Abstract
An investigation of a questionable species of the genus Alseodaphne led to the discovery of a new genus Alseodaphnopsis H. W. Li & J. Li, gen. nov., separated from Alseodaphne Nees, and a new species Alseodaphnopsis ximengensis H. W. Li & J. Li, sp. nov., endemic to Yunnan province, China. This new species is characterized by having big, axillary, paniculate inflorescences, as well as large, subglobose fruits. Based on DNA sequence data from two gene regions (nuclear ribosomal ITS and LEAFY intron II), we investigate its phylogenetic position within the Persea group. Phylogenies using maximum parsimony (MP) and Bayesian inference (BI) support the recognition of Alseodaphnopsis as a distinct genus but do not resolve well its relationship within the Persea group. The new genus is circumscribed, eight new combinations for its species are made, and a description and illustration of the new species are provided.
Introduction
The Persea group is a subset of the Lauraceae, including seven currently recognized genera, Alseodaphne Nees, Apollonias Nees, Dehaasia Bl., Machilus Rumph. ex Nees, Nothaphoebe Bl., Persea Mill., and Phoebe Nees, containing a total of 400 to 450 species [1–2]. Most of these species are native to tropical and subtropical Asia, whereas ~20% are distributed in warm-temperate to tropical regions of America [1–2]. According to the results of several recent molecular studies, the Persea group is monophyletic [1–5]. Some of them also showed that Persea subg. Eriodaphne, Machilus, Persea subg. Persea, and Phoebe formed well-supported monophyletic groups, and the genus Alseodaphne was not monophyletic [1–2]. However, the relationships of species within and among the Alseodaphne clades are still not well resolved.
In its current circumscription, Alseodaphne consists of fifty species or more, of which about 90% are distributed in tropical Asia, including Cambodia, China, India, Indonesia, Laos, Malaysia, Myanmar, the Philippines, Sri Lanka, Thailand, and Vietnam; ten species (seven endemic) are present in China, including A. andersonii (King ex Hook. f.) Kosterm., A. gracilis Kosterm., A. hainanensis Merr., A. hokouensis H. W. Li, A. huanglianshanensis H. W. Li & Y. M. Shui, A. marlipoensis (H. W. Li) H. W. Li, A. petiolaris (Meisn.) Hook. f., A. rugosa Merr. & Chun, A. sichourensis H. W. Li, and A. yunnanensis Kosterm., distributed in Guangdong, Yunnan, and Hainan [6–7]. Nine of these ten species (except A. hainanensis) are distributed in the northern marginal zone of the tropics in southwestern China.
The delimitation of Alseodaphne Nees historically has been difficult and circumscriptions have been variable since the genus was first described by Nees [8], who incorporated four species, of which A. semecarpifolia Nees only stands. The three other species belong to Phoebe and Litsea Lam. (Lauraceae), and Castanopsis (D. Don) Spach (Fagaceae). Meisn. [9] followed Nees and moved Phoebe excelsa Nees to Alseodaphne. Bentham & Hooker [10] treated Alseodaphne and Nothaphoebe as sections within Persea. Gamble [11] recognized Alseodaphne and Persea as two distinct genera, while Hooker [12] reduced Nothaphoebe to Alseodaphne, and Boerlage [13] moved all Malesian Nothaphoebe to Alseodaphne. Ridley [14], on the other hand, kept the two genera Nothaphoebe and Alseodaphne separate in his The Flora of the Malay Peninsula. Kostermans [15] included Alseodaphne and Nothaphoebe in Persea in his overview of all Lauraceae, but changed his opinion and recognized Alseodaphne and Nothaphoebe as independent genera later [16]. He also thought that these two genera possibly might be fused again in the future, because they are extremely close to each other. van der Werff [17] came to a similar conclusion. He found no significant difference between the two genera and included Nothaphoebe in Alseodaphne, estimating the number of species to about 90. He also expressed doubts about the delimitation between Alseodaphne and Dehaasia. Julia et al. [18], however, recognized Alseodaphne, Dehaasia and Nothaphoebe as distinct genera based on a combination of numerous morphological characters. In the light of the DNA phylogenetic results, Rohwer and Rudolph [5] found that Alseodaphne perakensis (Gamble) Kosterm. and Dehaasia cuneata (Bl.) Bl. formed a strongly supported clade. Rohwer et al. [1] found that the relationship of Nothaphoebe umbelliflora (Bl.) Bl. and some species of Alseodaphne was very close, but Alseodaphne did not appear monophyletic in their study, although with insufficient support. The work of Li et al. [2] showed that Alseodaphne was clearly a polyphyletic group. Its species were placed in two distinct clades, one of which included also Dehaasia (5 spp. examined) and Nothaphoebe umbelliflora. The species of this clade are mainly distributed in tropical Asia. The second clade (Clade III in Li et al., 2011) was poorly resolved at the base and included also the species of Phoebe as well as a few Neotropical species currently placed in Persea. The Alseodaphne species belonging to this clade are mainly distributed in the northern margin of tropics in southwestern China. In our study, we won’t discuss in depth the relationships of species within Alseodaphne, Nothaphoebe, and Dehaasia, as our samples from tropical SE Asia are limited, but aim at the species of Alseodaphne with their main distribution in southwestern China.
We conducted an exploratory trip to Yunnan, China, and collected a questionable plant possibly belonging to Alseodaphne, and this was the reason to study the species from SW China in more detail. In this study, the phylogenetic relationships of the genus Alseodaphne distributed in southwestern China are assessed by using DNA sequence data in addition to morphological data.
Materials and methods
Ethics statement
Collection of these species was conducted in compliance with existing regulations for plants defined as non-commercial, as determined by local government offices. In addition, these sample collections were performed in China with the written approval from the National Forest Bureau and relevant local governments, complying with Chinese and international regulations for the collection of native plant samples.
Morphological observations
Material of the questionable taxon was collected in November 2007, January 2008, August 2015 and November 2015 from the county of Ximeng (22°41′28.36″N, 99°38′59.68″E), Yunnan, China. The morphological description is based on fresh and pressed specimens. Details of the flowers were examined and photographed under a stereomicroscope (ZEISS Discovery V12.0). The morphological comparison with other closely related species is based on study of living plants in the field as well as herbarium specimens, supplemented by information gathered in the relevant literature [6–7,16]. The specimens examined have been deposited in the herbarium of HITBC. Inflorescence size data and fruit size data of the new taxon and known Alseodaphne species were collected from pressed specimens or literature searches, and compared using the independent samples t test. Significance was assumed at p<0.05. All statistical analyses were performed using the SPSS statistical software package 16.0 [19].
Taxon sampling
In the present study, the ingroup sampling included 66 samples (including the new taxon) representing most of the genera within the Persea group (Machilus, Phoebe, Dehaasia, Nothaphoebe, and Alseodaphne). As in the work of Li et al. [2], nine species from four closely related genera (Actiondaphne, Lindera, Litsea, Neolitsea) were selected as the outgroups. Voucher information and GenBank accession numbers are listed in S1 Table.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to IPNI, from where they will be made available to the Global Names Index. The IPNI LSIDs can be resolved and the associated information viewed through any standard web browser by appending the LSID contained in this publication to the prefix http://ipni.org/. The online version of this work is archived and available from the following digital repositories: PubMed Central, LOCKSS.
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from silica-gel dried leaf specimens using the Plant Genomic DNAKit (Tiangen Biotech, Beijing, China). The analyses presented here used the sequence data from two DNA regions, the internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) of the nuclear ribosomal DNA and LEAFY intron II. These regions have been shown to be valuable in phylogenetic studies within the Persea group [2]. The ITS and LEAFY intron II regions were amplified and sequenced following the work of Li et al. [2]. The amplified products of LEAFY intron II were purified using the EZNA Cycle-Pure Kit (Omega Bio-Tek, Georgia, USA) before cloning. Cloning was performed using the pEASY-T3 Cloning Kit (TransGen Biotech, Beijing, China). At least 6 positive clones from each individual sample were sequenced and up to 12 positive clones were sequenced for some samples. The sequence chromatogram output files were assembled and edited using Sequencer 4.5 (GeneCodes, Ann Arbor, Michigan, USA).
Sequence alignment and phylogenetic analyses
DNA sequences were aligned by the program Clustal X 1.81 [20] and edited manually using BioEdit 7.0.9.0 [21]. A single representative sequence was chosen randomly from multiple clones of each individual sample, as all clones from the same individual sample invariably formed a single clade in a preliminary analysis. Individual and combined datasets for the two markers were assembled as ITS, LEAFY intron II, and ITS + LEAFY intron II. Phylogenetic relationships based on the individual and combined datasets were inferred using unweighted maximum parsimony (MP) by the program PAUP*4.0b10 [22], and Bayesian inference (BI) analyses by the program MrBayes 3.1.2 [23–24].
In the MP analyses, a heuristic search was performed with 100 random addition sequence replicates, tree-bisection-reconnection (TBR) branch swapping, collapse of zero length branches, Multrees on and character state changes unordered and equally weighted. Each random addition sequence replicate was allowed to save up to 1000 trees. Bootstrap support values (BS) of the internal nodes were obtained with 100 bootstrap replicates, using the same options as described above.
In the BI analyses, the best-fit model of evolution was chosen for each dataset (ITS and LEAFY intron II) by the program Modeltest 3.7 [25–26] based on the Akaike information criterion (AIC). The Markov chain Monte Carlo (MCMC) algorithm was performed for 2,000,000 generations with one cold and three heated chains, starting from random trees and saving one tree each 100 generations. The first 5000 trees (25%) were discarded as burn-in after checking for stability on the log-likelihood curves, and the remaining 15,000 trees were used to construct the consensus tree. The branch support was determined as Bayesian Posterior Probabilities (BPP).
Results
Comparison of morphological characters
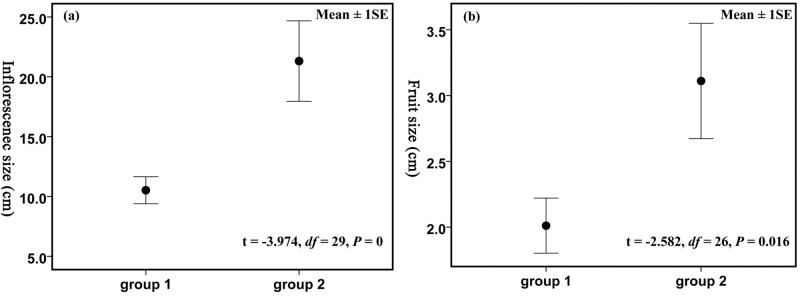
A comparison of morphological characters among the different species of Alseodaphne revealed two groups, consisting of species mainly distributed in tropical Asia (group 1) and species mainly distributed in the northern marginal zone of the tropics in southwestern China (group 2), respectively. Group 1 includes the type species, A. semecarpifolia, whereas the new taxon examined here is placed in group 2. The morphological differences between group 1 species and group 2 species are listed in Table 1. Most of these characters are quantitative and cannot distinguish these two group if considered separately, but in combination, they are effectively separating them. Among the more reliable differences are deciduous perianth lobes in young fruit, not perulate terminal buds as well as whitish twigs contrasting with blackish petioles in dried specimens in group 1 vs. persistent perianth lobes in young fruit, perulate terminal buds, not obviously whitish twigs in group 2. In addition, the error bar charts of the inflorescence size and fruit size between group 1 species and group 2 species showed that both the them were significant (p = 0 and p = 0.016 respectively) (Fig 1). The data of inflorescence size and fruit size are listed in S2 and S3 Tables, respectively.
Table 1. Morphological differences between group 1 and group 2.
| Group 1 | Group 2 | |
|---|---|---|
| Petiole | Thin, 1–1.5 mm | Thick, 2-4mm |
| Twig | Thin, 2.5–4.5 mm; obviously whitish in color | Thick, 4–11 mm; not obviously not whitish in color |
| Terminal bud | Not or rarely perulate | Usually perulate, rarely not perulate |
| Leaf texture | Variable (thinly chartaceous, chartaceous, thinly coriaceous or coriaceous) | Usually coriaceous, rarely chartaceous |
| Midrib upper surface | Raised or sunken | Usually sunken, sometimes flat |
| Inflorescences | Relatively small, 3–20 cm long; few-branched, 1–2 orders; few-flowered | Relatively large, 8.5–35 cm long; many-branched, 3–4 orders; many-flowered |
| Perianth lobes | Deciduous already in young fruit | ± Persistent at least in young fruit |
| Fruit | Small to medium size, 0.7–3.5.cm; some with ribs | Medium to big size, (1.3) 3–5 cm; without ribs |
Fig 1. Error bar charts of the inflorescence size and fruit size between group 1 and group 2.
Phylogenetic analyses
The two DNA loci, ITS and LEAFY intron II, included 600 and 769 aligned position respectively. Modeltest suggested that their evolution was best explained by the TVM + I + G and HKY + G evolutionary models, respectively. The topologies of the consensus trees obtained from the MP and Bayesian analyses, based on different datasets (ITS, LEAFY intron II and ITS + LEAFY intron II), were mostly congruent. Most of their major clades were identical, and only minor variation in the composition and relationships of a few terminal nodes were detected, possibly caused by insufficient phylogenetic signal in the data and insufficient sample size. Moreover, these inconsistencies received only very weak support. Here, only Bayesian consensus trees with bootstrap support (BS) values and posterior probability support (PPS) values are presented for demonstration.
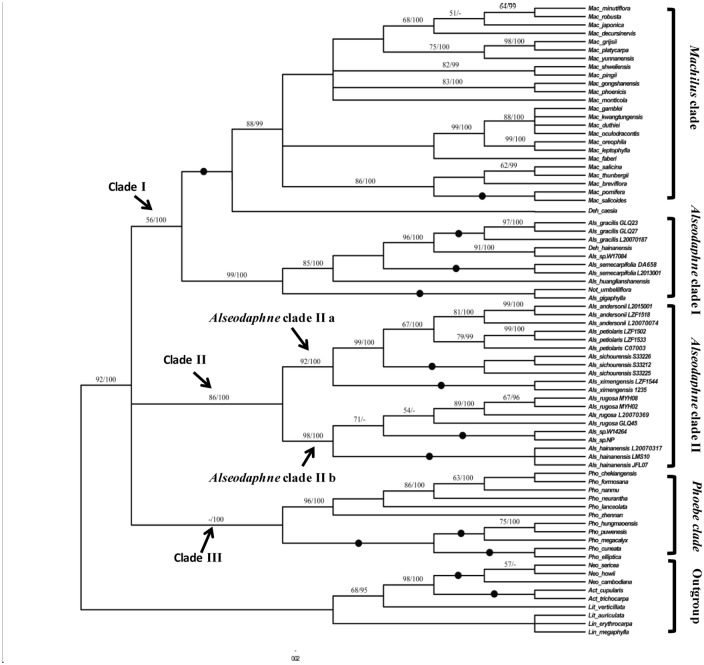
The Bayesian consensus tree obtained from the ITS dataset (S1 Fig) is largely congruent with the work of Li et al. [2]. The two principal clades within the Persea group are (1) the Machilus clade, and (2) a clade including species of Alseodaphne, Dehaasia, Nothaphoebe, and Phoebe. Some major clades in the ITS tree, which also appear in the ITS + LEAFY intron II tree (Fig 2), are labeled for comparison.
Fig 2. Bayesian consensus tree based on ITS + LEAFY intron II combined sequence dataset.
Bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 95%) are shown above branches. ● = both bootstrap value and Bayesian posterior probability 100%.
The Bayesian consensus tree obtained from the LEAFY intron II dataset (S2 Fig) is also largely compatible with the work of Li et al. [2]. The three principal clades within the Persea group are (1) Alseodaphne clade II a, (2) Alseodaphne clade II b+ Phoebe clade, and (3) the Machilus clade + a clade including Alseodaphne, Dehaasia, Nothaphoebe. As in S1 Fig, some major clades are labeled for comparison with the ITS + LEAFY intron II tree.
The Bayesian consensus tree obtained from the ITS+LEAFY intron II dataset (Fig 2) is largely congruent with phylogenies inferred from separate datasets but more resolved and better supported internally. Thus, the ITS + LEAFY intron II tree is used for the following discussion and its topology is described below.
Within the ITS + LEAFY intron II tree, all species (so far investigated) from the Persea group form a well-defined monophyletic clade (92% BS and 100% PPS). We defined three principal clades (clade I, II, and III) within the Persea group. Clade I consists of all Machilus and Dehaasia species included in the present study, and four Alseodaphne species distributed in tropical Asia (A. semecarpifolia and A. gigaphylla) and southwestern China (A. huanglianshanensis, A. gracilis), as well as Nothaphoebe umbelliflora. It receives 56% BS and 100% PPS, and has two principal subclades. The major component of the first principal subclade is the Machilus Clade, which comprises all representatives of Machilus included in the present study. It receives 88% BS and 99% PPS, and shows very little internal resolution. The second principal subclade is Alseodaphne cladeⅠ, which consists of four Alseodaphne species, Dehaasia hainanensis, and Nothaphoebe umbelliflora. The Alseodaphne clade I receives 99% BS and 100% PPS, and shows very good internal resolution. Clade II consists of Alseodaphne species distributed mainly in southwestern China, viz., A. andersonii, A. hainanensis, A. petiolaris, A. rugosa, A. sichourensis and the new species described below, plus two unidentified samples (Alseodaphne sp. NP, Alseodaphne sp. W14264) that also may represent new species. It receives 86% BS, 100% PPS and has two principal subclades, identical with the Alseodaphne clades II a and II b retrieved from the LEAFY intron II analysis. Clade III consist of all Phoebe species in this study and received 100% PPS but no significant BS. Its two principal subclades, however, are strongly supported in the MP as well as in the BI analyses.
Discussion
Machilus clade and Phoebe clade—These two clades have been retrieved in almost identical composition and topology in an earlier analysis [2], so that there is no need to discuss them here again.
Alseodaphne clade I and Alseodaphne clade II—Just as in the work of Li et al. [2], Alseodaphne appears polyphyletic within the Persea group, with at least two different origins. Of all the Alseodaphne species investigated, four species (A. gracilis, A. huanglianshanensis, A. semecarpifolia and A. gigaphylla) and one unidentified sample (Alseodaphne sp. W17084) fall into Alseodaphne clade I, whereas six species (A. andersonii, A. petiolaris, A. sichourensis, A. rugosa, A. hainanensis, and the new taxon described below) and two unidentified samples (Alseodaphne sp. NP, Alseodaphne sp. W14264) are found in an independent clade (Alseodaphne clade II). The Alseodaphne clade I includes the type species, A. semecarpifolia, which is most dry-resistant species in Alseodaphne [16], so that the name Alseodaphne will stay with this clade, which represents the traditionally recognized typical Alseodaphne species distributed mainly in tropical Asia.
The origin of the Alseodaphne clade II species is apparently different. Most of them are from southwestern China, and Alseodaphne sp. W14264 was collected in northern Vietnam, not far from the Chinese border. The earliest fossil of Alseodaphne was found in Changchang Basin of Hainan Island (China), and the extinct Alseodaphne changchangensis is closest to the living species A. hainanensis [27]. Among the extant species, A. hainanensis and A. rugosa occur in Hainan, and both are members of Alseodaphne clade II. Alseodaphne hainanensis also has been reported from northern Vietnam [7]. Hainan Island belongs to the same phytogeographical region as tropical southern China, and it may have been connected to northern Vietnam and Guangxi at least in the Eocene [28]. We also find that the morphological characters within the independent Alseodaphne clade II are mainly consistent with the molecular results. This clade is divided into two subclades receiving 92% BS, 100% PPS and 98% BS, 100% PPS respectively (Fig 2). In most of the species of Alseodaphne clade II b the lower surfaces of the leaves are distinctly glaucous (except Alseodaphne sp. NP), while all of the species of Alseodaphne clade II a have completely green lower leaf surfaces.
The fact that this independent Alseodaphne clade II differs also morphologically from the traditionally recognized Alseodaphne species has already been discussed by Rohwer et al. [1]. In this study, we also find some differences between these two clades (Table 1). We therefore think that the independent Alseodaphne clade II should be recognized as a new genus, which we call Alseodaphnopsis. A formal description of the new genus is provided below. The vegetative characters may be insufficient to distinguish the two genera independently, as most of them are quantitative characters, but in combination, they can be used to segregate the two genera. The principal characters to distinguish the two genera include: 1) twigs thick, 4–11 mm in diameter, not obviously whitish in color vs. thin, 2.5–4.5 mm in diameter, and obviously whitish in color; 2) terminal buds perulate vs. not perulate; 3) perianth lobes persistent at least in young fruit vs. early deciduous; 4) inflorescences relatively large, 8.5–35 cm long, generally many-flowered, with 3–4 order of branching vs. 3–20 cm long, few-flowered, with 1–2 orders of branching; and 5) mature fruit relatively large, 3–5 cm or < 2.5cm in diameter. We found that twigs are not obviously whitish in color and the terminal buds are perulate in almost all species of the new genus Alseodaphnopsis, whereas the twigs are obviously whitish in color and terminal buds are not perulate in most of the traditional Alseodaphne species. This may be an adaptation to the more seasonal climate in the area of distribution of Alseodaphnopsis, with colder winters than in the area of distribution of most Alseodaphne species. In addition, the perianth lobes are persistent in young fruit period in at least some species of Alseodaphnopsis. We have observed this character in A. andersonii, A. petiolaris and A. hainanensis, but not in the type species A. semecarpifolia, possibly also caused by the difference of climate. Although the size of panicles and fruits look as quantitative ones, perulate terminal buds as well as persistent perianth lobes look as more or less qualitative ones, these qualitative characters are better to be called as the key characters to define Alseodaphne clade II. Alseodaphne gracilis and A. huanglianshanensis, which are distributed in northern tropical Asia, are nested in Alseodaphne clade I, not in Alseodaphne clade II, as most of the other species from tropical China. Also morphologically, they are more similar to A. semecarpifolia, which is likewise located in Alseodaphne clade I, than to most other Chinese species. We have observed that A. gracilis has no perianth lobes in its young fruit, which matches with its placement in traditional Alseodaphne. In addition, A. gracilis, A. huanglianshanensis and A. semecarpifolia possess thinly leathery leaves and thin twigs with conspicuously whitish bark, whereas the Alseodaphnopsis species have leathery leaves and thick twigs that are green or brown, but not whitish in fresh material.
Based on both morphological and molecular evidence, we therefore propose a new genus Alseodaphnopsis, separated from the traditional genus Alseodaphne, to accommodate the independent Alseodaphne clade II including most of the species distributed in the northern margin of tropics in southwestern China (including the new species, Alseodaphnopsis ximengensis). In Yunnan (China), at least two further Alseodaphne species (A. hokouensis H. W. Li, A. marlipoensis (H. W. Li) H. W. Li) belongs to this clade, judged from morphology, so that we can expect this group to become larger with increasing taxon sampling.
Taxonomic treatment
1. New genus
Alseodaphnopsis H. W. Li & J. Li, gen. nov. [urn:lsid:ipni.org:names:77165677–1] Type: Alseodaphnopsis petiolaris (Meisn.) H. W. Li & J. Li (Nothaphoebe petiolaris Meisn., here designated)
Diagnosis: The new genus Alseodaphnopsis H. W. Li et J. Li is obviously very close to the genus Alseodaphne Nees (s. str.), but differs from the latter morphologically by 1) twigs thick, 4–11 mm in diam., and not obviously whitish in color; 2) terminal buds usually perulate; 3) perianth lobes ± persistent at least in young fruit; 4) inflorescences relatively large, 8.5–35 cm long, many-flowered, with 3–4 orders of branching; 5) fruits medium to large size (3–5 cm), without ribs.
Description: Trees evergreen. Terminal buds perulate. Twigs robust, 4–10 mm in diam., not whitish. Leaves alternate, pinninerved, always clustered at the ends of the branchlets, abaxial side glaucous or not. Inflorescence axillary, paniculate, bracts and bracteoles deciduous. Flowers bisexual, trimerous. Receptacle short; perianth lobes 6, subequal or extremely unequal, slightly dilated after anthesis and ± persistent at least in young fruit. Fertile stamens 9, in 3 whorls; filaments of 1st and 2nd whorls glandless, those of 3rd whorl each with 2 glands at base; anthers 4-locular; locules of 1st and 2nd whorls introrse, those of 3rd whorl extrorse or upper locules lateral and lower extrorse. Staminodes 3, of innermost whorl, small, clavate to nearly sagittate. Ovary partly immersed into shallow receptacle; style shorter than ovary; stigma discoid. Fruit medium to big size, 3–5 cm in diam., without ribs, black or purplish black when mature, oblong or subglobose; fruit stalk slightly enlarged or much enlarged, red, green, or yellow, nearly cylindric or obconical, fleshy or somewhat woody, always warty.
Etymology: Alseodaphnopsis alludes to the resemblance to traditional Alseodaphne (s. str.)
Distribution and habitat: Alseodaphnopsis includes nine species, mainly distributed in the northern marginal part of the tropical zone in southwestern China, but extending also to NE India, Laos, Myanmar, Thailand and Vietnam. As far as it is known, the species grow preferentially in forests on limestone mountains.
2. New combinations
Here, we make eight new combinations for the species in this new genus as follows:
1) Alseodaphnopsis andersonii (King ex Hook. f.) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165678–1] Type: Assam, fl., Jenkins s.n, (BO!, CAL, K)
Basionym: Cryptocarya andersonii King ex Hook. f., Fl. Brit. India. 5: 120. 1886 ≡ A. andersonii (King ex Hook. f.) Kosterm., Reinwardtia 6 (2): 159. 1962.
= Alseodaphne keenanii Gamble, Kew Bull. 1914: 188.
Distributed in China (SE & S Yunnan, SE Xizang); NE India, Laos, Myanmar, Thailand and Vietnam.
2) Alseodaphnopsis petiolaris (Meisn.) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165679–1] Type: Assam, Nuka Hills, Simons s.n. (BO!, CAL, K)
Basionym: Nothaphoebe petiolaris Meisn. in A. Candolle, Prodr. 15(1): 59. 1864.
≡ Alseodaphne petiolaris (Meisn.) Hook. f., Fl. Brit. India 5: 145. 1886; Persea petiolaris (Meisn.) Debarman, Bull. Bot. Surv. India 32: 257. 1962.
Distributed in China (S Yunnan); India and Myanmar.
3) Alseodaphnopsis sichourensis (H. W. Li) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165680–1] Type: China. Yunnan: Sichour, Tsaokuoshan, Wen-shan Exp. 61–077 (KUN!)
Basionym: Alseodaphne sichourensis H. W. Li, Act. Phytotax. Sin. 17 (2): 70. 1979.
Distributed in China (SE Yunnan).
4) Alseodaphnopsis marlipoensis (H. W. Li) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165681–1] Type: China. Yunnan: Marlipo, Tianbao Farm, S.C. Wang 81 (KUN!)
Basionym: Cinnamomum marlipoensis H. W. Li, Act. Phytotax. Sin. 13 (4): 48. 1975.
≡ Alseodaphne marlipoensis (H. W. Li) H. W. Li, Act. Phytotax. Sin. 17 (2): 71. 1979.
Distributed in China (SE Yunnan).
5) Alseodaphnopsis rugosa (Merr. & Chun) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165682–1] Type: China. Hainan: Chun & Tso 44254 (A!)
Basionym: Alseodaphne rugosa Merr. & Chun, Sunyatsentia 2: 232. 1935.
Distributed in China (Hainan, SE Yunnan).
6) Alseodaphnopsis hainanensis (Merr.) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165683–1] Type: China. Hainan: Tsang & Fung 766 = L.U.18300 (A!, BO!, DD, K, L)
Basionym: Alseodaphne hainanensis Merr., Lingnan Sci. J. 13: 57. 1934.
Distributed in China (Hainan); N Vietnam.
7) Alseodaphnopsis hokouensis (H. W. Li) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165684–1] Type: China. Yunnan: Hokou, K.H. Tsai 1039 (KUN!)
Basionym: Alseodaphne hokouensis H. W. Li, Act Phytotax. Sin. 17 (2): 71. 1979.
Distributed in China (SE Yunnan).
8) Alseodaphnopsis lanuginosa (Kosterm.) H. W. Li & J. Li, comb. nov. [urn:lsid:ipni.org:names:77165685–1] Type: Vietnam. Tonkin: Chapa, Petelot 3565(BO!, P)
Basionym: Alseodaphne lanuginosa Kosterm., Candollea 28: 116. 1973.
Distributed in N Vietnam.
Note: Four additional species from SW China and N Vietnam, A. hokouensis, A. yunnanensis, A. marlipoensis and A. lanuginosa are not contained in our phylogenetic analyses, but according to the morphological characters, A. hokouensis, A. marlipoensis and A. lanuginosa are similar to A. petiolaris, A. sichourensis and A. andersonii, which belong to Alseodaphnopsis [16, 29], while A. yunnanensis is similar to A. huanglianshanensis which belongs to Alseodaphne [30]. We therefore treat A. hokouensis, A. marlipoensis and A. lanuginosa in Alseodaphnopsis while A. yunnanensis is retained in Alseodaphne.
3. New species
Alseodaphnopsis ximengensis H. W. Li & J. Li, sp. nov. (Figs 3 and 4) [urn:lsid:ipni.org:names:77165686–1] Type: China. Yunnan Province: Pu’er City, Ximeng County, ca. 1300 m altitude, 22°41′28.36″N, 99°38′59.68″E, in seasonal rain forest, 20 November 2011, J. W. Li 1235 (fl.) (Holotype, Isotypes: HITBC!).
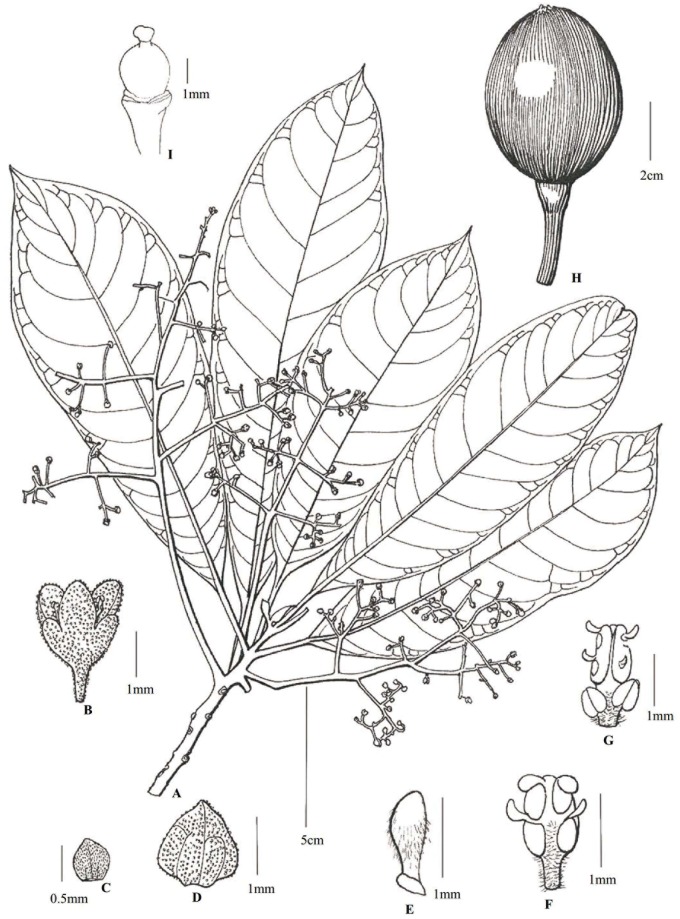
Fig 3. Alseodaphnopsis ximengensis H. W. Li & J. Li sp. nov.
A. Flowering branch; B. Flower, lateral view; C. Outer perianth lobes, outside view; D. Inner perianth lobes, inside view; E. A staminode; F. A fertile stamen of the 1st or 2nd whorl; G. A stamen of the third whorl; H. Fruit; I. Pistil. (drawn by L. Wang based on J. W. Li 1235 sampled from Ximeng County, Yunnan).
Fig 4. Alseodaphnopsis ximengensis H. W. Li & J. Li sp. nov.
A. Branchlet with inflorescences; B. Branchlet with immature infructescences; C. Branchlet with mature fruits; D-F. Mature fruits; G-H. Flowers. (Photos by J.W. Li).
Diagnosis: This new species shows a superficial similarity to Alseodaphnopsis petiolaris (Meisn.) H. W. Li & J. Li in its big leaves and elongated petioles, but differs by its glabrous twigs, leaves and panicles as well as subglobose big fruit.
Description: Trees evergreen. One-year-old branchlets robust, 8–11 mm in diam., yellowish-brown, glabrous, with elevated orbicular lenticels and large suborbicular leaf scars; current year branchlets slender, elongate, terete, 4–6 mm in diam., glabrous, green when young but all brown when dry. Terminal buds large, ca.1 cm, glabrous. Leaves clustered at apex of branchlet; petiole 2–4 mm thick, 2.5–4 cm long, concave-convex; leaf blade greenish on both surfaces, red-brown when young, oblong-oblanceolate, 17–30 × 6–11 cm, leathery, glabrous on both surfaces, midrib conspicuously elevated abaxially, impressed adaxially, lateral veins 13–17 pairs, elevated on both surfaces, arcuately connected at ends, base cuneate, apex acute to obtuse with a short acumen of 5–7 mm. Panicle axillary, glabrous, 20–30 cm long, with 6–14 lateral branches and 3–4 orders of branching; peduncle 3–7.5 cm. Flowers small, ca. 2 mm long; pedicels slender, 2–4 mm, dilated in fruit. Perianth lobes 6, broadly ovate, white pubescent on margin; outer ones smaller, ca. 0.5×0.5 mm, inner ones larger, ca. 1.5×1.5 mm, deciduous in mature fruit. Fertile stamens 9, ca. 1.5 mm in 1st whorl, ca. 1.7 mm in 2nd whorl, ca. 2 mm in 3rd whorl; filaments villous, very short, those of 3rd whorl each with 2 stalkless glands at base, others glandless; anthers of 1st and 2nd whorls elliptic, with 2 upper slightly smaller locules and 2 lower large locules, locules all introrse, anthers of 3rd whorl oblong, with 2 upper smaller locules and 2 lower large locules, locules all latrorse-extrorse. Ovary globose, ca. 2 mm long, glabrous, style short; stigma conspicuous. Fruit large, subglobose, green when young but brown or black when dry, ca. 4.7 cm in diam.; fruit stalk robust, 9–16 (29) cm long, dilated at the tip, up to 8 mm in diam. Fl. November, fr. July-August of next year.
Additional specimens examined (paratypes): China. Yunnan Province: Pu’er City, Ximeng County, ca. 1300 m altitude, in seasonal rain forest, 25 January 2011, J. W. Li 283 (fr.) (HITBC!, same tree with the holotype J. W. Li 1235); China. Yunnan Province: Pu’er City, Ximeng County, 10 November 1985, collector: Y. Y. Qian (No collecting number) (HITBC!, HITBC No. 110940, (fl.)).
Supporting information
Sequences starting with AY, FM, FJ, and HQ come from work of Li et al. (unpublished), Rohwer et al. (2009), Chen et al (2009), and Li et al. (2011) respectively.
(DOCX)
The data of inflorescence are gathered in the relevant literatures below the table, except for A. ximengensis H. W. Li et J. Li and A. sp. NP.
(DOCX)
The data of fruit are gathered in the relevant literatures below the table, except for A. ximengensis H. W. Li et J. Li and A. sp. NP.
(DOCX)
Bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 95%) are shown above branches. ● = both bootstrap value and Bayesian posterior probability 100%.
(PDF)
Bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 95%) are shown above branches. ● = both bootstrap value and Bayesian posterior probability 100%.
(PDF)
Acknowledgments
The authors would like to thank the Center for Integrative Conservation. We are grateful to Zhi-fang Liu, Jian-hua Xiao for collection assistance and the Plant Phylogenetics & Conservation Group members for previous data accumulation. Special thanks go to Deby Arifiani, Henk van der Werff and Yu Song for their kindly help with leaf materials, and to Bing Liu, Yong Yang along with two anonymous reviewers for their valuable comments on the manuscript. This work is funded by Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (Y4ZK111B01).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is funded by Southeast Asia Biodiversity Research Institute, Chinese Academy of Science(Y4ZK111B01).
References
- 1.Rohwer JG, Li J, Rudolph B, Schmidt SA, van der Werff H & Li HW. Is Persea (Lauraceae) monophyletic? evidence from nuclear ribosomal ITS sequences. Taxon, 2009; 58, 1153–1167. [Google Scholar]
- 2.Li L, Li J, Rohwer JG, van der Werff H, Wang ZH & Li HW. Molecular phylogenetic analysis of the Persea group (Lauraceae) and its biogeographic implications on the evolution of tropical and subtropical Amphi-Pacific disjunctions. American Journal of Botany, 2011; 98, 1520–1536. doi: 10.3732/ajb.1100006 [DOI] [PubMed] [Google Scholar]
- 3.Rohwer JG. Toward a phylogenetic classification of the Lauraceae: evidence from matK sequences. Systematic Botany, 2000; 25, 60–71. [Google Scholar]
- 4.Chanderbali AS, van der Werff H & Renner SS. Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Annals of the Missouri Botanical Garden, 2001; 88, 104–134. [Google Scholar]
- 5.Rohwer JG & Rudolph B. Jumping genera: the phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Annals of the Missouri Botanical Garden, 2005; 92, 153–178. [Google Scholar]
- 6.Li HW, PAI PY, LEE SK, WEI FN, Yang YC, Huang PH, et al. Lauraceae In: Li HW (Ed.), Flora Reipublicae Popularis Sinicae, vol. 31 Science Press, Beijing, China; 1982. [Google Scholar]
- 7.Li HW, Li J, Huang PH, Wei FN & van der Werff H. Lauraceae In: Wu ZY, Raven PH, Hong DY (Eds.), Flora of China, vol. 7 Science Press and Missouri Botanical Garden Press, Beijing, China, St. Louis, Missouri, USA; 2008. [Google Scholar]
- 8.Nees von Esenbeck CG. Lauraceae. Plantae Asiaticae Rariores, 1831 [Google Scholar]
- 9.Meissner C. Lauraceae. Prodromus Systematis Naturalis Regni Vegetabilis, 1864; 15, 1–260. [Google Scholar]
- 10.Bentham G & Hooker JD. Laurineae. Genera plantarum, 1880; 3, 146–165. [Google Scholar]
- 11.Gamble JS. Lauraceae: Materials for a flora of the Malayan Peninsula. Journal of the Asiatic Society of Bengal, 1912; 75, 35–202. [Google Scholar]
- 12.Hooker JD. Laurineae. Flora of British India, 1886; 5, 116–189. [Google Scholar]
- 13.Boerlage, JG. Handleiding tot de Kennis der Flora Nederlandsch Indië; 1900.
- 14.Ridley, HN. The Flora of The Malay Peninsula, Vol. V. 1925.
- 15.Kostermans AJGH. Lauraceae. Reinwardtia, 1957; 4, 275–277. [Google Scholar]
- 16.Kostermans AJGH. A synopsis of Alseodaphne Nees (Lauraceae). Candollea, 1973; 28, 93–136. [Google Scholar]
- 17.van der Werff H. An annotated key to the genera of Lauraceae in the Flora Malesiana region. Blumea, 2001; 46, 125–140. [Google Scholar]
- 18.Julia S, Soepadmo E & Yahud W. Problem in the generic delimitation between Alseodaphne, Dehaasia and Nothaphoebe (Lauraceae) in Borneo. Blumea, 2009; 54, 192–197. [Google Scholar]
- 19.Released, SI. SPSS for Windows, Version 16.0; 2007.
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F & Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 1997; 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series, 1999; 95–98.
- 22.Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland, Massachusetts, USA; 2003. [Google Scholar]
- 23.Huelsenbeck JP & Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics, 2001; 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F & Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 2003; 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- 25.Posada D & Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics, 1998; 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 26.Posada D & Buckley TR. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology, 2004; 53, 793–808. doi: 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 27.Li J, Qiu J, Liao W & Jin J. Eocene fossil Alseodaphne from Hainan Island of China and its paleoclimatic implications. Science in China Series D: Earth Sciences, 2009; 52, 1537–1542. [Google Scholar]
- 28.Zhu H. Biogeographical Evidences Help Revealing the Origin of Hainan Island. Plos One, 2016; 11 (4), e0151941 doi: 10.1371/journal.pone.0151941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SK, Wei FN, Wei YT & Li HW. Materiae ad floram lauracearum sinicarum (III). Acta Phytotaxonomica Sinica, 1979; 17, 45–74. [Google Scholar]
- 30.Li HW, Shui YM. Alseodaphne huanlianshanensis, a new species of the Lauraceae from Yunnan, China. Acta Phytotaxonomica Sinica, 2004; 42, 551–554. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences starting with AY, FM, FJ, and HQ come from work of Li et al. (unpublished), Rohwer et al. (2009), Chen et al (2009), and Li et al. (2011) respectively.
(DOCX)
The data of inflorescence are gathered in the relevant literatures below the table, except for A. ximengensis H. W. Li et J. Li and A. sp. NP.
(DOCX)
The data of fruit are gathered in the relevant literatures below the table, except for A. ximengensis H. W. Li et J. Li and A. sp. NP.
(DOCX)
Bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 95%) are shown above branches. ● = both bootstrap value and Bayesian posterior probability 100%.
(PDF)
Bootstrap values (≥ 50%) / Bayesian posterior probabilities (≥ 95%) are shown above branches. ● = both bootstrap value and Bayesian posterior probability 100%.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.