables, milk, shellfish, iced drinks | 10-100 | Hepatitis A | 18,19,20 |
| Hepatitis E | Raw/undercooked boar and deer meat, livers, and liver sausages | Unknown | Hepatitis E | 3,21,25 |
Norovirus
Norovirus is an RNA virus in the family Caliciviridae causing more than 90% of non-bacterial gastroenteritis outbreaks in the United States 10. They can be classified as five genotypes from GI to GV based on the nucleotide sequences of the capsid protein and polymerase. A total of eight genotypes out of 29 known genotypes of norovirus cause infection in humans. According to the U. S. Center for Disease Control and Prevention (CDC), noroviruses are responsible for 70% and 50% of waterborne and foodborne enteritis, respectively 11. The symptoms of foodborne norovirus include vomiting, gastralgia, fever, diarrhea, and death due to dehydration in children and elderly.
Norovirus is very stable in the environment and highly contagious, requiring only a low infectious dose, as doses smaller than 102 copies/mL may cause infection in humans 12. The virus strongly resists heat up to 60°C, as well as acid and chlorine in tap water up to 6.25 mg/L, which make its prevention and clearance difficult. Norovirus is transmitted through daily activities, water, and food. Contaminated food by food handlers is an important transmission route. The virus causes 19-21 million illness in the United States annually 13. It is reported that 50% of the infection occurs in food related locations, such as cruise ships, school cafeterias, and restaurants 11. Hence the detection of foodborne norovirus is critically important for human health.
Rotavirus
Rotavirus belongs to the Reoviridae family, it is a non-enveloped double-stranded RNA virus 14. It causes severe gastroenteritis and diarrhea in young children and infants. It is an important cause of childhood gastroenteritis, accounting for 1.5 million child deaths and 130 million cases of diarrhea worldwide annually Rotavirus is also associated with infantile diarrhea disease 15,16. Rotaviruses cause an estimated 4 million infections, 70,000 hospitalization, and more than 100 deaths in the United States annually 16.
The viruses belong to the group of zoonotic emerging viruses transmitted through fecal-oral route, which can be transmitted between multiple animal species and humans 17. They cause serious economic losses, particularly in domestic animals 16. Even though the rotavirus strains are distinct between human and animals, cross-species infection occurred from time to time because some human strains are similar to animal strains. Very low doses of the viruses can be infectious, such as 1 plaque forming unit (PFU); as such a few virus particles in food or water can cause infection in humans. Outbreaks of waterborne and foodborne rotavirus infection have been noticed in several countries 16. Therefore, they demonstrate the need for highly sensitive detection technology to ensure food and water safety as related to these viruses.
Hepatitis Viruses
Hepatitis viruses related to foodborne infection include hepatitis A, B, and E viruses, with the E viruses causing the most food infections in North America.
Hepatitis A Virus
Hepatitis A virus is a small, single stranded, and non-enveloped RNA virus of the Picornaviridae family 18. It can be grouped into I, II and III genotypes based on its genomic characterisation. Hepatitis A virus infections are the leading cause of viral hepatitis, with 1.4 million of new cases worldwide annually 19. The cost of foodborne hepatitis A is estimated to be more than 36,000 U. S. dollars per individual in the United States.
Hepatitis A virus can be infectious at low doses of 10-100 viral particles 20. It is transmitted through the fecal-oral route in humans, as well as intake of contaminated water and foods such as fruits, uncooked vegetable, and shellfish. Waterborne and foodborne infection of hepatitis A viruses accounts for 2-7% of the total disease burden, and foodborne infection is common, often resulting in a larger and prolonged outbreak.
Hepatitis B Virus
Hepatitis B virus is a partially double stranded, enveloped, and parenterally transmitted circular DNA virus 21. Hepatitis B virus infection is the tenth leading cause of death and a major health threat globally 22. As the major cause of chronic hepatitis, cirrhosis and hepatocellular carcinoma, the infection is spreading to more than 350 million people as its carriers disburse globally. It has infected about 30% of the world population, causing more than 780,000 death annually from complications. A total hepatitis B virus related cost is more than 600 million Euro in western countries annually.
Hepatitis B virus is transmitted by contact with infected individuals through blood or other body fluids 23. It is at high risk when the viral concentration reaches 105 copies/mL, which is associated with liver diseases including cancer. The exposure to contaminated blood or body fluids is considered to be the major transmission routes for the virus. Antibodies against the surface antigens of the virus are produced to high levels following hepatitis B virus infection.
Hepatitis E Virus
Hepatitis E virus is a single stranded RNA virus with positive sense in the family Hepeviridae 24. Four genotypes with several subtypes of the virus are defined, and genotypes 3 and 4 are zoonotic viruses infecting humans, pigs, and other animals. Acute hepatitis E virus outbreaks occurred multiple times in different locations of the world. It is mostly self-limiting with low mortality rate in juveniles and adults; but mortality rates in pregnant women, with those in the third trimester reach 25% 24.
Hepatitis E virus spreads via the fecal-oral route from contaminated food and water 25. It usually causes acute infection with increasing numbers of autochthonous events occurring globally, including in Europe. Hepatitis E virus can be transmitted by contaminated food, as increasingly reported in small outbreaks and sporadic cases. Infection is related to the consumption of undercooked or raw meat from deer and boar, and commercial livers and liver sausages. It has also been detected in commercial livers, sausages, and pork derived food from grocery stores. Hepatitis E virus contamination rates in commercial foods are 2-31% depending on the locations and food types.
Other Foodborne Viruses
Other foodborne viruses, besides the above mentioned common ones, are SARS, avian influenza virus, and Nipah virus. SARS coronavirus can be spread to humans through the consumption and preparation of food animals or wild animals, which may have been derived from a different reservoir, for example, bats 8. Avian influenza virus, such as the highly pathogenic H5N1 virus, has been spread to humans from the preparation and consumption of duck and chicken food products 26. Unlike most foodborne viruses which are non-enveloped particles and stable on food, Nipah virus has lipid envelopes and thus may have reduced stability in the environment and on food 9.
While the above foodborne viruses belong to relatively rare cases and hardly cause large-scale outbreaks, they capture attention and raise questions regarding whether foodborne pathogens represent a potential route for introducing new viral diseases to human populations and their future 27. Virologists expressed the probability that new viruses may emerge from food animals or by evolution of old viruses. This will inevitably make changes to our economy, demography, and sociology, such as the changes made in living animal transportation globally, in exotic food consuming habits, and in human population expansion.
Point of Care Testing and Its Scope in Foodborne Virus Detection
Based on the needs and requirements in current food safety monitoring, a point-of-care (POC) concept has been introduced to food testing technology and devices. The POC refers to biochemical technology whenever the medical care is needed at or near the site of patient care, which results in immediate information regarding an individual's conditions for treatment or further testing decisions 28. Its advantages are minimal sample volumes, widening accessibility, rapid analysis time, and reduced costs. Ideally POC technology and device can be used on-site and real-time to provide the first result in minutes using a simple protocol with a few steps to analyse samples 29. The application of POC technology and devices in food safety and pathogen detection is emerging at its infancy stage, particularly in foodborne virus detection.
POC Testing and Technology
In the food safety area, POC testing and technology are experiencing rapid development. Development of low-cost and reliable diagnostic testing with on-site application will have considerable advantages over laboratory-based testing, because lab-based testing requires expensive equipment and technological personnel which are mostly unavailable on-site 29. POC testing requires that all of the analytical processes, from sample collection to result communication, should be performable in one or a few simple steps to reduce time and costs between the test and treatment.
Current POC testing and technologies include microfluidic, nanomaterials, biosensors, and other technologies such as antibodies, enzymes, aptamers, nanozymes, and antigens 72. One of the predominant technologies is molecular detection, including enzyme assay, immunoassay, polymerase chain reaction, and microarray, which link the POC testing with proteins in the form of antigen and antibody, DNA, and RNA detection and measurement 30. Molecular detection technologies cut test time significantly to increase POC testing speed and accuracy. Some of these technologies have been developed into POC devices that can be used on-site for real-time testing.
POC Device
Current development of POC devices contains several characteristics, such as wireless cell phone based technologies, lab-on-a-chip, and paper based devices with long-term reagent storage and novel assay format strategies (Figure 1) 29, 31. Various cell phone based devices are commercially available or under intensive development to provide cost-effective mobile POC detection (Figure 2) 32. Paper based, and lab-on-a-chip platforms pave the way to simplified, automated, robust, and low-cost detection of foodborne viruses.
Figure 1.
POC paper device 31. The paper test strip is immersed in the urine sample for a few seconds and after a few minutes; the colour resulting from the reaction can be visually compared against the chromatic scale provided.
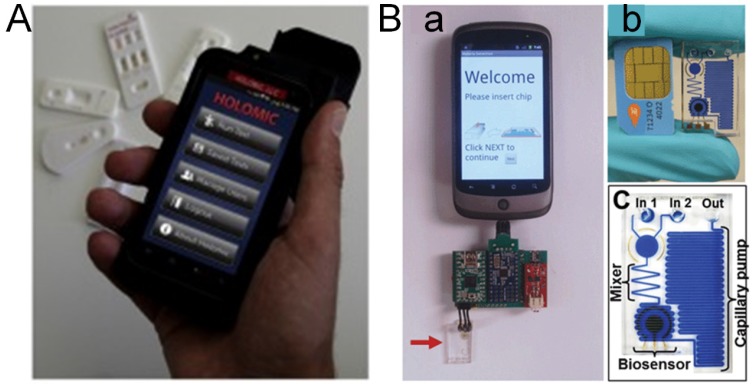
Figure 2.
Cell phone based POC technology 32. (A) Holomic Rapid Diagnostic Reader (HRDR-200) used in lateral flow assay (LFA). (B): (a) Electrochemical sensor based on cell phone technology; The microfluidic chip is marked by the arrow. (b) SIM Card from a mobile phone compared with a microfluidic chip. (c) Diagram of a microfluidic chip showing components after dye filling for better visualization.
Nevertheless, key challenges and pending issues remain to be solved on the way to successful commercialisation and implementation of these POC device and platforms. The foremost challenge is how to demonstrate high levels of analytical precision and reproductively when large numbers of samples are handled and analysed without major interference from nonspecific materials 33. The POC technologies need to be validated by accredited standards and guidelines. Such quality control and batch to batch variability need to be set up effectively and efficiently. Additionally, the integration and miniaturising of large devices, such as microfluidic devices, remain challenging; even though recent advances in acousto-microfluidic technology shows promise to address some of these challenges 34. Furthermore, the emerging nanomaterials enhance sensitivity and analytical performance, but their application on cytotoxicity remains controversial and requires further address.
Current progress on the development of POC technologies and devices focuses on foodborne bacterial detection, which is far more advanced than foodborne virus detection 35. Since technologies of bacterial detection cannot be applied directly to virus detection, (refer to Section 8.5 Current Challenges and Future Trends for more detail) this review focuses on the current limited foodborne virus detection, and the possibilities of adopting technologies and devices indirectly from bacterial studies, basic virus studies, and human clinical virus studies. This needs intensive exploration of these indirectly related technologies to make them fit the specific requirements of foodborne virus detection and measurement.
Microfluidic Technologies in POC of Foodborne Viruses
Microfluidics are one of the most promising technologies in foodborne virus detection. Their main advantages, compared to other POC technologies, are portability, low consumption of reagent, and extremely low requirement of sample amount 36. These advantages make them easy to use, rapid, and inexpensive when compared to conventional testing. The small sample amount and short diffusional distance make them significantly more rapid than other macroscale equivalents (Figure 3) 72. The potential to complete all necessary steps within one device and a single reaction makes them an ideal candidate of POC testing.
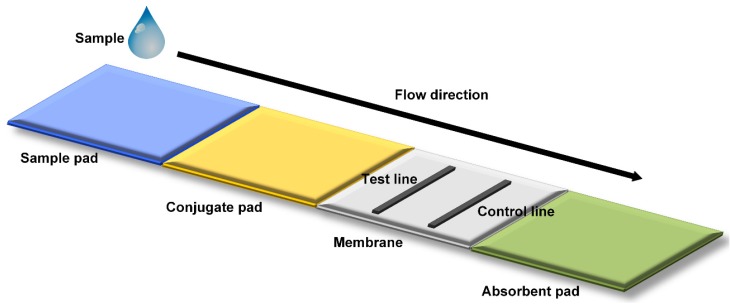
Figure 3.
Mechanism of microfluidic POC Technology 31. Schematic representation of a lateral flow strip is shown. A liquid sample is deposited on to the sample pad, migrating through a conjugate pad and a porous membrane for detection in a final absorbent pad. In most strip tests, the appearance of the control line indicates a valid test, while the appearance of a second test line indicates a positive test result.
Microfluidic Technology and Systems
Microfluidic technology requires extremely small amount of fluid as microliters or even nanolitres and measures ten to hundreds of micrometres of foods using interconnected networks 37, 38. Microfluidics has been ongoing in its evolution with microfabrication technology since its invention in the early 1990s. Fluid for testing is transported either passively through capillary forces or actively through pumping to achieve laminar fluid flow. Automated control of all steps can reduce human errors and increase accuracy, reproducibility, and reliability of test results. The opportunities for testing different samples simultaneously make microfluidics a perfect candidate in multiplexing. Two types of microfluidic systems are currently available in foodborne virus detection: micro total analysis system and the paper based analytical system.
Micro total analysis system is mostly designed as devices with lab-on-a-chip features 39, 40. Its advantages are using fluid as a working medium and integrating different functionalities on a scale, which allow complicated molecular techniques to be completed at once, such as immunoassays and polymerase chain reaction (PCR). Materials of silicon, glass, and polymers, particularly thermoplastic polymers are most often used in microfluidic devices.
Paper based microfluidic systems keep the advantages of conventional microfluidics, such as size, speed and reduced sample amount, but adds an inexpensive multiplexed setting 41. Paper is considerably easy to source, cheap, and biodegradable, and, most importantly, easy to modify chemically. Other advantages of paper devices include requiring no external power sources, a high ratio of surface to volume, and minimal technical expertise requirements. Paper based microfluidics, compared to microfluidics with lab-on-a-chip formats, has significant advantages: simpler technology and reduced costs; as well as disadvantages: issues in sample retention and evaporation making it less suitable for very small volumes.
Microfluidic Application for Foodborne Virus Detection
Microfluidic technology has been successfully applied to waterborne and foodborne virus detection. Microfluidic lab-on-a-chip modules have been used to detect murine norovirus with drop-based technology 42. Human and rodent noroviruses may exchange RNA between the virus genomes in an infected host, an important driver of genetic diversity, which is critically important in norovirus infection diagnosis and future infectious strain prediction. The successful application of drop-based microfluidics on two murine noroviruses co-infected mice offers a platform for amplification, detection, and recovery of individual recombinants in co-infection.
Lab-on-a-chip microfluidics was used to detect murine norovirus contaminated oysters with micro-bead beating 43. Food sample preparation has been challenging in foodborne virus detection. The study demonstrates that microfluidics can be applied to foodborne norovirus detection using a shape switchable sample preparation chamber. Noroviruses in the sample were absorbed on microbeads by bead beating lysis and electrostatic physisorption. The extracted RNA was amplified and detected in a detection chamber. This microfluidic technology is ready to be applied to detecting noroviruses in large volumes of environmental and food samples.
Microfluidic quantitative PCR has been used to simultaneously quantify eleven major human viruses in environmental water samples, such as enterovirus, Aichi virus, adenovirus, astrovirus, sapovirus, rotavirus, norovirus and hepatitis A and E viruses 44. River water contaminated with wastewater was used as validation samples. Quantitative information can be obtained as high throughput with detection limits of 2 copies/µl of DNA or cDNA. The results were similar to those detected by quantitative PCR.
Nanomaterials in POC of Foodborne Viruses
Overview of Nanomaterials
Nanomaterial based technology has been applied to foodborne virus detection to ensure food safety 45. A variety of nanomaterials can be used, such as graphene and carbon nanotubes (CNTs); quantum dots (QDs); and metal nanoparticles including gold (Au), silver (Ag), and titanium based nanomaterials 46, 47. Nanomaterials play a major role in biosensor design for foodborne virus assay, and enable faster, sensitive, specific, and cost-effective detection outside of laboratory. This review describes their advantages, principles, and limitations with regard to their multiplexing capability, sensitivity, and simplicity (Table 2).
Table 2.
Nanomaterials available for foodborne virus detection
| Technologies | Analytes | Detection limits | References |
|---|---|---|---|
| Cellulose nanocrystals | Norovirus | 0.01 ng/mL | 52 |
| Graphene-gold hybrids | Norovirus | 100 pg/mL | 55 |
| Phage nanoparticles | Norovirus | 107 copies/mL | 56 |
| Silver nanorod | Rotavirus | 0.1 fM | 57 |
| Nanocrystals | Rotavirus | 6.35 µg/mL | 60 |
| Gold nanoparticles | Hepatitis B virus | 102 copies/mL | 62 |
| Gold nanoparticles | Hepatitis E virus | 10 copies/mL | 24 |
| Silica nanoparticles | Hepatitis A virus | 8.6 pM | 108 |
| Graphene nanoparticles | Hepatitis B virus | 50 aM | 67 |
| Immunomagnetic nanobeads | Avian influenza virus | 0.128 HA unit | 173 |
| Carbon nanotube | Human influenza virus | 3.4 PFU/mL | 70 |
In nanomaterials, gold nanoparticles and quantum dots are the most commonly used materials. However, new forms of nanomaterials are being developed to represent more molecular entities and to be more amenable to nanoscale fabrication 48. Another promising material is magnetic nanoparticles, with advantages of easy manipulation and cost-effectiveness. They can be combined with a capturing agent, such as a DNA probe, a glycan molecule, or an antibody, to target DNA or protein molecules of interest. For foodborne viruses, the earlier detection is to determine viral nucleic acid; and viral protein measurement is a later detection. In particular, novel metal nanomaterials such as Au NPs and Ag NPs have been highlighted due to their plasmonic properties 24.
The application of plasmonic nanomaterials can be categorised into plasmonic and non-plasmonic systems 49. In plasmonic systems, metal nanoparticles (NPs) are considered as a plasmonic probes which have a suitable inter-particle distance, smaller than the diameter of particles to generate plasmonic coupling of particles. A visible color from red to blue can be produced variously with colorimetric detectability (Figure 4). An advanced example of plasmonic nanomaterials is direct aggregation, without specific ligands between a single-stranded primer DNA, and its adsorption on citrate-coated Au gold nanoparticles. On the other hand, indirect aggregation can be used to detect viruses through modifying virus targeting molecules on virus surfaces. The challenging key to this technology is controlling particle aggregation in a reproducible manner. This can be improved using a protein-glycan pairing relying on the multivalence properties of glycan to improve weak protein detection of viral surface proteins. Another advanced improvement is signal amplification using catalytic or DNA enzymatic assays, such as alkaline phosphatase catalyst, and DNA enzymes to provide rapid detection of virus sequences.
Figure 4.
Schematic depictions of solution based plasmonic detection methods 49. A) Colorimetry of directly aggregated nanoparticles upon successful RNA amplification; and B) colorimetry of indirectly aggregated nanoparticles after the interaction with influenza virus.
In non-plasmonic systems, nanomaterials can functionalized with fluorescent labels and use as fluorescent probes rather than plasmonic probes. Non-plasmonic probes consider smaller metal nanoclusters, such as Ag nanoclusters with red or near-infrared fluorescent labels, for use as a probe. Such smaller metal nanoclusters can be stabilised by nucleic acid strands binding on a graphene oxide sheet. Other non-plasmonic nanomaterials are also used, such as BaGdF5:Yd/Er NPs conjugated oligonucleotide with target linked Au nanoparticles, immunomagnitic nanobeads with biometallisation and electrochemistry features to detect viruses of 0.011 ng/mL 49.
Nanomaterial-based technology in virus detection involves two steps: isolation and capture of target viral molecules from the sample, and recognition and enhancement of the signals. Signal detection of nanomaterials includes optical and electrochemical approaches 51. Optical detection with fluorescent properties is highly sensitive with low detection limits. Electrochemical detection, compared to optical detection, is often used with graphene or carbon nanotubes.
Nanomaterials in Detection of Foodborne Viruses
Nanomaterials have been used to detect foodborne viruses including hepatitis viruses, norovirus, rotavirus, and avian influenza viruses.
Nanomaterials Detecting Norovirus
Nanomaterials can be used to detect norovirus, or alternatively, norovirus-like particles. Cationic polymer brush-modified cellulose nanocrystals (CNCs) were used to bind norovirus-like particles effectively 52. The modified CNCs were verified by dynamic light scattering and electromicroscopy. Another nanomaterial method to detect norovirus-like particles is a SPR-assisted immunosensor with CdSe-ZnS-based quantum dot fluorescent labels 53. The SPR enhancement, intensity of auto-fluorescence, and excitation efficiency of quantum dots were evaluated and maximized to optimize the single-to-noise ratio to reach the minimum detection sensitivity of 0.01 ng/mL, corresponding to 100 virus-like particles.
A location-specific nanoplasmonic sensing of lipid bilayer membrane binding with label-free and real-time features was used to detect noroviruses 54. It confirms that noroviruses interacted with glycosphingolipids accumulating in invaginations, which enables the induction of negative membrane curvature. This represents a new application of nanoplasmonic sensors impossible for conventional SPR or other planar surface techniques, but its limit is that it requires an ordinary spectrophotometer in virus detection.
A nanoparticle based assay with size-controlled preparation of peroxidase-like graphene-Au gold hybrids is used to detect norovirus-like particles 55. The graphene-Au hybrid structure enhances Raman intensity and catalyzes the peroxidase substrate to produce a detectable blue color, which is confirmed using antibody conjugated graphene-Au nanoparticles. This graphene-Au assay can reach a detection limit of 100 pg/mL, which is 100 times more sensitive compared to conventional ELISA method.
Traditional lateral-flow assay (LFA) has the drawback of lower sensitivity due to its colored particles, such as gold nanoparticles or blue latex. A novel LFA using phage nanoparticles as reporters is used to detect norovirus-like particles based on an optimized antibody sandwich pair identified from ELISA 56. The use of the antibody pair is validated by both gold nanoparticle and bacteriophage and LFA. The phage nanoparticle LFA is rapid and highly sensitive with a detection limit of 107 virus copies/mL, which is 100-fold lower than the gold nanoparticle LFA alone with the same antibody pair.
Nanomaterials Detecting Rotavirus
Nanomaterials, such as silver nanorods, graphene oxide nanoparticles, and nanocrystal particles, can be used to detect rotaviruses. An aligned silver nanorod array based on oblique angle deposition of SER-active substrates is used to detect and differentiate a reporter molecule and rotaviruses 57. The silver nanorod assay depends on the polarization of excitation light, the substrate coating, and the incident angle. The study shows that the well-established chemometric techniques in SER can be used to determine subtle spectral differences allowing analyte classification 58. Its detection limit of the reporter molecule is lower than 0.1 fM.
A plasmonic magnetic hybrid three dimensional (3D) architectures is developed based on an antibody core, and a graphene oxide nanoparticle shell with polyethylene glycol as H2N-PEG-NH2 chemistry to detect rotaviruses 15. The initial 3D plasmonic magnetic nanoparticles were built with an iron oxide core and plasmonic gold shell HAuCl4 59. They use surface-enhanced Raman SER imaging and antibody-attached 3D network to bind strongly to and efficiently remove rotaviruses from infected drinking water. The results are confirmed using SER and fluorescent imaging with RT-PCR.
A label-free hydrogel based nanocrystal photonic system is used to detect rotaviruses without any sample pre-treatment of commercial rotavirus antigen samples 60. Its sensitivity can reach 6.35 µg/mL that is analysed quantitatively by measuring peak wavelength and confirmed by ELISA. To target the virus accessibility to the inner sensor structure and enhance virus sensing, the hydrogel based inverse opal structure enlarges the nanopores by etching with O2 plasma.
Nanomaterials Detecting Hepatitis Viruses
Nanomaterials can be used to detect hepatitis A, B, and E foodborne viruses. Nanomaterials of gold nanorods can be used to visualize virus replication and detect virus particles 46, 61. This is used to detect hepatitis B viruses to increase surface area and immobilisation of single-stranded DNA with electrochemical biosensor 47. Scanning electron and atomic-force microscopy can evaluate nanorod morphology, which can detect picomolar level of DNA and distinguish complementary DNA from mismatched and non-complementary DNA sequences. Another gold nanoparticle, combined with an anodic aluminum oxide (AAO) film, is used to detect hepatitis B viruses targeting single-stranded DNA without PCR, with a detection level up to 102 copies/mL 62.
A reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay is used to combine with Au gold nanoparticles to detect hepatitis E viruses 24. The single tube and one step RT-LAMP with colorimetric labeling can be used to replace conventional detection methods and remove the need for expensive equipment as required by POC. It is simple, rapid, and highly sensitive with a detection limit of 101 RNA copies.
A one-tube colorimetric and surface plasmon resonance (SPR) active nanoparticle platform is used to detect hepatitis A and B viruses simultaneously and quantitatively with picomolar sensitivity, which is archived by eliminating spectral overlap through non-negative matrix factorisation 63. Another nanomaterial platform combines sandwich immunoassay, fluorescent labeling, and magnetic separation together to detect hepatitis A and B virus DNA with high sensitively, easy operation, quantitative quality, and picomolar detection up to 0.1 pM 51. An additional Au gold probe is used to measure hepatitis A and B viruses with restriction-enzyme-encoded DNA-modified magnetic in dark filed image with sensitivity up to 0.1 pM 64. It has a great versatility and multiplexed possibility due to the availability of more than 2,000 restriction enzyme choices.
A nanomaterial of amino functionalised carbon coated magnetic nanoparticles (NH2-CC-MNPs) is used to detect hepatitis B viruses with electrochemical detection of DNA hybridization 65. The morphology of NH2-CC-MNPs is confirmed by dispersive X-ray spectroscopy, fourier transform infrared spectroscopy, and scanning electron microscopy. The quinine oxidation signal from DNA hybridisation was detected using PGE combined with pulse voltammetry techniques reaching high sensitivity of picomolar level.
Graphene oxide based nanoparticle and immunosensor can be used for food safety screening of viruses 66. As a novel technology, reduced graphene oxide (RGO) nanogrids are used to detect hepatitis B virus based on a nanoporous silicon oxide template 67. Scanning electron microscopy, surface profilometry, Raman, and conductance measurement are used to verify the RGO nanogrid structure. The combined effects of quantum dot like transport behaviours, insignificant line edges, and improved interaction of molecules within the nanopores make this biosensor ultrasensitive, able to detect up to 50 aM, which is currently the highest detection level of graphene oxide nanogrid technology.
Nanomaterials Detecting Avian Influenza Virus
Nanomaterials have been applied to the detection of avian and human influenza viruses using immunomagnetic nanobeads, and hybrid gold nanoparticles. One of the approaches uses immunomagnetic nanobeads, monoclonal antibodies, and fluorescent quantum dots to detect avian influenza viruses without sample pre-treatment 68. The sensitivity is optimized by high-luminance quantum dots and biotin-streptavidin conjugation to reach a detection limit of 60 virus copies/200 µL. The approach has a high specificity and reproducibility with variability of 1.35% and 3.0% between assays.
An assay based on probes of gold nanoparticles and colorimetric anti-hemagglutinin antibodies is used to detect human H3N2 influenza viruses 69. Plasmon coupling is used to aggregate the antibody-gold nanoparticle and to release a visible light emission, which is detected by a UV spectrophotometer. The assay is a single step and enzyme-free virus detection method applicable to other virus pathogens. Another assay based on colorimetric hybrid gold nanoparticles binding to carbon nanotube surface is used to detect human H3N2 influenza viruses 70. The nanohybrid architecture is confirmed by transmission electron microscopy, and UV-visible spectrophotometer has a catalytic ability to oxidise tetramethyl-benzidine by H2O2 and yield a blue color in aqueous solution depending on the virus concentration. The assay sensitivity can reach a detection limit of 3.4 PFU/mL which is 380 times more sensitive than the conventional ELISA assay.
A polyethylene glycol (PEG) gold nanoparticle assay is used to detect human H3N2 influenza virus with α2,6-thio-linked sialic acid derivative and synthesised thiolated trivalent 71. A ratio of 75:25 of hemagglutinin to PEG ligand and trivalent α2,6-thio-linked sialic acid ligand is optimized by testing various ratios in the detection of non-purified influenza viruses. Importantly the assay can be used to differentiate human H3N2 and avian H5N1 influenza viruses using trivalent thio-linked sialic acid ligands with the highly binding specificity as α2,6-binding and α2,3-binding, respectively.
Biosensors in POC of Foodborne Viruses
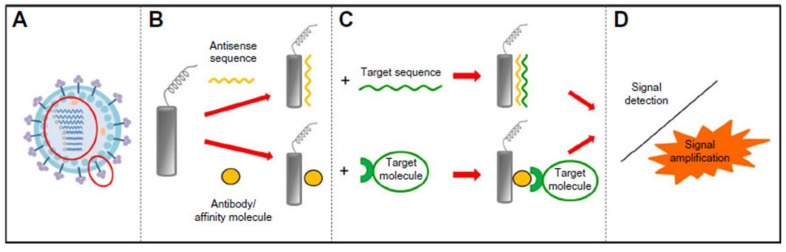
Biosensors have been widely applied to detection method research of foodborne viruses. A biosensor is a device that converts a biological response to an input signal detected by a bioreceptor and converted and amplified by a transducer to a recognisable and measurable output signal with the essential waveform features (Figure 5) 71. The output signal will be displayed, stored, and analysed to generate useful diagnostic information 72.
Figure 5.
Schematic diagram of a biosensor 72. Its bioreceptor recognizes target analytes. The transducer converts biological responses into equivalent electrical signals. Amplifier amplifies the low generated signal into a large output signal that contains essential waveform features.
Overview of Biosensors
A biosensor can be categorised by its various bioreceptors or transducers 72. When categorising by bioreceptors, biosensors can be categorised into six categories:antibody/antigen, enzyme, nucleic acid, phage, cell, and biomimetic. The most commonly used bioreceptors are antibody, enzyme, and nucleic acid, in which the enzymes are mostly used as biorecognition elements and labels other than its actual bioreceptor function. When categorising by transducers, biosensors can be categorised into three categories: electrochemical-based, optical-based, and SPR-based categories (Table 3).
Table 3.
Biosensors available for foodborne virus detection
| Technologies | Analytes | Detection limits | References |
|---|---|---|---|
| Electrochemical with gold & ConA | Norovirus | 35 copies/mL | 11 |
| Silicon nanowire field-effect transistor | Norovirus | 1 fM | 99 |
| Label-free G-quadruplex nanotweezer | Norovirus | 4 nM | 100 |
| Thioglycolic CdZnSeS quantum dot | Norovirus | 8.2 copies/mL | 101 |
| Label-free electrochemical nanogold | Rotavirus | 2.3 PFU/mL | 121 |
| Electrochemical chip-based | Hepatitis A, B, C, D, and E viruses | 1.0 ng/mL | 106 |
| Label-free micro-electro cantilevers | Hepatitis A & C viruses | 1.66 pM | 107 |
| Resonance light scattering | Hepatitis A virus | 8.6 pM | 108 |
| Impedimetric zeolite nanocrystals | Hepatitis B virus | 50 copies/mL | 109 |
| SPR methacrylamide biointerface | Hepatitis B virus | 0.3 ng/mL | 110 |
| Label-free electrochemical nanohybrid | Hepatitis B virus | 0.03 ng/mL | 22 |
| Electrochemical double -deck gold | Hepatitis B virus | 3.3 fM | 111 |
| Microarray-based nanogold | Hepatitis E virus | 100 fM | 112 |
| Electrochemical magnetic beads | Avian influenza virus | 1 ng/mL | 113 |
| Dual-color homogenous | Human enterovirus | 12 µg/mL | 114 |
| SPR organic light emitting portable | Human enterovirus | 4.8 pg/mL | 115 |
Biosensors under Bioreceptor Categories
Antibody-/-antigen bioreceptor based biosensors are one of the major categories of biosensors because of the high specificity of antibody-antigen interaction 73. Biosensors using antibody bioreceptors as a sensing element for foodborne pathogen detection include SPR sensors, nanowire direct charge biosensors, magnetoelastic resonance biosensors, and immunosensors. More detail of the antibody biosensors is given in the Antibody Section below.
Enzymatic bioreceptor based biosensors are the second major categories of biosensors. They are well-developed and widely used in foodborne pathogen detection 74. Enzyme-substrate binding is highly specific with special catalytic activities and provides sufficient electron transfer to an electrode of a biosensor. Three enzymes have been most popularly used: alkaline phosphatase, horseradish peroxidase, and beta-galactosidase. More detail is given in the enzyme sections below.
Nucleic acid bioreceptor based biosensors are the third major category of biosensors based on the complementary property of nucleic acid base pairs 75. Nucleic acid bioreceptors are more specific and sensitive compared to antibody-antigen bioreceptors, while the latter is faster and more robust. Nucleic acids, mostly DNA, exist widely in virus pathogens, and their highly specific complementary property can be used for detection due to their unique DNA sequences. Compared to enzyme and antibody bioreceptors, nucleic acid bioreceptors can be regenerated and synthesised with simpler, faster, and cheaper features. Furthermore, DNA microarray technology using single-stranded nucleic acid aptamers can be used to identify and quantify foodborne pathogens with high speed and accuracy. Nucleic acid biosensors are based mostly on optical, electrochemical, electro-chemiluminescence, quartz crystal, and SPR conducers 76, 173.
Phage bioreceptor based biosensors are based on bacteriophages employed as biorecognition elements to identify foodborne pathogen viruses 182. They are engineered viruses binding to specific receptors on targeting pathogen viruses with high specificity. They can be used in conjunction with different sensing platforms to achieve the detection goals. Designed peptides displayed by phages can be used to bind specific antibodies or viral antigens with strong recognition capacity for biosensor fabrication 77, 78.
Biomimetic bioreceptor based biosensors are designed and fabricated to mimic antibody, enzyme, cell, or nucleic acid bioreceptors are emerging as molecular imprinting tools attractive to and highly accepted for artificial recognition agents 79. Glycol codes with a variety of carbohydrate residues provide highly specific affinity lock-in properties, and can be used to detect a wide ranges of virus pathogens 80. Artificial molecularly imprinted receptors can recognise and bind various targeting molecules with high affinity and specificity 81. They have advantages compared to their biological counterparts, such as resistance to microbial spoilage, long-term stability, custom synthesis, and potential reusability. They do not require animal inoculation for virus detection and their rebinding properties are useful and exceptionally successful.
Biosensors under Transducer Categories
Optical transducer based biosensors have a high potential to deliver POC assays because of their high sensitivity and capability to detect single molecules 82. But optical biosensors require a fluorophore molecule to be attached to the target to enable fluorescent detection and increase sensitivity. An optical fluorescent retrovirus detection is tagged with a pH-sensor genetically encoded to enable simultaneous measurement of pH drop and resulting virus-endosome fusion 83. An engineered nanoscale ZnO with enhanced fluorescence can be used to detect cDNA without the need for DNA amplification, which is developed by ultrasonic spray pyrolysis with four fluorophores labeling in one single system 84. Optical biosensors can also be used as label-free chemi-resistive sensors by a lithographical nanowire electrodeposition assay based on a batch-fabricated polypyrole nanoribbon 85.
Electrochemical transducer based biosensors are easy-to-use platforms with their unique properties, such as compatibility with micro-fabricated technology and easily modifiable surface, making them particularly suitable for virus detection 86,87. Two major features of electrochemical bioreceptors are modifiable probes or antibodies followed by target isolation, and amplified signal detection (Figure 6) 128. The amplified signals can be either quantitative potentinmetric, amperometric, or impedimetric; and the signal can be either labeled or label-free, although labels are often required because electrochemical signals are usually less sensitive 88. Other features such as hybridization, and amplified or changed signal intensity can be applied to electrochemical biosensors. Moreover, electrochemical impedance biosensors have recently been used in virus detection with hybridization or redox systems. The combination with hybridization is easily measured because of high sensitivity and duration without the requirement of electrodes compared to its conventional counterpart 89. Redox system based on the Faradaic or non-Faradaic process may reduce protein layers; and its combination with a genosensor can achieve label-free virus detection 90.
Figure 6.
Four steps needed for the electrochemical biosensing of viral pathogens 50. A) Virus elements targeted; B) electrode (sensor) being modified by biorecognition element; C) itargets are isolated; and D) detection of signal or detection of signal after amplification.
SPR transducer based biosensors are one of the most advanced real time and label-free sensors 91. SPR uses collective oscillation with permittivity of opposite signs presenting at the interface of a metal-dielectric. It measures the refractive index on a metal surface with very thin nanomaterial layers. Its most common application is detecting affinity parameters between antigen-antibody, protein-DNA, and ligand-receptor interactions 92. Other interactions can also be detected, such as enzyme-substrate and complementary DNA-DNA binding actions. SPR biosensors can be combined with hybridization, immunosensors, and aptamer techniques to enhance their applicability to virus detection 93-95 .
Biosensors in Detection of Foodborne Viruses
Various biosensors have been applied to foodborne virus detection, such as norovirus, rotavirus, and hepatitis viruses. Electrochemical biosensors have advantages over analytical transducing biosensors due to their high quantification, fast speed, and high sensitivities. Most importantly, they fit the POC purpose with their features of ease of data logging and miniaturisation 96, 97. Label-free biosensors are under development to target simplified and low-cost POC detection of foodborne viruses.
Biosensors Detecting Norovirus
In the emerging biosensor technologies, silicon microring resonate photonics is gaining attention due to the applicability of its chip-scale integration, low-cost production, sensitivity, and multiplexed detection using silicon fabrication 98. A platform has been designed using an organophosphonate vinyl sulfone linker and coating surface with silicon resonator to detect noroviruses. It bears multiple regenerations and reproducible binding in high-pH and high-salt solutions, and after storage in ambient conditions for one month. Its durability and stability facilitate its use as POC device in prolonging lifetime, variable sensing conditions, and minimising storage and delivery costs.
An electrochemical biosensor uses a concanavalin A (ConA) conjugated and nanostructured gold electrode to capture noroviruses selectively 11. Cyclic voltammetry confirmed the sensitivity and a detection limit of 35 copies/mL in solution extracted from lettuce, and its selectivity as 98% because the signals detected from hepatitis A and E viruses are only 2-2.8% of the norovirus signals at an identical concentration. The sensor fits POC requirements, as it is sensitive, selective, fast, and most importantly, does not require a pretreated sample. Additionally, it is relatively cheap, as much as 1/50 times cheaper than using antibodies alone.
A silicon nanowire field-effect transistor biosensor can detect norovirus DNA single nucleotide polymorphism (SNP) using HNA (HF+HNO3+CH3COOH) to define the nanowire and top-down fabrication 99. Its sensitivity is increased by covalently modifying the nanowire with DNA probes; and its detection limit reaches 1 fM with a few second response time. The biosensor design facilitates mass manufacturing with its production potency in conventional silicon industry.
A label-free split G-quadruplex nano-tweezer can detect norovirus mRNA to detect peroxidase activity and directly produce a signal 100. The nano-tweezer, with a single signal-transducing molecule, can self-assemble from three single-stranded DNA molecules by simple mixing without any washing steps. It recognises norovirus mRNA specifically and causes the structure to produce a change, which restores its peroxidase activity and produces the detectable signal. Its sensitivity in detection limits is 4 nM, and its design is simple without the need of delicate optimization.
Another norovirus RNA detecting biosensor has been developed using a beacon probe conjugated alloyed thioglycolic-capped CdZnSeS quantum dot 101. A modified silanisation method is applied to encapsulate the thiol-capped quantum dot, leading to a high photo-luminescence quantum yield of 98%. Its sensitivity of detection limit is 8.2 copies/mL.
A label-free gold-immobilised synthetic peptide-based electrochemical biosensor is used to detect noroviruses 102. The cysteine incorporated recognition peptides and amino acid substituted are isolated from a phage displaying and selecting process. Its sensitivity of detection limit is 99.8 nM for recombinant noroviral capsid proteins; and 7.8 copies/mL for human noroviruses. The concept of affinity peptides as molecular binders can be applied to miniaturised micro-device as a diagnostic tool for POC testing.
Biosensors Detecting Rotavirus
A label-free photonic crystal biosensor can detect waterborne rotaviruses using surface functionalization with anti-rotavirus antibodies 103. A partially processed water sample with a 30 minute assay confirmed its sensitivity as 36 copies/mL, focusing on forming units without using any external reagents. The biosensor is suitable for virus contamination monitoring on site and real time.
A spectroscopic assay based on SERS can be used to detect human rotaviruses for direct structural characterisation of the viruses 104. The SERS spectra can identify and qualitatively classify virus strain fingerprints according to their G and P genotypes with > 96% accuracy. This technique is useful for virus laboratories and healthcare centres for pathogen detection and monitoring.
A label-free biosensor with photonic Fabry-Perot transducers based on relative optical power can detect human rotaviruses 105. Its sensitivity can be optimised by selecting optimum specific wavelength or spectral band and selecting its relative optical power variation based on the biomolecule concentration. This system has tested for its capability in the detection of the immobilization of rotavirus antigen and its antibodies. Its flexibility and adjustability make this biosensor a remarkable candidate for lab-on-a-chip devices in POC development.
Biosensors Detecting Hepatitis Viruses
Electrochemical immunosensors are applied to hepatitis virus detection, and one of them is a chip-based device immobilizing anti-virus antibodies onto an electrochemical biosensor using protein A and nanogold particles as matrices 106. The label-free device can capture five types of antigens from hepatitis viruses simultaneously, such as hepatitis A, B, C, D, and E viruses, as a one-step assay within 5 minutes. It uses a POC 2-electrode system to capture the virus antigens based on the molecular change prior to and post the antigen-antibody reaction with a detection limit of 1.0 ng/mL.
Another label-free device is designed as a resonant micro-cantilevers array based on anti-virus antibodies and micro-electro-mechanical system (MEMS) techniques 107. The device can detect both hepatitis A and C virus antigens at different concentrations without drying the cantilevers and without electrical connection, allowing disposable chips to be used. Actuation can be accomplished by utilizing laser illuminated optical sensing and an electromagnet sensing facilitating a detection limit of 0.1 ng/mL or 1.66 pM. The results are comparable to labeled detection using ELISA method.
A novel resonance light scattering sensor is designed to detect hepatitis A viruses using a mussel inspired polydopamine-coated virus imprinted polymer 108. The polymer is introduced on a surface of SiO2 nanoparticles by one-step, and the viruses are captured selectively by the imprinted polymer films to increase intensity and sensitivity. A simple fluorescent spectrophotometer is employed to detect the viruses with a detection limit of 8.6 pM.
Several novel biosensors have been developed in the detection of hepatitis B viruses based on impedimetric genosensor, plasmonic biosensor, and electrochemical immunosensor techniques. An impedimetric genosensor is developed using zeolite nanocrystals and multiwalled carbon nanotubes to detect PCR amplified single-stranded virus DNA 109. The DNA hybridisation is detected as the nanocrystals are placed on a glass electrode constructed from fluorine doped tin oxide, while the nanotubes are incorporated to enhance electro-conductivity. The detection results are compared to RT-PCR detection with 97% accuracy; and the sensitivity of detection limit is 50 copies/mL. The electrode can be reused by dipping in 0.1 M NaOH for three minutes with a loss of its initial activity as 50% in four weeks.
A fabricated SPR technique is applied to hepatitis B virus detection for surface antigen-antibody reaction in complex media 110. It uses a biointerface system inspired by poly[(N-(2-hydroxypropyl) methacrylamide) - co - (carboxybetaine methacrylamide)] brushes enhanced with bioreceptors. It allows rapid detection with the discrimination of anti-virus antibodies in positive and negative samples in 10 minutes confirmed by ELISA method. Its sensitivity of detection limit is 0.3 ng/mL.
A label-free electrochemical immunosensor is designed to detect hepatitis B viruses using a glassy carbon electrode and a nanohybrid surface assembled consisting of amino carbon nanotubes 22. The anti-hepatitis B core antigen-antibody reaction is detected by the use of label or chemical mediators and square wave voltammetry. The carbon nanotubes are used to enhance the hyaluronic acid to reach a limit of detection as 0.03 ng/mL.
Another electrochemical immnosensor is designed to detect hepatitis B virus surface antigen-antibody reaction using a double-deck gold film with thionine unites and platinum nanovire inlaid globular SBA-15 as an electrical probe 111. The design can reduce the spatial limitations of loading secondary antibodies, provide pore accessibility of guest species from outside, and offer catalytically active sites on a large scale. The presence of thionine enhances the electrical conductivity between the gold film and the nanowire to facilitate linear detection with a detection limit of 3.3 fM.
The development in biosensor techniques of hepatitis E viruses is relatively rare compared to other hepatitis viruses. A microarray-based biosensor is designed to detect hepatitis E viruses using nanogold labeled oligonucleotide probes 112. The NH2 end-modified oligonucleotide probes are immobilised on the chip surface to capture the cDNA amplified from the virus RNA. The SH end-modified oligonucleotide with nanogold colloid is used as detection probe. The detection time is shortened to two minutes with a limit of detection of 100 fM.
Biosensor techniques have been developed to detect other viruses, such as avian influenza viruses and enteroviruses. A bienzymatic electrochemical immunosensor based on a magnetic bead is designed to detect H9N2 avian influenza viruses using a gold Au electrode 113. The bienzymatic detection is facilitated using the 1st enzyme tagged on magnetic beads on the gold electrode accumulated; and the 2nd enzyme as immobilised layer by layer on the gold electrode. It has high selectivity to neglect other viruses such as H5N1 virus, Newcastle disease virus, Banna virus and Pseudorabies virus has a limit of detection as 1 ng/mL.
A dual-color fluorescent and homogenous immunosensor is designed to detect human enterovirus 71 antigen-antibody reaction using quantum dot and antibody-gold conjugation 114. It can be modified by replacing the antibodies and used to detect any viruses or proteins with a limit of detection of 12 µg/mL. Another biosensor for human enterovirus 71 is developed as a portable SPR device to run on low power in a miniaturised platform with a color tunable organic light emitting diode 115. Its signals are optimised by applying optimum signal to noise ratio enhancement to facilitate an assay in several minutes with a limit of detection as 67 copies/mL or 4.8 pg/mL.
Other Advanced Technologies in POC of Foodborne Viruses
Other advanced technologies available for POC detection of foodborne viruses include detecting immune reactions between antibodies and antigens, aptamers to detect nucleotides and peptides, enzymes used as labels other than actual bioreceptors, and nanozymes as artificial enzymes based on nanomaterials aiming to imitate natural enzymes.
Antibodies/antigens
Antibodies, as common bioreceptors of biosensors, can be polyclonal or monoclonal, and natural or recombinant 116. Monoclonal antibodies have better specificity than polyclonals because they target their antigens specifically; but polyclonal antibodies target more than one antigens. Recombinant antibodies can be designed with man-made features to increase specificity and functions.
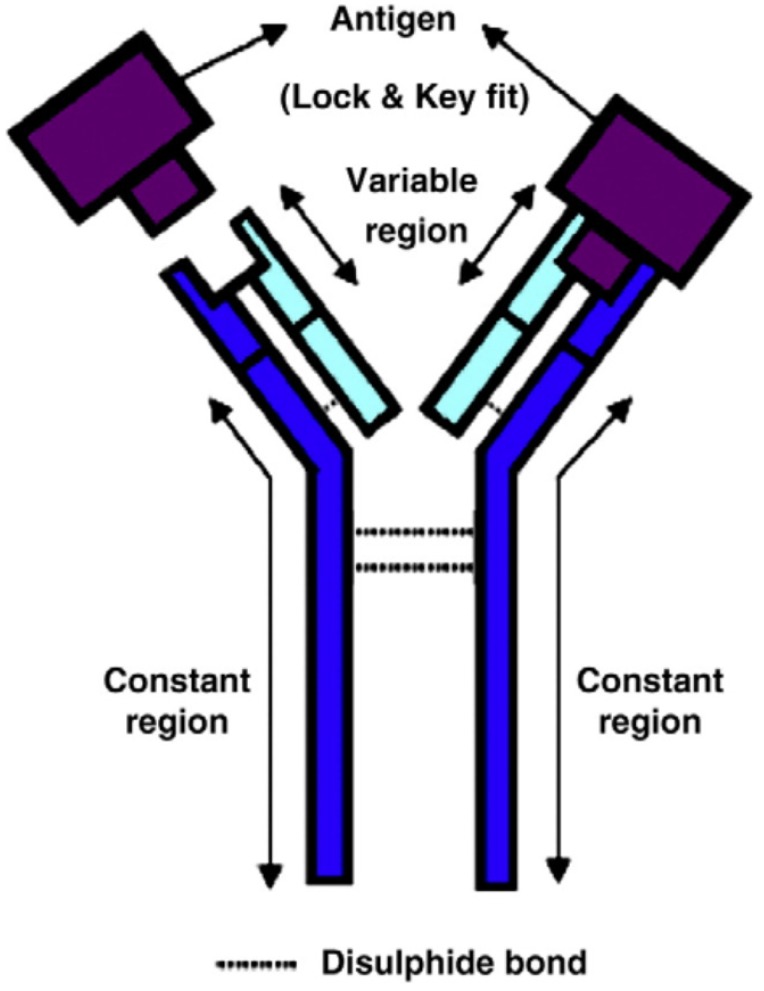
Antigen-antibody interaction is highly specific close to a lock-and-key fit with high affinity and low non-specificity (Figure 7) 72. It is possible to find an antibody to fit almost any unique antigens; thus allowing a huge diversity of a large variety of molecule shapes. This unique antigen-antibody specific property makes antibody biosensors one of the most applicable and powerful tools in the detection of foodborne viruses 73.
Figure 7.
The basic structure of an antibody, and antigen-antibody lock and key fit 72. The antigen binding site of antibody binds specifically with complementary target antigen.
Examples of antibody facilitating biosensors in foodborne pathogen detections include SPR biosensors 117, nanowire direct charge biosensors 118, magnetoelastic resonance biosensors 119, and immunosensors 120. In these detections, antibodies can be either directly or indirectly labeled to generate detectable signals, with direct label being preferred in most detections because it saves time and increases sensitivity compared to indirectly labeled antibodies. Indirectly labeled antibodies rely on a non-labeled primary antibody together with a labeled secondary antibody binding to the primary antibody (Table 4).
Table 4.
Differences in antibody labeling methods
| Methods | Advantages | Disadvantages |
|---|---|---|
| Direct | 1. Simple & rapid; 2. Specific and no cross-reaction. |
1. Immune reactivity may be reduced due to high specificity; 2. Time consuming & expensive if more than a few antibodies used. |
| Indirect | 1. Higher reactivity due to non-specific binding; 2. Better antibody and label availabilities. |
1. Non-specific signal and cross-reactivity; 2. Longer procedures and times. |
| Non- Labelled | 1. Simplest and fastest; 2. Specific without cross-reaction. |
1. Reduced immune reactivity; 2. Complex if more antibodies needed. |
Novel antibody related virus detection has been developed for simpler and faster POC assays. For example, label-free or reagent-less strategies based on antibodies with engineered or natural binding proteins can be used to detect pathogens without any additional labeling 73. For example, a design with label-free electrochemical impedance spectroscopy can detect rotoviruses fabricated by cysteamine on gold sononanoparticles as self-assembled monolayers 121. The glassy surface of carbon electrode is immobilised by anti-rotavirus antibodies on cysteamine cross-linked glutaraldehyde. The binding of the antibodies to rotaviruses leads to a signal in the interface of impedance spectra. The detection limit by this immunosensor is 2.3 PFU/mL and the total analysis time is 55 minutes. It is selective to exclude hepatitis A virus and enteroviruses.
A magnetic near-field enforced illumination biosensor is developed to detect noroviruses in contaminated water using two different antibodies 122. One antibody is linked to polystyrene beads and the other antibody is linked to magnetic beads to generate a sandwich when binding to viruses. The activated magnetic beads propel the reaction of antigen-antibodies and the emission of optical signals from the polystyrene beads. This sensor allows the virus detection limit of 40 copies/100 µL without a washing step in contaminated water.
A molecular motor F0F1-ATPase biosensor is designed to detect foodborne noroviruses based on probe-RNA specific binding 123. A specific probe is designed to encompass a conservative region of noroviruses; and F0F1-ATPase in chromatophore is constructed as a molecular motor to drive the biosensor in noroviruses capturing. Its detection limit is 0.005 ng/mL of norovirus RNA with one hour of processing time, and without cross-reaction to other foodborne viruses.
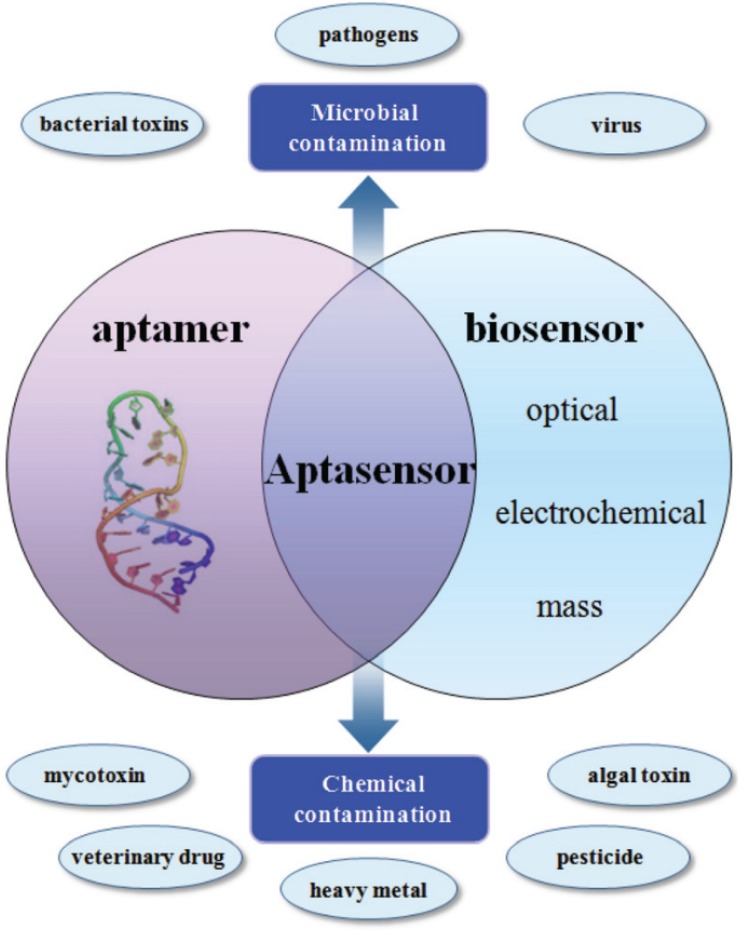
Aptamers
Aptamers are single-stranded oligo nucleotides such as DNA or RNA, or peptides, which bind specifically with high affinity to foodborne viruses 124. Aptamers can be selected using systematic ligand evolution of ligands by exponential enrichment (SELEX) from a pathogen nucleic acid or peptide library containing at least 1015 different targeting sequences 125. A typical SELEX process is to create a random synthesised library of nucleotides, with each nucleotide containing a random central region flanked by different PCR primer binding sequences to facilitate target selection. After selection, the chosen aptamers can recognise and bind to targeting sequences through several properties, such as molecular shape complementarities, hydrogen bonding, electrostatic interactions, or stacking of aromatic rings.
Compared with antigen-antibody reaction, aptamers bear tremendous advantages. For example, they can be synthesised easily; their bindings are faster, higher specificity and affinity, and more stable; and they can be used for wider range of targets like small molecules, drugs, and metal ions 126. Different chemical tags can be added to aptamers to increase flexibility without affecting affinity. Their lower molecular weight enhances the stability to sustain repetitious denaturation and renaturation. Aptasensor, due to its advantages, is increasingly explored, particularly in intact virus detection using SPR and other platforms 127.
Aptasensors are biosensors utilising aptamers as bioreceptors to combine different conductors for selective molecular application and recognition 128. The combination of multi-purpose aptamers and multi-function conductors, such as electrochemical, optical and mass conductors, facilitates the use of aptasensors in both pathogen and chemical detection (Figure 8). Aptamer techniques have been widely applied to foodborne virus detection.
Figure 8.
Application of aptamers in biosensing technologies for the food analysis 128.
Aptamers fit the requirements of foodborne virus POC detection to be fast, accurate, easy to use, and affordable 129. They provide higher discriminating capacity of serotypes and faster adaptation to continuously evolving virus strains. For example, a DNA aptamer strategy has been introduced to detect murine and human noroviruses; and a promising aptamer has been selected post nine-rounds of SELEX with very high affinity to both virus types 130. The binding affinity of the selected aptamer AG3 is a million-fold higher than a non-specific DNA control sequence, and has a million-fold higher selectivity from a similar virus, feline calicivirus. The aptasensor combining AG3 and biosensor technique using gold nanoparticles can detect norovirus particles with a limit of detection as 180 copies/mL in environmental samples.
In the SELEX process, nitrocellulose membranes, agarose gels, and adsorptive microplates are used as supports to immobilise virus targets, but the subsequent separation under these supports is time-consuming and tedious. To eliminate the carrier defect, a study applies carrier magnetic nanoparticles to immobilise the surface antigen of hepatitis B viruses 131. DNA aptamers are selected from a library after 13 selection rounds and used to build an aptasensor based on chemiluminescence immunoassay and magnetic separation. The derived aptasensor can be used to detect the virus surface antigen from purified or actual samples with a limit of detection limit as 0.1 ng/mL, which is lower than the limit of 0.5 ng/mL from ELISA measurement.
Human noroviruses are ideal candidates for aptamer detection because their significant antigenic diversity limits its detection using antibodies. An aptamer study targeting ssDNA in foodborne human norovirus GII.4 identifies an aptamer #25 showing affinity to both GII.2 and GII.4 strains, which is confirmed by ELISA 132. Its binding affinity is equivalent to a commercial anti-norovirus antibody. A magnetic capture method using aptamer combines the #25 aptamer and RT-PCR to detect norovirus in contaminated lettuce. The capture efficiency is 2.5-36% with a limit of detection as 10 copies/3 mL in a lettuce sample.
Another aptamer study targets the P domain protein of human GII.4 norovirus strain using E. coli to express and purify the P protein 133. After SELEX, an aptamer M6-2 is selected and confirmed by ELISA targeting GI.7, GII.2, two GII.4, and GII.7 strains with low to moderate binding affinity. Aptamer magnetic capture and RT-PCR are combined with the selected M6-2 aptamer to capture noroviruses with a detection limit of 4.88 log10 input genomic copies. The M6-2 derived aptasensor was successfully tested for its capturing capability of purified GII.4 New Orleans outbreak noroviruses.
A miniaturised and portable MEMS-based electrochemical aptasensor is developed to detect human noroviruses by combining aptamer and micro fabrication technique 134. The electrode is modified by drop casting a virus-specific and fluorescent labeled aptamer to be immobilised on the surface of a gold electrode. The binding capability between the aptamers and on-chip sensing electrode is evaluated by testing the sensor responses to different titers of murine noroviruses as a model of human noroviruses. The MEMS aptasensor exhibits rapid and clear response to different virus titers with a limit of detection of 50 PFU/mL.
Enzymes
Enzymes as a bio-recognition element in biosensor research are widely studied and well developed. Almost all enzymes are proteins except a few that are ribonucleoproteins with their catalytic activities in the RNA part 72. Enzymes are chosen based on their specific catalytic activity and binding capability to substrates. Enzyme selection can provide sufficient electro transfer to a working electrode with a suitable substrate. Using enzymes as bioreceptors to detect foodborne viruses provides a high specificity; and their catalytic activities can enhance the sensitivity of detected signals.
In most cases, enzymes are not used as the actual bioreceptors, but as labels, such as labeling antigens, antibodies, and nucleotide probes. The improvement in enzyme labeling techniques facilitates sensitive detection, particularly in immunoassay detection 74. The advantages of enzyme labeling are its specificity, stability, and the possibility of visual recognition. Its advantages over radioisotope and fluorescent tag labeling are higher sensitivity and stability without health hazards. Enzymes are stable in storage for years; and their visibility reduces the need for expensive and complicated equipment. Furthermore, small enzymes can be used to label antigens and antibodies to pass cell membranes to facilitate the observation of protein localisation and tissue structure. The disadvantages of enzyme labeling are the possibility of endogenous enzyme interference in samples and multiple assay steps.
The commonly used enzymes in pathogen detection are alkaline phosphatase, beta-galactosidase, and horseradish peroxidase (HRP). A colorimetric assay based on alkaline phosphatase induced metallisation is applied to the detection of H9N2 avian influenza virus particles linked to immnomagnetic separation 135. The color change induced when enzymes are combined with gold nanoparticle-induced silver deposition to enhance the optical signals as 4-6 folds higher than similar metal nanoparticle colorimetric methods. It is coupled with a magnetic bead-based sandwich immunoassay to detect the viruses in complex samples, such as chicken serum, without pretreatment and with a limit of detection as 17.5 pg/mL.
Nanozymes
Nanozymes are artificial enzymes based on nanomaterials to replicate the essential and generic properties of natural enzymes, which were named by Scrimin and colleagues to describe gold clusters protected by a thiol monolayer with ribonuclease activity 136. The advantageous characteristics of nanoenzymes over natural enzymes are their excellent robustness and stability, long-term preservation capability, and low-cost productivity by facile scale up 137. The nanomaterials used as nanozymes can be categorised as metal oxide-based nanomaterials, cerium oxide-based nanomaterials, carbon-based nanomaterials, and metal-based nanomaterials, and 138. These nanomaterials are found to bear unexpectedly enzyme-like activities which are taken as advantages in nanozyme development with wide application in foodborne virus detection.
Cerium oxide-based nanomaterials with enzyme-mimic material of cerium oxide (ceria) produce highly catalytic activity due to the mixture of valence states of Ce3+ and Ce4+ together with the presence of oxygen vacancies 139. The dominant Ce3+ and oxygen vacancy on nanoceria surface are due to its large surface to volume ratio 140. Oxygen vacancies lead to the reduction of positive charge by Ce3+ to stabilise the Ce3+ oxidation state. The two oxidation states can be switched with each other between the Ce3+ and Ce4+ recycle processes to generate catalytic energy and activity. Its application on enzyme mimics includes catalase, oxidase, and superoxide oxidase (SOD), mimics.
Metal oxide-based nanomaterials include iron oxide, cobalt oxide, copper oxide, manganese dioxide, and vanadium pentoxide exhibiting an intrinsic peroxidase-like activity 138,141. Cobalt oxide nanomaterials are a dual intrinsic enzyme mimics using cobalt oxide to provide peroxidase and catalase activities; for example, cubic Co3O4 nanoparticles 142. Copper oxide nanomaterials such as CuO nanoparticles show peroxidase mimic activity with higher affinity to its substrate and H2O2 compared to HRP in colorimetric assay 143. Manganese dioxide nanomaterials (MnO2 NPs) can be made as nanosheets, nanosticks, nanospheres, or nanowires with high and stable peroxidase mimic activity comparable to HRP 144. Furthermore, vanadium pentoxide nanomaterials such as V2O5 nanowires have peroxidase mimic catalytic activity comparable to natural vanadium-dependent haloperoxidase 145.
Metal-based nanomaterials include gold nanomaterials, platinum nanomaterials, and bimetal nanomaterials. Gold nanoparticles with either negative or positive surface charges show mimic activity of peroxidase, which can be tuned by adjusting the affinities between the substrates and nanozymes 146. Platinum nanomaterials are encapsulated with apo-ferritin, exhibiting SOD mimics with long-term stability 147. Bimetal nanomaterials are a combination of multiple metals such as AuM, which is a combination of Au, Bi, Pd, and Pt based on gold nanoparticles to display peroxidase mimics 148.
Carbon-based nanomaterials include graphene and derivatives, and fullerene and derivatives. Fullerene and its derivatives can be considered as a radical sponge; when they are made soluble they can be modified with biomolecules in solution with SOD mimics 149. Graphene oxide and its derivatives exhibit higher affinity to organic substrates with peroxidase mimic activity but lower affinity to H2O2 compared to HRP 150. Graphene with hemin can be synthesised and functionalised as hybrid nanosheets to display higher affinity to H2O2 but lower affinity to substrates compared to HRP.
Nanozyme techniques have been widely applied to foodborne virus detection including some novel viruses with potential food contamination possibilities. Ebola viruses, for example, caused threatening outbreaks in West Africa recently, is considered a potential foodborne pathogen with possibilities to infect farmed animals such as pigs 151. A nanozyme technique is applied to Ebola virus detection using Fe3O4 magnetic nanoparticle based immuno-chromatographic strip 152. It detects the glycoprotein of the viruses with a limit of detection as 1ng/mL within 30 minutes, which is 100-fold more sensitive than the ELISA detection.
Enzyme-free and label-free bimetal nanoparticles of PtAu NPs displaying peroxidase mimic activity can act as barcode in catalyzing oxidation of amplex red substrate into stable fluorescent signal, which can be used to detect hepatitis C virus DNA 153. The integration of the PtAu NPs nanoparticles and biosensors can distinguish single-base mismatched mutant DNA. Its detection limit can reach 5 pM with linear increases when the virus concentration was increased from 10 pM to 500 pM.
Technology Applications & Current Challenges in Foodborne Virus Diagnosis
Modern trends in foodborne bacterial and viral diseases call for POC detection devices for the following purposes: (1) environmental monitoring of infectious agents; (2) self-applied quality control by food production companies and farms; (3) regular safety inspection by governmental agencies; and (4) pathogen screening and cluster identification in clinical emergency settings 30.
Early, on-site POC detection devices are critical in food safety insurance processes such as diagnosis and monitoring 29, 31, 154. Previous POC device development focused on medical diagnosis, but not food safety. However, the emerging growth of POC device research facilitates its implementation in foodborne pathogen monitoring. The key requirements for POC device in foodborne virus diagnosis and monitoring include the precise manipulation of fluids, the capability of handling micro-amount of samples, the capacity for highly sensitive detection, and diagnostic outputs with easy-read and interpretation features 29, 30.
Current expectations of POC devices in foodborne virus monitoring are platforms with combination and hybridised functions, such as the capability for superior algorithmic complexity, computational power, wireless, and portable 155, 156. Based on the expectations, the POC devices and technologies, which fit the requirements, are microfluidic devices, hydrophilic substrate-based detection devices, paper-based analytical devices, and smartphone-based wireless technologies.
Microfluidic Devices
As described in the earlier section, microfluidic technologies and devices are mostly under intensive study and product development. Microfluidic technologies involve extremely small amounts of fluid, from tens to hundreds of microliters, passive or active fluid transport through capillary forces or pumping mechanisms, and microfabrication with shorter diffusional distances 31, 36. These features make microfluidics ideal technologies being able to complete all necessary steps within one device and in a single reaction, which allows sample pretreatment, analysis, and signal capture and interpretation. Microfluidic devices are one of the most likely types in the future to meet all requirements of POC devices.
Up to date, a low amount of novel microfluidic POC devices reach the market for foodborne virus detection, despite the fact that thousands of POC diagnostic applications and platforms are published and usable for food safety diagnosis and monitoring 157. Surprisingly few lab-on-a-chip microfluidic technologies have been translated to commercial products in North America and Europe, where the market is expected to grow to $8.78 billion by 2021 158. The key promoter for the growth is the need for early diagnosis and personalised treatment in human clinics, and this will also definitely promote the growth of microfluidic device in foodborne virus detection.
Hydrophilic Substrate-Based Detection Devices
Hydrophilic substrate-based devices are conceptual devices mostly based on lignocellulose detection using materials such as paper (as described next), bamboo, thread and cotton with potency in foodborne pathogen detection. Bamboo lignocellulose materials are highly promising in the form of stirrers as both bamboo alone and bamboo-wood hybrid for rapid bioanalysis in food and water safety, such as soup, and meat safety monitoring 159.
Thread lignocellulose materials possess fibrous and porous qualities ready to be made into devices for quantitative detection 160. Sewing techniques are applied to thread based devices to weave, twist and encapsulate knots, tape and gel incorporated to provide microfluidic capacity. Thread materials can also be combined with plasma oxidation or other materials such as cotton to generate adequate fluid transportation.
Cotton lignocellulose materials attract researchers due to their absorptive capacity in potential POC application 161. Cotton based device with fluid paths, hydrophilic interior and hydrophobic exterior has been developed to deliver sensitive and semi-quantitative results, and to mitigate possible contamination. The conceptual hydrophilic substrate-based devices require future development with promising potency and valuable capacity.
Paper-Based Analytical Devices
Paper lignocellulose materials are the most featured and fabricated material in POC devices for pathogen detection 162. Paper based materials are combined with lateral flow assays, dipstick assays, and microfluidic assays to generate paper-based analytical devices. Referred to as lab-on-paper, microfluidic paper based devices uses micro-flow path to allow microliter-scale sample analysis 163, 164. The advantages of paper materials over other microfluidic materials such as glass, plastic and silicon are that paper is lightweight, biodegradable, self-powering, easily manufactured and affordable, which makes it convenient materials for domestic, in field and resource-free applications.
Paper based devices can be designed as 2D and 3D patterns with leveraging intrinsic capillary for hydrophilic channels to provide wicking micro-scale fluids. A wax-patterned paper material is combined with 3D-slip design to overcome fluidic limitation and increase sensitivity in human norovirus detection 165. The paper based material enables different microfluidic paths for sequential fluid delivery without the need for peripheral equipment. The assay results are naked eye visible with a test time of 60 minutes and a detection limit of 9.5 x 104 copies/mL. The resulting paper based device can be used in one single step and is suitable for unskilled users.
Smartphone-Based Wireless Technology
Smartphone-based wireless technology is a combination of computational processors, optical sensors, GPS receivers, touch-screen displays, Wi-Fi adaptors, wireless networks, and rechargeable batteries 166, 167. All of the components can be integrated into a multi-functional, pocket-size and portable device to create an ideal candidate in pathogen detection. The combination of smartphone technology and POC devices can meet the requirements in scientific measurement and commercial control of foodborne virus outbreak and food chain inspection and monitoring. They are user friendly and deployable in healthcare facilities, commercial sectors, and remote rural areas.
Smartphone wireless technology can be used to combat current challenges in on-site and remote detection of foodborne pathogens to avoid the needs of conventional laboratory based and resource intensive platforms 168, 169. It can be designed as either a colorimetric or a fluorometric reading format to be used in the detection of various pathogens and their nucleic acid and proteins. A recent application trend is developing the use of smartphones as microscopy devices, which, together with other modern and creative applications, widens the future applicable possibilities of smartphone technology.
Current application of smartphone based devices include a colorimetric assay developing a device with reproducible color quantification of up to 12 color regions despite different lighting conditions 170. The smartphone-based technology in foodborne pathogen detection is mostly applied to foodborne bacterial detection but not virus detection. For example, a smartphone based paper strip modified with polystyrene microparticles and antibodies is developed to detect salmonella in food samples 183. The microparticles and antibodies are agglutinated in the presence of and related to the concentrations of the bacteria. Reading information can be further processed by an application installed on the smartphone with 1 minute time length and a 10 CFU/mL detection limit. Smartphone wireless technology is applied to our research laboratory in foodborne virus detection and the results will be published in the future.
Current Challenges and Future Trends
The research and commercialisation of POC devices for foodborne bacterial detection are far more advanced compared to those for foodborne virus detection. The opportunity to apply bacterial detection techniques to virus is low because bacteria are easier to be cultured and amplified, but virus culture and amplification are difficult and cannot be enriched in most cases 35. Moreover viruses cannot replicate in host free environments and is almost always found in low numbers in the complex food matrices. The size of viruses ranges from 20 to 400 nm, while the size of bacteria ranges from 1 micron to 5 microns. Due to the extremely smaller size of viruses, the sensing platforms developed for bacteria need to be extensively modified for the detection of virus particles. In addition, the composition of cell surface (peptidoglycan) proteins of bacteria is not comparable with the hemagglutinin and neuraminidase compositions of viruses, and the biorecognition elements has to be tailor made to create the binding with the proteins. Along with smaller sizes, absence of cell wall and ribosomes in the viruses in comparison to the bacteria also allows for changes in the localized microenvironment towards the development of biosensing platforms. Considering the factors of bacterial detection techniques that cannot be used for viral detection, and that viruses often present with lower copy numbers than bacteria in food samples, this situation demands superior sensitivity of the virus detection devices to be at least attomolar or picomolar level.
For real-time and on-site applications, the U. S. Food and Drug Administration (FDA) and the Canadian Food Inspection Agency (CFIA) require the food pathogen tests to be conducted using 100 mL of food solution volumes, or at-least 25 gram of solid food samples 171, 172. This requires that any virus detection devices should be developed to meet the requirements of pathogen handling in either 100 mL of food liquid or 25 grams of solid food samples, indicating a definitive requirement of high-throughput setting in the system design of real-time and on-site detection of foodborne viruses.
Food is a complex sample matrix and viral particles are usually found only in low concentrations. Hence, separation, concentration and purification of viral pathogens is essential towards sensitive virus detection. Due to the heterogeneity in genome and surface structures among viruses, a universal viral extraction technique would be extremely difficult for rapid and real-time food-borne viral biosensing detection. Among the four commonly applied viral concentrations techniques namely precipitation with glass wool filtration, polyethylene glycol, monolithic adsorption filtration and organic flocculation with skim milk, Hjelmsø et al., (2017) found the precipitation with polyethylene glycol technique, to be an efficient protocol for highest viral specificity 174.
Iron electrocoagulation was found to be a superior technique in comparison to chemical coagulation pretreatment for virus removal from water and wastewater samples using microfiltration membranes 175. The adsorption of the negatively charged viruses to the positively charged iron flocs and subsequent removal of the flocs by microfiltration membranes might aid in the enhanced sample pre-treatment processes. Addition of Concanavalin A (Con A) to sample solutions containing virus and reacting the added Con A with virus in the solution to form a virus-Con A conjugate, followed by separating the Virus-ConA conjugate from the same solution has been proposed as an economic method of concentrating virus and detecting the virus within a short time 176.
Out of four virus recovery methods namely the ultrafiltration, immunomagnetic separation, ultracentrifugation and PEG precipitation techniques, Summa et al., (2012) 177 found that ultracentrifugation yielded the highest recovery of noroviruses from lettuce and ham; while PEG precipitation recovered the best yield of virus from raspberries.
There is currently no harmonization in the consensus towards selection of sample size or the number of samples to be analyzed from a suspected batch for viral detection. Hence, there is an absolute and immediate need for harmonization of detection of viruses from food matrices 178. A rapid real-time sensitive virus biosensing platform would significantly benefit from development of a reference detection method such as proposed in ISO/TS 15216. 179-181.
Major challenges for POC platform development in current foodborne virus detection include (1) POC platform facilitating multiplex testing; (2) enhancing the signal-to-noise ratio to reduce background noises and increase detectable signals (3) enhancing detection specificity and sensitivity; (4) improving storage duration; (5) allowing user-friendly and unskilled operation 30. Other challenges include contamination issues in smartphone based devices before they can be used for foodborne pathogen detection; and data security and handling to meet the requirements in widespread use and communication of electronic data 29.
With the changing of the global demographic and epidemiologic structure, as well as food processing and harvesting systems, we can expect new foodborne viruses to emerge in society through both animal and plant derived foods. A rapid and sensitive detection system can reduce ongoing transmission of pathogens as well as play crucial role in preventing pathogen transmission through early detection and control of foodborne illness outbreaks. However, more attention should be given to develop a simple, cheap, sensitive, selective, and POC detector combining with modern technology.
In recent years, advancements in nanotechnology and in biological fields enable us to combine different nanoscale materials such as Au NPs, CNTs, MNPs, and QDs in single superstructure and open the possibility to make heterostructuring which would provide us new aspects to (a) modify or enhance optical properties of single nanomaterials; (b) enhance electrical properties in comparison to single nanoparticles; (c) combine different properties in one entity i.e. fluorescence and magnetic properties; or plasmonic and magnetic properties; or plasmonic and fluorescence properties in one nanostructure. In the near future, such multi-functional nanohybrids could be applied to detect and separate different foodborne viruses simultaneously from complex biological matrices.
Colorimetric detection systems are some of the most practical, real life applicable and promising techniques. The introduction of multi nanozymes in one nanostructure with different shapes may provide more surface to volume ratio to bind with substrate, and enhance detection sensitivity due to different enzymatic activity. We believe hetero structuring by metallic nanomaterials will garner more attention in the near future and will provide us with a new horizon in bioanalytical detection systems.
Concluding Remarks
Foodborne viruses and their detection are related to human life and safety, and they are relatively new, receiving more attention due to emerging contamination events and small scale outbreaks. The current research and commercialisation of POC devices for foodborne virus detection are far behind those for foodborne bacterial detection. Since the direct transfer of developed bacterial detection methods to virus detection is unlikely, the current situation in device and technology development is challenging. Efforts in research and development are needed to adopt technologies from virus detection in healthcare fields and foodborne bacterial detection to fulfill the goals of foodborne virus detection in food safety.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (400705) and the Ontario Ministry of Research and Innovation (300512).
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- AAO
anodic aluminum oxide
- ATPase
adenylpyrophosphatase
- CDC
Disease Control and Prevention
- CFIA
Canadian Food Inspection Agency
- CFU
colony-forming units
- CNCs
cellulose nanocrystals
- DNA
deoxyribonucleic acid
- ELISA
enzyme-linked immunosorbent assay
- FDA
Food and Drug Administration
- GPS
Global Positioning System
- HRP
horseradish peroxidase
- LFA
lateral-flow assay
- MEMS
Micro-Electro-Mechanical Systems
- PCR
polymerase chain reaction
- PEG
polyethylene glycol
- PFU
plaque forming unit
- POC
point of care
- RGO
reduced graphene oxide
- RNA
ribonucleic acid
- RT-PCR
Reverse transcription polymerase chain reaction
- SELEX
systematic ligand evolution of ligands by exponential enrichment
- SOD
superoxide oxidase
- SPR
surface plasmon resonance.
References
- 1.Callejón RM, Rodríguez-Naranjo MI, Ubeda C. et al. Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog. Dis. 2015;12(1):32–38. doi: 10.1089/fpd.2014.1821. [DOI] [PubMed] [Google Scholar]
- 2.Kozak GK, MacDonald D, Landry L. et al. Foodborne outbreaks in Canada linked to produce: 2001 through 2009. J. Food Prot. 2013;76(1):173–183. doi: 10.4315/0362-028X.JFP-12-126. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm B, Leblanc D, Leger D. et al. Farm-level prevalence and risk factors for detection of hepatitis E virus, porcine enteric calicivirus, and rotavirus in Canadian finisher pigs. Can. J. Vet. Res. 2016;80(2):95–105. [PMC free article] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ. et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeDuc P, Agaba M, Cheng CM. et al. Beyond disease, how biomedical engineering can improve global health. Sci Transl Med. 2014;6(266):266fs48. doi: 10.1126/scitranslmed.3009067. [DOI] [PubMed] [Google Scholar]
- 6.Newell DG, Koopmans M, Verhoef L. et al. Food-borne diseases - the challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol. 2010;139(Suppl 1):S3–15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acheson DW. Foodborne infections. Curr Opin Gastroenterol. 1999;15(6):538–45. doi: 10.1097/00001574-199911000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Wit E, Prescott J, Falzarano D. et al. Foodborne transmission of Nipah virus in Syrian hamsters. PLOS Pathogens. 2014;10(3):e1004001. doi: 10.1371/journal.ppat.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullis L, Saif LJ, Zhang Y. et al. Stability of bovine coronavirus on lettuce surfaces under household refrigeration conditions. Food Microbiol. 2012;28(2):232–254. doi: 10.1016/j.fm.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koo HL, Ajami N, Atmar RL. et al. Noroviruses: The leading cause of gastroenteritis worldwide. Discov Med. 2010;10(50):61–70. [PMC free article] [PubMed] [Google Scholar]
- 11.Hong SA, Kwon J, Kim D. et al. A rapid, sensitive and selective electrochemical biosensor with concanavalin A for the preemptive detection of norovirus. Biosens. Bioelectron. 2015;64:338–344. doi: 10.1016/j.bios.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 12.Lee HM, Kwon J, Choi JS. et al. Rapid detection of norovirus from fresh lettuce using immunomagnetic separation and a quantum dots assay. J Food Prot. 2013;76(4):707–11. doi: 10.4315/0362-028X.JFP-12-343. [DOI] [PubMed] [Google Scholar]
- 13.Hall AJ, Lopman BA, Payne DC. et al. Norovirus disease in the United States. Emerg Infect Dis. 2013;19(8):1198–205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esona MD, Gautam R. Rotavirus. Clin Lab Med. 2015;35(2):363–91. doi: 10.1016/j.cll.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, Yust B, Nellore BP. et al. Accurate identification and selective removal of rotavirus using a plasmonic-magnetic 3D Ggraphene oxide architecture. J Phys Chem Lett. 2014;5(18):3216–21. doi: 10.1021/jz501402b. [DOI] [PubMed] [Google Scholar]
- 16.Greening GE, Cannon JL. Human and animal viruses in food (including taxonomy of enteric viruses). In Viruses in foods (pp. 5-57) New York, NY: Springer; 2016. [Google Scholar]
- 17.White PA, Netzler NE, Hansman GS. Foodborne viral pathogens. Florida, USA: CRC Press; 2017. [Google Scholar]
- 18.Sánchez G. Analytical methods for detecting the hepatitis A virus in food. In Hepatitis A Virus in Food (pp. 9-20) New York, NY: Springer; 2013. [Google Scholar]
- 19.Adefisoye MA, Nwodo UU, Green E. et al. Quantitative PCR detection and characterisation of human adenovirus, rotavirus and hepatitis A virus in discharged effluents of two wastewater treatment facilities in the Eastern Cape, South Africa. Food Environ. Virol. 2016;8(4):262–274. doi: 10.1007/s12560-016-9246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez G. Hepatitis A virus in food: detection and inactivation methods (p. 47) New York, NY: Springer; 2013. [Google Scholar]
- 21.Jagya N, Varma SP, Thakral D. et al. RNA-seq based transcriptome analysis of hepatitis E virus (HEV) and hepatitis B virus (HBV) replicon transfected Huh-7 cells. PLoS One. 2014;9(2):e87835. doi: 10.1371/journal.pone.0087835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabral DG, Lima EC, Moura P. et al. A label-free electrochemical immunosensor for hepatitis B based on hyaluronic acid-carbon nanotube hybrid film. Talanta. 2016;148:209–215. doi: 10.1016/j.talanta.2015.10.083. [DOI] [PubMed] [Google Scholar]
- 23.Yao CY, Fu WL. Biosensors for hepatitis B virus detection. World J Gastroenterol. 2014;20(35):12485–92. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Yuan L, Wan J. et al. Colorimetric detection of hepatitis E virus based on reverse transcription loop mediated isothermal amplification (RT-LAMP) assay. J. Virol. Methods. 2014;197:29–33. doi: 10.1016/j.jviromet.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Di Bartolo I, Angeloni G, Ponterio E. et al. Detection of hepatitis E virus in pork liver sausages. Int. J. Food Microbiol. 2015;193:29–33. doi: 10.1016/j.ijfoodmicro.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Mena I, Ma J. et al. Newcastle disease virus-vectored H7 and H5 live vaccines protect chickens from challenge with H7N9 or H5N1 avian influenza viruses. J. Virol. 2015;89(14):7401–7408. doi: 10.1128/JVI.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown C. Emerging zoonoses and pathogens of public health significance — an overview. Rev Sci Tech. 2004;3:435–442. doi: 10.20506/rst.23.2.1495. [DOI] [PubMed] [Google Scholar]
- 28.Wu MY, Hsu M, Chen S. et al. Point-of-care detection devices for food safety monitoring: Proactive disease prevention. Trends Biotechnol. 2017;35(4):288–300. doi: 10.1016/j.tibtech.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Vashist SK, Luppa PB, Yeo LY. et al. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015;33(11):692–705. doi: 10.1016/j.tibtech.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Mangal M, Bansal S, Sharma SK. et al. Molecular detection of foodborne pathogens: A rapid and accurate answer to food safety. Crit. Rev. Food Sci. Nutr. 2016;56(9):1568–1584. doi: 10.1080/10408398.2013.782483. [DOI] [PubMed] [Google Scholar]
- 31.Busin V, Wells B, Kersaudy-Kerhoas M. et al. Opportunities and challenges for the application of microfluidic technologies in point-of-care veterinary diagnostics. Mol. Cell. Probes. 2016;30(5):331–341. doi: 10.1016/j.mcp.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Vashist SK, Schneider EM, Luong JHT. Commercial smartphone-based devices and smart applications for personalized healthcare monitoring and management. Diagnostics. 2014;4(3):104–28. doi: 10.3390/diagnostics4030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li CZ, Vandenberg K, Prabhulkar S. et al. Paper based point-of-care testing disc for multiplex whole cell bacteria analysis. Biosens. Bioelectron. 2011;26:4342–4348. doi: 10.1016/j.bios.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 34.Gracioso Martins AM, Glass NR, Harrison S. et al. Toward complete miniaturisation of flow injection analysis systems: microfluidic enhancement of chemiluminescent detection. Anal. Chem. 2014;86(21):10812–10819. doi: 10.1021/ac502878p. [DOI] [PubMed] [Google Scholar]
- 35.Chhabra P, Vinjé J. Molecular detection methods of foodborne viruses. In viruses in foods (pp. 303-333) New York, NY: Springer; 2016. [Google Scholar]
- 36.Gervais L, de Rooij N, Delamarche E. Microfluidic chips for point-of-care immunodiagnostics. Adv Mater. 2011;23(24):H151–76. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 37.Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL. et al. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS) Lab Chip. 2011;11(5):845–50. doi: 10.1039/c0lc00403k. [DOI] [PubMed] [Google Scholar]
- 38.Oh SJ, Park BH, Choi G. et al. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip. 2016;16(10):1917–26. doi: 10.1039/c6lc00326e. [DOI] [PubMed] [Google Scholar]
- 39.Connelly JT, Kondapalli S, Skoupi M. et al. Micro-total analysis system for virus detection: microfluidic pre-concentration coupled to liposome-based detection. Anal Bioanal Chem. 2012;402(1):315–23. doi: 10.1007/s00216-011-5381-9. [DOI] [PubMed] [Google Scholar]
- 40.Tao Y, Rotem A, Zhang H. et al. Rapid, targeted and culture-free viral infectivity assay in drop-based microfluidics. Lab Chip. 2015;15(19):3934–40. doi: 10.1039/c5lc00556f. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Ballerini DR, Shen W. A perspective on paper-based microfluidics: Current status and future trends. Biomicrofluidics. 2012;6(1):11301–1130113. doi: 10.1063/1.3687398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Cockrell SK, Kolawole AO. et al. Isolation and Analysis of Rare Norovirus Recombinants from Coinfected Mice Using Drop-Based Microfluidics. J Virol. 2015;89(15):7722–34. doi: 10.1128/JVI.01137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung SH, Baek C, Cong VT. et al. The microfluidic chip module for the detection of murine norovirus in oysters using charge switchable micro-bead beating. Biosens Bioelectron. 2015;67:625–33. doi: 10.1016/j.bios.2014.09.083. [DOI] [PubMed] [Google Scholar]
- 44.Ishii S, Kitamura G, Segawa T. et al. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl Environ Microbiol. 2014;80(24):7505–11. doi: 10.1128/AEM.02578-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inbaraj BS, Chen BH. Nanomaterial-based sensors for detection of foodborne bacterial pathogens and toxins as well as pork adulteration in meat products. J. Food Drug Anal. 2016;24(1):15–28. doi: 10.1016/j.jfda.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeh HY, Yates MV, Mulchandani A. et al. Molecular beacon-quantum dot-Au nanoparticle hybrid nanoprobes for visualizing virus replication in living cells. Chem Commun (Camb) 2010;46(22):3914–6. doi: 10.1039/c001553a. [DOI] [PubMed] [Google Scholar]
- 47.Zeng Q, Zhang Y, Liu X. et al. Multiple homogeneous immunoassays based on a quantum dots-gold nanorods FRET nanoplatform. Chem Commun (Camb) 2012;48(12):1781–3. doi: 10.1039/c2cc16271g. [DOI] [PubMed] [Google Scholar]
- 48.Vaculovicova M, Michalek P, Krizkova S. et al. Nanotechnology-based analytical approaches for detection of viruses. Anal. Methods. 2017;1:1. [Google Scholar]
- 49.Park JE, Kim K, Jung Y. et al. Metal nanoparticles for virus detection. Chem Nano Mat. 2016;2(10):927–936. [Google Scholar]
- 50.Krejcova L, Michalek P, Rodrigo MM. et al. Nanoscale virus biosensors: state of the art. Dovepress. 2015;2015:47–66. [Google Scholar]
- 51.Liu Y, Zhang X, Yifeng E. et al. Sandwich immunoassays of multicomponent subtrace pathogenic DNA based on magnetic fluorescent encoded nanoparticles. Biomed Res Int. 2016;2016:7324384. doi: 10.1155/2016/7324384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosilo H, McKee JR, Kontturi E. et al. Cationic polymer brush-modified cellulose nanocrystals for high-affinity virus binding. Nanoscale. 2014;6(20):11871–81. doi: 10.1039/c4nr03584d. [DOI] [PubMed] [Google Scholar]
- 53.Ashiba H, Sugiyama Y, Wang X. et al. Detection of norovirus virus-like particles using a surface plasmon resonance-assisted fluoroimmunosensor optimized for quantum dot fluorescent labels. Biosens Bioelectron. 2016 doi: 10.1016/j.bios.2016.08.099. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 54.Junesch J, Emilsson G, Xiong K. et al. Location-specific nanoplasmonic sensing of biomolecular binding to lipid membranes with negative curvature. Nanoscale. 2015;7(37):15080–5. doi: 10.1039/c5nr04208a. [DOI] [PubMed] [Google Scholar]
- 55.Ahmed SR, Takemeura K, Li TC. et al. Size-controlled preparation of peroxidase-like graphene-gold nanoparticle hybrids for the visible detection of norovirus-like particles. Biosens Bioelectron. 2017;87:558–565. doi: 10.1016/j.bios.2016.08.101. [DOI] [PubMed] [Google Scholar]
- 56.Hagström AE, Garvey G, Paterson AS. et al. Sensitive detection of norovirus using phage nanoparticle reporters in lateral-flow assay. PLoS One. 2015;10(5):e0126571. doi: 10.1371/journal.pone.0126571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Driskell JD, Shanmukh S, Liu YJ. et al. Infectious agent detection with SERS-active silver nanorod arrays prepared by oblique angle deposition. IEEE Sens J. 2008;8(6):863–70. [Google Scholar]
- 58.Jarvis RM, Brooker A, Goodacre R. Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss. 2006;132:281–92. doi: 10.1039/b506413a. [DOI] [PubMed] [Google Scholar]
- 59.Fan Z, Senapati D, Khan SA. et al. Popcorn-shaped magnetic core-plasmonic shell multifunctional nanoparticles for the targeted magnetic separation and enrichment, label-free SERS imaging, and photothermal destruction of multidrug-resistant bacteria. Chemistry. 2013;19(8):2839–47. doi: 10.1002/chem.201202948. [DOI] [PubMed] [Google Scholar]
- 60.Maeng B, Park Y, Park J. Direct label-free detection of Rotavirus using a hydrogel based nanoporous photonic crystal. RSC Adv. 2016;6(9):7384–7390. [Google Scholar]
- 61.Shakoori Z, Salimian S, Kharrazi S. et al. Electrochemical DNA biosensor based on gold nanorods for detecting hepatitis B virus. Anal Bioanal Chem. 2015;407(2):455–61. doi: 10.1007/s00216-014-8303-9. [DOI] [PubMed] [Google Scholar]
- 62.Chen CC, Lai ZL, Wang GJ. et al. Polymerase chain reaction-free detection of hepatitis B virus DNA using a nanostructured impedance biosensor. Biosens Bioelectron. 2016;77:603–8. doi: 10.1016/j.bios.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Fang W, Wu Z. et al. A one-tube multiplexed colorimetric strategy based on plasmonic nanoparticles combined with non-negative matrix factorization. Talanta. 2014;128:305–10. doi: 10.1016/j.talanta.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 64.Jang KJ, Lee H, Jin HL. et al. Restriction-enzyme-coded gold-nanoparticle probes for multiplexed DNA detection. Small. 2009;5(23):2665–8. doi: 10.1002/smll.200901105. [DOI] [PubMed] [Google Scholar]
- 65.Altay C, Senay RH, Eksin E. et al. Development of amino functionalized carbon coated magnetic nanoparticles and their application to electrochemical detection of hybridization of nucleic acids. Talanta. 2017;164:175–182. doi: 10.1016/j.talanta.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Jung JH, Cheon DS, Liu F. et al. A graphene oxide based immuno-biosensor for pathogen detection. Angew Chem Int Ed Engl. 2010;49(33):5708–11. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]
- 67.Basu J, RoyChaudhuri C. Attomolar sensitivity of FET biosensor based on smooth and reliable graphene nanogrids. IEEE Electron Device Lett. 2016;37(4):492–495. [Google Scholar]
- 68.Zhao W, Zhang WP, Zhang ZL. et al. Robust and highly sensitive fluorescence approach for point-of-care virus detection based on immunomagnetic separation. Anal Chem. 2012;84(5):2358–65. doi: 10.1021/ac203102u. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y, Zhang L, Wei W. et al. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst. 2015;140(12):3989–95. doi: 10.1039/c5an00407a. [DOI] [PubMed] [Google Scholar]
- 70.Ahmed SR, Kim J, Suzuki T. et al. Enhanced catalytic activity of gold nanoparticle-carbon nanotube hybrids for influenza virus detection. Biosens Bioelectron. 2016;85:503–8. doi: 10.1016/j.bios.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 71.Marín MJ, Rashid A, Rejzek M. et al. Glyconanoparticles for the plasmonic detection and discrimination between human and avian influenza virus. Org. Biomol. Chem. 2013;11(41):7101–7107. doi: 10.1039/c3ob41703d. [DOI] [PubMed] [Google Scholar]
- 72.Velusamy V, Arshak K, Korostynska O. et al. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv. 2010;28(2):232–54. doi: 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Ueda H, Dong J. From fluorescence polarization to Quenchbody: Recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. Biochim. Biophys. Acta, Proteins Proteomics. 2014;1844(11):1951–1959. doi: 10.1016/j.bbapap.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Chemburu S, Wilkins E, Abdel-Hamid I. Detection of pathogenic bacteria in food samples using highly-dispersed carbon particles. Biosens Bioelectron. 2005;21(3):491–9. doi: 10.1016/j.bios.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 75.Palecek E, Bartosik M. Electrochemistry of nucleic acids. Chem. Rev. 2012;112(6):3427–3481. doi: 10.1021/cr200303p. [DOI] [PubMed] [Google Scholar]
- 76.Kumar H, Rani R. Development of biosensors for the detection of biological warfare agents: its issues and challenges. Sci Prog. 2013;96(Pt 3):294–308. doi: 10.3184/003685013X13777066241280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pavan S, Berti F. Short peptides as biosensor transducers. Anal Bioanal Chem. 2012;402(10):3055–70. doi: 10.1007/s00216-011-5589-8. [DOI] [PubMed] [Google Scholar]
- 78.Su W, Cho M, Nam JD. et al. Highly sensitive electrochemical lead ion sensor harnessing peptide probe molecules on porous gold electrodes. Biosensors and Bioelectronics. 2013;48:263–9. doi: 10.1016/j.bios.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 79.Karimian N, Turner AP, Tiwari A. Electrochemical evaluation of troponin T imprinted polymer receptor. Biosens Bioelectron. 2014;59:160–5. doi: 10.1016/j.bios.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 80.Chabre YM, Roy R. Multivalent glycoconjugate syntheses and applications using aromatic scaffolds. Chemical Society Reviews. 2013;42(11):4657–708. doi: 10.1039/c3cs35483k. [DOI] [PubMed] [Google Scholar]
- 81.Whitcombe MJ. Molecularly imprinted polymers: smart hydrogel crystal gardens. Nat Chem. 2011;3(9):657–658. doi: 10.1038/nchem.1125. [DOI] [PubMed] [Google Scholar]
- 82.Sin ML, Mach KE, Wong PK. et al. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14(2):225–44. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Padilla-Parra S, Matos PM, Kondo N. et al. Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proceedings of the National Academy of Sciences. 2012;109(43):17627–32. doi: 10.1073/pnas.1211714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iyer MA, Oza G, Velumani S. et al. Scanning fluorescence-based ultrasensitive detection of dengue viral DNA on ZnO thin films. Sens. Actuators B. 2014;202:1338–1348. [Google Scholar]
- 85.Chartuprayoon N, Rheem Y, Ng JC. et al. Polypyrrole nanoribbon based chemiresistive immunosensors for viral plant pathogen detection. Anal. Methods. 2013;5(14):3497–502. [Google Scholar]
- 86.Li F, Feng Y, Dong P. et al. Gold nanoparticles modified electrode via a mercapto-diazoaminobenzene monolayer and its development in DNA electrochemical biosensor. Biosens Bioelectron. 2010;25(9):2084–8. doi: 10.1016/j.bios.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Adam V, Huska D, Hubalek J. et al. Easy to use and rapid isolation and detection of a vira l nucleic acid by using paramagnetic microparticles and carbon nanotubes-based screen-printed electrodes. Microfluid. Nanofluid. 2010;8(3):329–39. [Google Scholar]
- 88.Uliana CV, Tognolli JO, Yamanaka H. Application of factorial design experiments to the development of a disposable amperometric DNA biosensor. Electroanalysis. 2011;23(11):2607–15. [Google Scholar]
- 89.Eksin E, Erdem A. Chitosan-carbon Nanofiber Modified Single-use Graphite Electrodes Developed for Electrochemical Detection of DNA Hybridization Related to Hepatitis B Virus. Electroanalysis. 2016;28:2514–2521. [Google Scholar]
- 90.Malecka K, Grabowska I, Radecki J. et al. Electrochemical detection of avian influenza virus genotype using amino-ssDNA probe modified gold electrodes. Electroanalysis. 2013;25(8):1871–8. [Google Scholar]
- 91.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42(2):150–61. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 92.Abbas A, Linman MJ, Cheng Q. New trends in instrumental design for surface plasmon resonance-based biosensors. Biosens Bioelectron. 2011;26(5):1815–24. doi: 10.1016/j.bios.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu J, Wan JY, Yang ST. et al. A surface plasmon resonance biosensor for direct detection of the rabies virus. Acta Veterinaria Brno. 2012;81(2):107–11. [Google Scholar]
- 94.Wang R, Zhao J, Jiang T. et al. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods. 2013;189(2):362–369. doi: 10.1016/j.jviromet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 95.Bai H, Wang R, Hargis B. et al. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors (Basel) 2012;12(9):12506–18. doi: 10.3390/s120912506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mandal SS, Navratna V, Sharma P. et al. Titania nanotube-modified screen printed carbon electrodes enhance the sensitivity in the electrochemical detection of proteins. Bioelectrochemistry. 2014;98:46–52. doi: 10.1016/j.bioelechem.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Fu C, Liu L. et al. Electrochemical measurement of Clostridium tetani using a reduced graphene oxide modified electrode and polyaniline-gold nanoparticle-labelled probe. Analytical Methods. 2014;6(15):5771–6. [Google Scholar]
- 98.Shang J, Cheng F, Dubey M. et al. An organophosphonate strategy for functionalizing silicon photonic biosensors. Langmuir. 2012;28(6):3338–44. doi: 10.1021/la2043153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong L, Yu XM. Highly sensitive detection of DNA with HNA defined silicon nanowire fet. 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences 27-31 October; 2013. Freiburg, Germany. [Google Scholar]
- 100.Nakatsuka K, Shigeto H, Kuroda A. et al. A split G-quadruplex-based DNA nano-tweezers structure as a signal-transducing molecule for the homogeneous detection of specific nucleic acids. Biosens Bioelectron. 2015;74:222–6. doi: 10.1016/j.bios.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 101.Adegoke O, Seo MW, Kato T. et al. An ultrasensitive SiO2-encapsulated alloyed CdZnSeS quantum dot-molecular beacon nanobiosensor for norovirus. Biosens Bioelectron. 2016;86:135–42. doi: 10.1016/j.bios.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 102.Hwang HJ, Ryu MY, Park CY. et al. High sensitive and selective electrochemical biosensor: Label-free detection of human norovirus using affinity peptide as molecular binder. Biosens Bioelectron. 2017;87:164–170. doi: 10.1016/j.bios.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 103.Pineda MF, Chan LLY, Kuhlenschmidt T. et al. Rapid specific and label-free detection of porcine rotavirus using photonic crystal biosensors. IEEE Sens. J. 2009;9(4):470–477. [Google Scholar]
- 104.Driskell JD, Zhu Y, Kirkwood CD. et al. Rapid and sensitive detection of rotavirus molecular signatures using surface enhanced Raman spectroscopy. PLoS One. 2010;5(4):e10222. doi: 10.1371/journal.pone.0010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holgado M, Sanza FJ, López A. et al. Description of an advantageous optical label-free biosensing interferometric read-out method to measure biological species. Sensors. 2014;14(2):3675–3689. doi: 10.3390/s140203675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tang D, Tang J, Su B. et al. Simultaneous determination of five-type hepatitis virus antigens in 5min using an integrated automatic electrochemical immunosensor array. Biosensors and Bioelectronics. 2010;25(7):1658–62. doi: 10.1016/j.bios.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 107.Timurdogan E, Alaca BE, Kavakli IH. et al. MEMS biosensor for detection of Hepatitis A and C viruses in serum. Biosens Bioelectron. 2011;28(1):189–94. doi: 10.1016/j.bios.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 108.Yang B, Gong H, Chen C. et al. A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus. Biosens Bioelectron. 2017;87:679–685. doi: 10.1016/j.bios.2016.08.087. [DOI] [PubMed] [Google Scholar]
- 109.Narang J, Singhal C, Malhotra N. et al. Impedimetric genosensor for ultratrace detection of hepatitis B virus DNA in patient samples assisted by zeolites and MWCNT nano-composites. Biosens Bioelectron. 2016;86:566–74. doi: 10.1016/j.bios.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 110.Riedel T, Surman F, Hageneder S. et al. Hepatitis B plasmonic biosensor for the analysis of clinical serum samples. Biosens Bioelectron. 2016;85:272–9. doi: 10.1016/j.bios.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 111.Wang P, Li M, Pei F. et al. An ultrasensitive sandwich-type electrochemical immunosensor based on the signal amplification system of double-deck gold film and thionine unite with platinum nanowire inlaid globular SBA-15 microsphere. Biosens Bioelectron. 2017;91:424–430. doi: 10.1016/j.bios.2016.12.057. [DOI] [PubMed] [Google Scholar]
- 112.Liu HH, Cao X, Yang Y. et al. Array-based nano-amplification technique was applied in detection of hepatitis E virus. J Biochem Mol Biol. 2006;39(3):247–52. doi: 10.5483/bmbrep.2006.39.3.247. [DOI] [PubMed] [Google Scholar]
- 113.Zhou CH, Long YM, Qi BP. et al. A magnetic bead-based bienzymatic electrochemical immunosensor for determination of H9N2 avian influenza virus. Electrochem. Commun. 2013;31:129–32. [Google Scholar]
- 114.Chen L, Zhang X, Zhang C. et al. Dual-color fluorescence and homogeneous immunoassay for the determination of human enterovirus 71. Anal Chem. 2011;83(19):7316–22. doi: 10.1021/ac201129d. [DOI] [PubMed] [Google Scholar]
- 115.Prabowo BA, Wang RY, Secario MK. et al. Rapid detection and quantification of Enterovirus 71 by a portable surface plasmon resonance biosensor. Biosens Bioelectron. 2017;92:186–191. doi: 10.1016/j.bios.2017.01.043. [DOI] [PubMed] [Google Scholar]
- 116.Lazcka O, Del Campo FJ, Muñoz FX. Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron. 2007;22(7):1205–17. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 117.Waswa J, Irudayaraj J, DebRoy C. Direct detection of E. coli O157: H7 in selected food systems by a surface plasmon resonance biosensor. LWT Food Sci. Technol. 2007;40(2):187–92. [Google Scholar]
- 118.Pal S, Alocilja EC, Downes FP. Nanowire labeled direct-charge transfer biosensor for detecting Bacillus species. Biosens Bioelectron. 2007;22(9-10):2329–36. doi: 10.1016/j.bios.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 119.Guntupalli R, Hu J, Lakshmanan RS. et al. A magnetoelastic resonance biosensor immobilized with polyclonal antibody for the detection of Salmonella typhimurium. Biosens Bioelectron. 2007;22(7):1474–9. doi: 10.1016/j.bios.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 120.Tokarskyy O, Marshall DL. Immunosensors for rapid detection of Escherichia coli O157:H7 - perspectives for use in the meat processing industry. Food Microbiol. 2008;25(1):1–12. doi: 10.1016/j.fm.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 121.Attar A, Mandli J, Ennaji MM. et al. Label-free electrochemical impedance detection of rotavirus based on immobilized antibodies on gold sononanoparticles. Electroanalysis. 2016;28(8):1839–46. [Google Scholar]
- 122.Yasuura M, Fujimaki M. Detection of extremely low concentrations of biological substances using near-field illumination. Sci. Rep. 2016;6:1. doi: 10.1038/srep39241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao Z, Zhang J, Xu ML. et al. A rapidly new-typed detection of norovirus based on F0F1-ATPase molecular motor biosensor. Biotechnol. Bioprocess Eng. 2016;21(1):128–133. doi: 10.1007/s12257-015-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee JF, Stovall GM, Ellington AD. Aptamer therapeutics advance. Curr Opin Chem Biol. 2006;10(3):282–9. doi: 10.1016/j.cbpa.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 125.Dong Y, Xu Y, Yong W. et al. Aptamer and its potential applications for food safety. Crit Rev Food Sci Nutr. 2014;54(12):1548–61. doi: 10.1080/10408398.2011.642905. [DOI] [PubMed] [Google Scholar]
- 126.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4(8):588–96. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 127.Kumar PK. Monitoring Intact Viruses Using Aptamers. Biosensors (Basel); 2016. p. 6. (3). pii: E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Duan N, Wu S, Dai S. et al. Advances in aptasensors for the detection of food contaminants. Analyst. 2016;141(13):3942–3961. doi: 10.1039/c6an00952b. [DOI] [PubMed] [Google Scholar]
- 129.van den Kieboom CH, van der Beek SL, Mészáros T. et al. Aptasensors for viral diagnostics. TrAC, Trends Anal. Chem. 2015;74:58–67. doi: 10.1016/j.trac.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Giamberardino A, Labib M, Hassan EM. et al. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS One. 2013;8(11):e79087. doi: 10.1371/journal.pone.0079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xi Z, Huang R, Li Z. et al. Selection of HBsAg-specific DNA aptamers based on carboxylated magnetic nanoparticles and their application in the rapid and simple detection of hepatitis B virus infection. ACS Appl. Mater. Interfaces. 2015;7(21):11215–11223. doi: 10.1021/acsami.5b01180. [DOI] [PubMed] [Google Scholar]
- 132.Escudero-Abarca BI, Suh SH, Moore MD. et al. Selection, characterization and application of nucleic acid aptamers for the capture and detection of human norovirus strains. PLoS One. 2014;9(9):e106805. doi: 10.1371/journal.pone.0106805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moore MD, Escudero-Abarca BI, Suh SH. et al. Generation and characterization of nucleic acid aptamers targeting the capsid P domain of a human norovirus GII.4 strain. J Biotechnol. 2015;209:41–9. doi: 10.1016/j.jbiotec.2015.06.389. [DOI] [PubMed] [Google Scholar]
- 134.Wang N, Kitajima M, Mani K, Miniaturized electrochemical sensor modified with aptamers for rapid norovirus detection. InNano/Micro Engineered and Molecular Systems (NEMS), 2016 IEEE 11th Annual International Conference on; 2016. Apr 17 (pp. 587-590). IEEE. [Google Scholar]
- 135.Zhou CH, Zhao JY, Pang DW. et al. Enzyme-induced metallization as a signal amplification strategy for highly sensitive colorimetric detection of avian influenza virus particles. Anal Chem. 2014;86(5):2752–9. doi: 10.1021/ac404177c. [DOI] [PubMed] [Google Scholar]
- 136.Manea F, Houillon FB, Pasquato L. et al. Nanozymes: gold-nanoparticle-based transphosphorylation catalysts. Angew Chem Int Ed Engl. 2004;43(45):6165–9. doi: 10.1002/anie.200460649. [DOI] [PubMed] [Google Scholar]
- 137.Shin HY, Yoon TY, Kim MI. Recent Advances in Nanozyme Research for Disease Diagnostics. KSBB J. 2015;30(1):1–10. [Google Scholar]
- 138.Wei H, Wang E. Nanomaterials with enzyme-like characteristics (nanozymes): next-generation artificial enzymes. Chem Soc Rev. 2013;42(14):6060–93. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 139.Wang X, Hu Y, Wei H. Nanozymes in bionanotechnology: from sensing to therapeutics and beyond. Inorg. Chem. Front. 2016;3(1):41–60. [Google Scholar]
- 140.Celardo I, Pedersen JZ, Traversa E. et al. Pharmacological potential of cerium oxide nanoparticles. Nanoscale. 2011;3(4):1411–1420. doi: 10.1039/c0nr00875c. [DOI] [PubMed] [Google Scholar]
- 141.Gao L, Yan X. Nanozymes: an emerging field bridging nanotechnology and biology. Sci. China: Life Sci. 2016;59(4):400–402. doi: 10.1007/s11427-016-5044-3. [DOI] [PubMed] [Google Scholar]
- 142.Mu J, Wang Y, Zhao M. et al. Intrinsic peroxidase-like activity and catalase-like activity of Co3O4 nanoparticles. Chem Commun (Camb) 2012;48(19):2540–2. doi: 10.1039/c2cc17013b. [DOI] [PubMed] [Google Scholar]
- 143.Chen W, Chen J, Liu AL. et al. Peroxidase-like activity of cupric oxide nanoparticle. ChemCatChem. 2011;3(7):1151–4. [Google Scholar]
- 144.Wan Y, Qi P, Zhang D. et al. Manganese oxide nanowire-mediated enzyme-linked immunosorbent assay. Biosens Bioelectron. 2012;33(1):69–74. doi: 10.1016/j.bios.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 145.André R, Natálio F, Humanes M. et al. V2O5 nanowires with an intrinsic peroxidase-like activity. Advanced Functional Materials. 2011;21(3):501–9. [Google Scholar]
- 146.Long YJ, Li YF, Liu Y. et al. Visual observation of the mercury-stimulated peroxidase mimetic activity of gold nanoparticles. Chem Commun (Camb) 2011;47(43):11939–41. doi: 10.1039/c1cc14294a. [DOI] [PubMed] [Google Scholar]
- 147.Zhang L, Laug L, Münchgesang W. et al. Reducing stress on cells with apoferritin-encapsulated platinum nanoparticles. Nano Lett. 2010;10(1):219–23. doi: 10.1021/nl903313r. [DOI] [PubMed] [Google Scholar]
- 148.Lien CW, Huang CC, Chang HT. Peroxidase-mimic bismuth-gold nanoparticles for determining the activity of thrombin and drug screening. Chem Commun (Camb) 2012;48(64):7952–4. doi: 10.1039/c2cc32833j. [DOI] [PubMed] [Google Scholar]
- 149.Narayanan KB, Park HH. Pleiotropic functions of antioxidant nanoparticles for longevity and medicine. Adv Colloid Interface Sci. 2013;201-202:30–42. doi: 10.1016/j.cis.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 150.Guo Y, Deng L, Li J. et al. Hemin-graphene hybrid nanosheets with intrinsic peroxidase-like activity for label-free colorimetric detection of single-nucleotide polymorphism. ACS Nano. 2011;5(2):1282–90. doi: 10.1021/nn1029586. [DOI] [PubMed] [Google Scholar]
- 151.Bausch DG. Ebola virus as a foodborne pathogen? Cause for consideration, but not panic. J Infect Dis. 2011;204(2):179–81. doi: 10.1093/infdis/jir201. [DOI] [PubMed] [Google Scholar]
- 152.Duan D, Fan K, Zhang D. et al. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens Bioelectron. 2015;74:134–41. doi: 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 153.Lin X, Liu Y, Tao Z. et al. Nanozyme-based bio-barcode assay for high sensitive and logic-controlled specific detection of multiple DNAs. Biosens Bioelectron. 2017;1:1. doi: 10.1016/j.bios.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 154.Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29(5):240–50. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ozcan A. Mobile phones democratize and cultivate next-generation imaging, diagnostics and measurement tools. Lab Chip. 2014;14(17):3187–94. doi: 10.1039/c4lc00010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Berg B, Cortazar B, Tseng D. et al. Cellphone-based hand-held microplate reader for Point-of-Care testing of Enzyme-linked immunosorbent assays. ACS Nano. 2015;9(8):7857–66. doi: 10.1021/acsnano.5b03203. [DOI] [PubMed] [Google Scholar]
- 157.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507(7491):181–9. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 158.Markets and Markets. Microfluidics Market by Component (Microfluidic Chips, Micropump, Microneedle), Material (Polymer, Glass, Silicon), Application (Genomics, Proteomics, Capillary Electrophoresis, POC, Clinical, Environmental, Drug Delivery) - Global Forecast to 2021. http://www.marketsandmarkets.com/Market-Reports/microfluidics-market-1305.html?gclid=CLbmuYqrx9ICFYF-fgods-gFEA.
- 159.Kuan CM, York RL, Cheng CM. Lignocellulose-based analytical devices: bamboo as an analytical platform for chemical detection. Sci Rep. 2015;5:18570. doi: 10.1038/srep18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Reches M, Mirica KA, Dasgupta R. et al. Thread as a matrix for biomedical assays. ACS Appl Mater Interfaces. 2010;2(6):1722–8. doi: 10.1021/am1002266. [DOI] [PubMed] [Google Scholar]
- 161.Lin SC, Hsu MY, Kuan CM. et al. Cotton-based diagnostic devices. Sci Rep. 2014;4:6976. doi: 10.1038/srep06976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu J, Wang S, Wang L. et al. Advances in paper-based point-of-care diagnostics. Biosens Bioelectron. 2014;54:585–97. doi: 10.1016/j.bios.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 163.Martinez AW, Phillips ST, Butte MJ. et al. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed Engl. 2007;46(8):1318–20. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wei X, Tian T, Jia S. et al. Target-responsive DNA hydrogel mediated "stop-flow" microfluidic paper-based analytic device for rapid, portable and visual detection of multiple targets. Anal Chem. 2015;87(8):4275–82. doi: 10.1021/acs.analchem.5b00532. [DOI] [PubMed] [Google Scholar]
- 165.Han KN, Choi JS, Kwon J. Three-dimensional paper-based slip device for one-step point-of-care testing. Sci Rep. 2016;6:25710. doi: 10.1038/srep25710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu X, Lin TY, Lillehoj PB. Smartphones for cell and biomolecular detection. Ann Biomed Eng. 2014;42(11):2205–17. doi: 10.1007/s10439-014-1055-z. [DOI] [PubMed] [Google Scholar]
- 167.Yang K, Peretz-Soroka H, Liu Y. et al. Novel developments in mobile sensing based on the integration of microfluidic devices and smartphones. Lab Chip. 2016;16(6):943–58. doi: 10.1039/c5lc01524c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Martinez AW, Phillips ST, Carrilho E. et al. Simple telemedicine for developing regions: camera phones and paper-based microfluidic devices for real-time, off-site diagnosis. Anal Chem. 2008;80(10):3699–707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lu Y, Shi W, Qin J. et al. Low cost, portable detection of gold nanoparticle-labeled microfluidic immunoassay with camera cell phone. Electrophoresis. 2009;30(4):579–82. doi: 10.1002/elps.200800586. [DOI] [PubMed] [Google Scholar]
- 170.Shen L, Hagen JA, Papautsky I. Point-of-care colorimetric detection with a smartphone. Lab Chip. 2012;12(21):4240–3. doi: 10.1039/c2lc40741h. [DOI] [PubMed] [Google Scholar]
- 171.US FDA. BAM: Detection and Enumeration of Listeria monocytogenes. https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm071400.htm.
- 172.CFIA. Policy on Listeria monocytogenes in Ready-to-Eat Foods. http://www.hc-sc.gc.ca/fn-an/legislation/pol/policy_listeria_monocytogenes_2011-eng.php.
- 173.Li D, Wang J, Wang R, Li Y, Abi-Ghanem D, Berghman L, Hargis B, Lu H. A nanobeads amplified QCM immunosensor for the detection of avian influenza virus H5N1. Biosens. Bioelectron. 2011;26(10):4146–54. doi: 10.1016/j.bios.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 174.Hjelmsø MH, Hellmér M, Fernandez-Cassi X, Timoneda N, Lukjancenko O, Seidel M, Abril J F. Evaluation of methods for the concentration and extraction of viruses from Sewage in the context of metagenomic sequencing. PloS One. 2017;12(1):e0170199. doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu B, Clifford DA, Chellam S. Comparison of electrocoagulation and chemical coagulation pretreatment for enhanced virus removal using microfiltration membranes. Water Research. 2005;39(13):3098–108. doi: 10.1016/j.watres.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 176.Kwon J, Choi JS, Kim D, Lee HM. Method for the concentration and detection of virus. United States Patent US 9,322,071; 2016. [Google Scholar]
- 177.Summa M, von Bonsdorff CH, Maunula L. Evaluation of four virus recovery methods for detecting noroviruses on fresh lettuce, sliced ham, and frozen raspberries. J. Virologic. Methods. 2012;183(2):154–60. doi: 10.1016/j.jviromet.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 178.Stals A, Baert L, Van Coillie E, Uyttendaele M. Extraction of food-borne viruses from food samples: a review. Int. J. Food Microbio. 2012;153(1):1–9. doi: 10.1016/j.ijfoodmicro.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 179.Butot S, Zuber S, Baert L. Sample preparation prior to molecular amplification: complexities and opportunities. Current Opinion in Virology. 2014;28(4):66–70. doi: 10.1016/j.coviro.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 180.ISO/TS15216-2. Microbiology of food and animal feed - Horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR - Part 2: Method for qualitative detection (2013)
- 181.Goyal SM, Aboubakr HA. Methods for virus recovery from foods; Viruses in Foods 2016; 231-276; Springer International Publishing. [Google Scholar]
- 182.Zourob M, Ripp S. Bacteriophage-Based Biosensors. In: Recognition Receptors in Biosensors; 2009. pp. 415–448. Springer International Publishing. [Google Scholar]
- 183.Park TS, Li W, McCracken KE. et al. Smartphone quantifies Salmonella from paper microfluidics. Lab Chip. 2013;13:4832–4840. doi: 10.1039/c3lc50976a. [DOI] [PubMed] [Google Scholar]