Abstract
MicroRNAs (miRs) have been demonstrated to be important in the establishment and progression of colon cancer. However the underlying molecular mechanisms remain to be fully elucidated. Polypeptide N-acetylgalactosaminyltransferase4 (GALNT4) participates in numerous cellular processes, including tumorigenesis. The present study used reverse transcription-quantitative polymerase chain reaction and western blotting to investigate the expression levels of miR-4262 and GALNT4 in tissues and cells. In addition, MTS and colony formation assays, and cell cycle analysis were performed to evaluate the effect of miR-4262 on cell proliferation and the cell cycle. The findings demonstrated that miR-4262 was a direct target of GALNT4 mRNA. Overexpression of miR-4262 was demonstrated to decrease GALNT4 mRNA and protein expression levels, and thereby suppressed cell viability, growth and cell-cycle progression in SW480 and SW620 colon cancer cells. In addition, knockdown of miR-4262 significantly increased the cell viability, growth, and cell-cycle progression of SW480 and SW620 cells. The expression level of miR-4262 was observed to be downregulated as the expression of GALNT4 was upregulated in colon cancer tissues and cell lines. In conclusion, the results demonstrated that miR-4262 may be involved in the development of colon cancer via targeting of GALNT4. The miR-4262/GALNT4 axis may be a novel target for diagnosing and understanding the underlying molecular mechanism of colon cancer.
Keywords: microRNA-4262, polypeptide N-acetyl galactosaminyl-transferase 4, colon cancer, SW480, SW620
Introduction
Colon cancer is the third most common malignancy and a leading cause of cancer-associated deaths worldwide (1). A total of 1.2 million cases are diagnosed each year and ~0.6 million deaths occur annually worldwide (2). The 5 year survival rate ranges from 40–60% (3). The high incidence and poor prognosis of colon cancer suggests that elucidation of the underlying mechanism is a primary concern, in order to improve diagnostic methods and therapies.
Polypeptide N-acetylgalactosaminyltransferase4 (GALNT4), is a member of the mucin gene family, which belongs to the family of N-acetylgalactosaminyl transferases, catalyzing the transfer of GalNAc to serine or threonine residues in the initial step of mucin-type O-linked protein glycosylation. This type of glycosylation is the most complex post-translational modification of proteins, important during cellular proliferation, differentiation and numerous pathological disorders (4). It has been previously demonstrated that GALNT was deregulated in colorectal cancer and various neoplasms (5).
MicroRNAs (miRs) are non-coding small RNAs that have established functions in the regulation of protein translation by base-pairing with the 3′-untranslated region (3′-UTR) of the target mRNAs (6,7). It has previously been suggested that miRNAs regulate carcinogenesis (8,9). To date, few investigations have been performed into the effects of miR-4262. A recent study reported that miR-4262 mediates the effects of angiotensin-converting enzyme 2 on the induction of apoptosis of pulmonary endothelial cells during acute lung injury (10). miR-4262 was also demonstrated to inhibit the translation of osteopontin, (OPN), which is a phosphorylated glycoprotein and an important mediator of the invasion of osteosarcoma, by directly targeting the 3′UTR of the OPN mRNA(10). As microarray data from our laboratory (data not shown) suggested that the expression levels of miR-4262 were reduced in colon cancer, the present study aimed to investigate the role of miR-4262 in this cancer type.
The present study demonstrated that miR-4262 was downregulated in colon cancer tissues and cell lines. Restoration of miR-4262 expression reduced cell viability and growth in vitro, whereas knockdown of miR-4262 increased cell viability and growth. In addition, GALNT4 was identified as a direct target of miR-4262 using a 3′UTR luciferase assay. Furthermore, it was observed that GALNT4 was upregulated in colon cancer tissues and cells. The results of the present study, therefore, suggested that miR-4262 acts as a tumor suppressor via regulation of GALNT4 expression and may thus be a promising therapeutic target for the treatment of colon cancer.
Materials and methods
Human colon cancer tissues and cell culture
A total of 20 paired fresh surgically resected osteosarcoma tumor tissues and adjacent non-tumor tissues, diagnosed by an independent pathologist, were collected from the People's Hospital of Weifang (Weifang, China) between November 2013 and November 2015. A total of 10 male and 10 female patients with a median age of 37.6 years were included. Patients did not receive chemotherapy or radiotherapy prior to surgery. Specimens were freshly frozen in liquid nitrogen immediately following collection and stored at −80°C until use. The tumor and non-tumor samples were confirmed by pathological examination. All samples were obtained with written informed consent and the investigation was approved by the Research Ethics Committee of the People's Hospital of Weifang.
The human HT-29, HCT116, SW480, SW620 and LoVo colon cancer cell lines, and the FHC and NCM460 normal colonic epithelial cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in RPMI-1640 medium (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Ambion; Thermo Fisher Scientific, Inc.) and 2 mM L-Glutamine in a humidified incubator at 37°C, in an atmosphere containing 5% CO2.
RNA oligonucleotides, plasmids and transfection
The miR-4262 mimic (catalog no. HMI1216; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), mimic control (ctrl; catalog no. HMI0003; Sigma-Aldrich; Merck KGaA), miR-4262 inhibitor (catalog no. HSTUD1216; Sigma-Aldrich; Merck KGaA) and negative control (NC; catalog no. HMC0002; Sigma-Aldrich; Merck KGaA) were utilized at a final concentration of 100 nM. Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for the transfection when the cells were grown to 70–80% confluence. The medium was replaced with fresh medium supplemented with 10% fetal bovine serum, 4 h following transfection.
Luciferase reporter gene assay
The 3′UTR of GALNT4 in the pMir-GLO reporter vector was purchased from Ambion; Thermo Fisher Scientific, Inc. (catalog no. AM5795). The miR-4262 binding site was mutated using QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA, USA), according to the manufacturer's protocol. Wildtype (GALNT4 3′UTR wt) and mutant constructs (GALNT4 3′UTR mut) were transfected into SW480 and SW620 cells concurrently with miR-4262 mimic or mimics ctrl. Cells were analyzed for relative luciferase activity using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA), 48 h following transfection. The experiment was conducted in triplicate.
MTS assay
Cell viability was measured using a CellTiter Aqueous assay with MTS tetrazolium (Promega Corporation), in which viable cells convert MTS tetrazolium into a formazan-colored product at an absorbance of 490 nm. A total of 2.5×103 cells/well were seeded in a 96-well plate and incubated for 24 h prior to attachment to the bottom of the well. The cell growth was then measured at 48 and 96 h after the cellular adhesion. Following the manufacturer's protocol, 20 µl MTS solution was added to 100 µl of culture media and incubated for 1 h at 37°C and the absorbance was measured at a wavelength of 490 nm. Five independent experiments were performed in quadruplicate.
Colony formation assay
SW480 and SW620 cells (200 cells per well) were seeded in 6-well plates. Cells were transfected with 100 nM miR-4262 mimic, 100 nM miR-4262 inhibitor and controls using Lipofectamine 2000. The medium was replaced with fresh media and incubated for 7–10 days, 48 h after transfection, at 37°C. When colonies became visible cells were stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) in PBS for 15 min at room temperature, and 10 cells were counted in five different fields of view were selected and visualized under light microscopy. The colony formation rate was measured as follows: cell colony number/seeded cell number ×100%. Experiments were performed at least three times.
Cell cycle analysis
Transfected SW480 and SW620 cells (~1×106 cells) were seeded into 6-well plates for 24 h in complete medium prior to cell deprivation of serum for 48 h and subsequently returned to complete medium for an additional 24 h. All cells were collected via centrifugation at room temperature at 400 × g for 10 mins, fixed in 75% ethanol, incubated at −20°C for 16 h, washed twice in cold PBS and finally stained with propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.). Following a final incubation on ice for 30 min, cells were analyzed using a FACS Calibur flow cytometer and CellQuest™ software version 3.0 (Beckman Coulter, Inc., Brea, CA, USA). A total of 10,000 events were counted for each sample.
Western blot analysis
Western blotting was performed according to the standard methods aspreviously described (11) using a rabbit anti-GALNT4 primary antibody (1:200; catalog no. ab151168; Abcam, Cambridge, MA, USA) or a rabbit anti-actin primary antibody (1:5,000; catalog no. A2066; Sigma-Aldrich; Merck Millipore), which served as a control. Membranes were subsequently probed with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:10,000; catalog no. ab6721; Abcam). Experiments were repeated three times.
RNA preparation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was obtained via reverse transcription using M-MLV (Promega Corporation) from 1 µg of obtained RNA. The level of miR-4262 was detected by stem-loop RT-qPCR with the following thermos cycling conditions: denaturing of DNA at 94°C for 4 min, followed by 40 cycles of 94°C for 60 sec, 58°C for 60 sec and 72°C for 60 sec. Primers were as follows: forward, 5′-TGCGGGACATTCAGA-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′ for miR-4262; forward, 5′-AAAGATTGCTGGGTCACACC-3′ and reverse, 5′-TGTCAAAAGGGAAATGAGGC-3′ for GALNT4; forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′ for U6; forward, 5′-CCACCTTCGATGCCGGGGCT-3′ and reverse, 5′-GGGGCCGAGTTGGGATAGGG-3′ for GAPDH. U6 snRNA served as an endogenous control. qPCR was performed on an ABI7500 thermo cycler Applied Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix ExTaq (Perfect Real Time) kits (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's protocol. The expression levels of miR-4262 and GALNT4 were normalized to that of the internal control U6 or GAPDH using the comparative delta Cq (2−ΔΔCq) method (12). Each sample was analyzed in triplicate and the mean expression level was calculated.
Statistical analysis
Statistical analyses were performed using SPSS software version 11.5 (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. Data was analyzed by a one-way analysis of variance followed by the Schefffe post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-4262 expression is downregulated in colon cancer tissues and cells
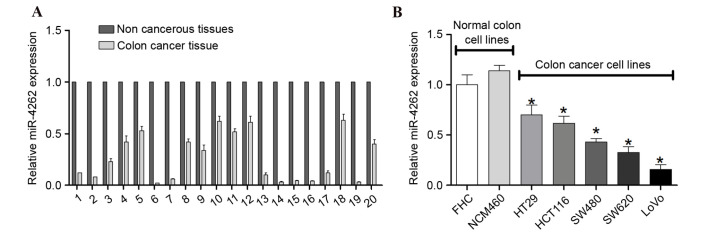
To investigate the role of miR-4262 in colon cancer, the expression levels of miR-4262 in 20 pairs of colon cancer tissues and adjacent non-tumor tissues were investigated. RT-qPCR revealed that miR-4262 was downregulated in colon cancer tissues compared with non-tumor tissues (Fig. 1A). Furthermore, the expression of miR-4262 in different human colon cancer cell lines was examined with RT-qPCR, which demonstrated that miR-4262 was additionally downregulated in HT29, HCT116, SW480, SW620 and LoVo colon cancer cell lines, compared with normal colon cell lines (P<0.05; Fig. 1B). These results suggested that miR-4262 exhibits anti-tumor properties during the development of colon cancer.
Figure 1.
Expression level of miR-4262 in colon cancer tissues and cell lines. Expression of miR-4262 in (A) 20 pairs of colon cancer and non-tumor tissues and (B) colon cancer cell lines and normal colon cell lines was assessed by reverse transcription-quantitative polymerase chain reaction, and normalized to small nuclear RNA U6 expression. Experiments were performed in triplicate. *P<0.05 vs. FHC. miR, microRNA.
Restoration of miR-4262 expression inhibits colon cancer cell viability, growth and cell cycle progression
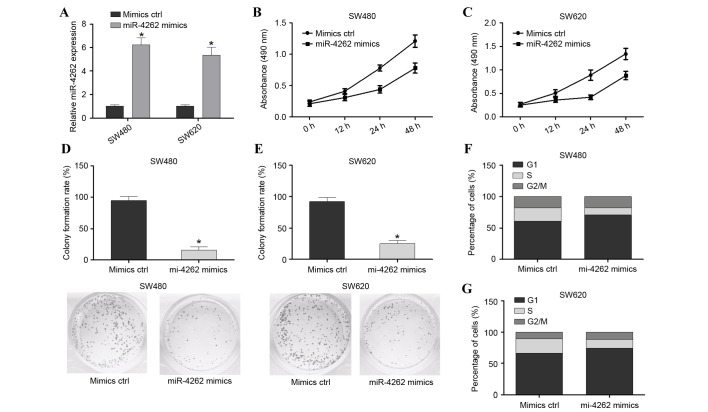
In order to investigate the effect of miR-4262 on colon cancer cells, SW480 and SW620 cell lines were transfected with mimic ctrl or miR-4262 mimics. Cells were harvested after 48 h and successful overexpression of miR-4262 was confirmed by RT-qPCR (P<0.05; Fig. 2A). The MTS assay revealed that the viability of SW480 and SW620 cells was significantly decreased in the miR-4262 mimic group compared with the mimic ctrl (Fig. 2B and C, respectively). The colony formation assay demonstrated that inhibition of miR-4262 significantly enhanced the colony formation rate (P<0.05; Fig. 2D). Furthermore, the effect of miR-4262 on the cell cycle was investigated, and it was observed that overexpression of miR-4262 in the SW480 and SW620 cell lines reduced the number of S-phase cells and enhanced the number of G2/M-phase cells compared with mimics ctrl; however, this was not statistically significant (Fig. 2E and F, respectively). In conclusion, overexpression of miR-4262 suppressed the viability, growth and cell cycle progression of colon cancer cells in vitro.
Figure 2.
miR-4262 overexpression suppresses cell viability, growth and cell cycle progression in SW480 and SW620 cells. (A) Transfection efficiency of miR-4262 mimics was assessed in SW480 and SW620 cells by reverse transcription-quantitative polymerase chain reaction relative to small nuclear RNA U6 expression. MTS assay assessing cell viability in (B) SW480 and (C) SW620 cells transfected with miR-4262 mimics or mimics ctrl. Colony formation assay of (D) SW480 and (E) SW620 cells transfected with miR-4262 mimics or mimics ctrl. Cell cycle analysis was performed on (F) SW480 and (G) SW620 cells transfected with miR-4262 mimics or mimics ctrl. *P<0.05 vs. mimics ctrl. miR, microRNA; ctrl, control.
Knockdown of miR-4262 promotes colon cancer cell viability, growth and cell cycle progression
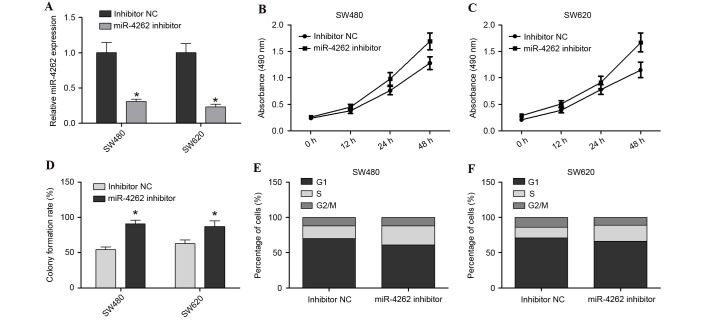
To further investigate the effect of miR-4262 downregulation on colon cancer cells, SW480 and SW620 were co-transfected with miR-4262 inhibitor or inhibitor NC. Cells were harvested after 48 h and successful transfection and subsequent downregulation of the expression of miR-4262 was confirmed by RT-qPCR (P<0.05; Fig. 3A). The cell viability of SW480 and SW620 was increased in the miR-4262 inhibitor group compared with inhibitor NC group (Fig. 3B and C, respectively). Furthermore, the colony formation assay revealed that the miR-4262 inhibitor promoted the growth of SW480 and SW620 cells compared with inhibitor NC (P<0.05; Fig. 3D). Cell cycle analysis revealed that the miR-4262 inhibitor led to an increase in the number of S-phase cells and decreased G2/M-phase cells, compared with inhibitor NC, in the SW480 and SW620 cells (Fig. 3E and F, respectively). In conclusion, knockdown of miR-4262 promoted the viability, growth and cell cycle progression of colon cancer cells.
Figure 3.
miR-4262 downregulation promotes cell viability, growth and cell cycle progression in SW480 and SW620 cells. (A) Transfection efficiency of miR-4262 inhibitor was assessed in SW480 and SW620 cells by reverse transcription-quantitative polymerase chain reaction relative to small nuclear RNA U6 expression. MTS assay assessing cell viability in (B) SW480 and (C) SW620 cells transfected with miR-4262 inhibitor or inhibitor NC. (D) Colony formation assay of SW480 and SW620 cells transfected with miR-4262 inhibitor or inhibitor NC. Cell cycle analysis was performed with (E) SW480 and (F) SW620 cells transfected with miR-4262 inhibitor or inhibitor NC. *P<0.05 vs. inhibitor NC. miR, microRNA; NC, negative control.
GALNT4 is a target of miR-4262
To investigate the potential target of miR-4262, we performed bioinformatics analysis with TargetScan (www.targetscan.org/vert_71/). The data of TargetScan confirmed the presence of an miR-4262 binding site within the 3′UTR of GALNT4, which is conserved across several species (Fig. 4A), suggesting that GALNT4 may be a target of miR-4262. To validate this hypothesis, a dual-luciferase reporter system was employed. Constructs containing wildtype (GALNT4 3′UTR wt) or mutated (GALNT4 3′UTR mut) 3′UTR sequences were cloned into the pmiRGLO luciferase vector, and co-transfected with the miR-4262 mimics/mimics ctrl into SW480 cells. The results demonstrated that overexpression of miR-4262 significantly suppressed the luciferase activity of GALNT4 3′UTR wt compared with mimics ctrl (P<0.05; Fig. 4B), while no effect was on the luciferase activity of GALNT4 3′UTR mut was observed from overexpression of miR-4262 compared with mimics ctrl (P>0.05; Fig. 4B). This indicated miR-4262 binding site specificity within the 3′UTR of GALNT4 (Fig. 4B). To further confirm the effect of miR-4262 on the endogenous level of GALNT4, RT-qPCR and western blotting were performed in cells transfected with miR-4262 mimics or mimics ctrl. The results identified that miR-4262 mimics resulted in decreased GALNT4 mRNA (Fig. 4C and D) and protein expression levels (Fig. 4E) in SW480 and SW620 cell lines, compared with cells transfected with mimics ctrl.
Figure 4.
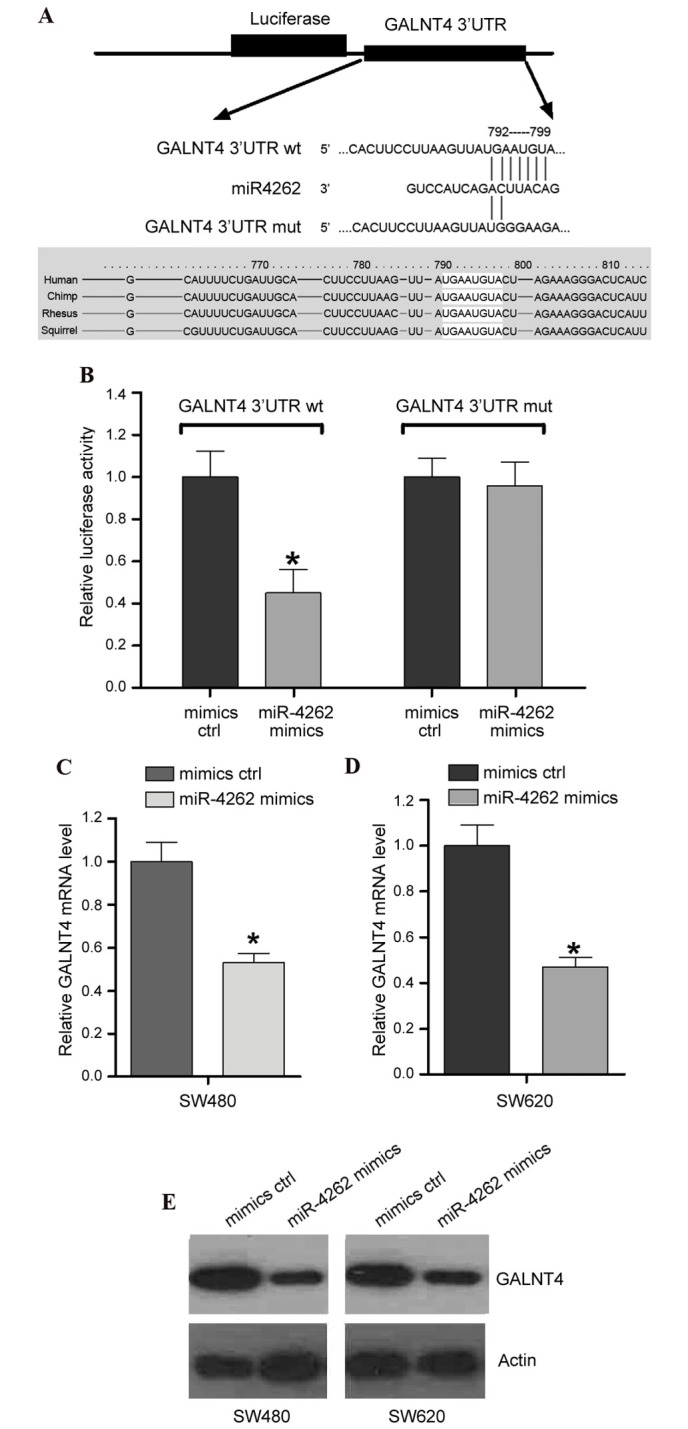
miR-4262 directly targets GALNT4 and suppresses its expression. (A) Bioinformatic analysis revealed an miR-4262 binding site within GALNT4 3′UTR wt that was mutated to create GALNT4 3′UTR mut. (B) Luciferase activity in cells co-transfected with GALNT4 3′UTR wt/mut and miR-4262 mimics/mimics ctrl. mRNA expression levels of GALNT4 in (C) SW480 and (D) SW620 cells transfected with miR-4262 mimics or mimics ctrl, assessed by reverse transcription-quantitative polymerase chain reaction. (E) Protein expression levels of GALNT4 in SW480 and SW620 cells transfected with miR-4262 mimics or mimics ctrl were detected by western blot analysis. *P<0.05 vs. mimics ctrl. miR, micro RNA; GALNT4, polypeptide N-acetylgalactosaminyltransferase 4; UTR, untranslated region; wt, wildtype; mut, mutated; ctrl, control.
GALNT4 is upregulated in colon cancer tissues and cells
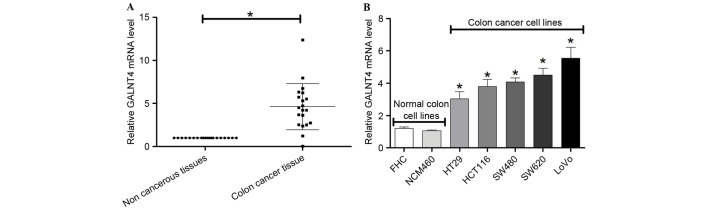
The present study further investigated the expression of GALNT4 with RT-qPCR in colon cancer tissues and cells. GALNT4 mRNA expression was significantly upregulated in 20 colon cancer tissues compared with non-tumor tissues (P<0.05; Fig. 5A). In addition, GALNT4 mRNA was upregulated in HT29, HCT116, SW480, SW620 and LoVo colon cancer cell lines compared with FHC and NCM460 normal colon cell lines (Fig. 5B).
Figure 5.
mRNA expression levels of GALNT4 in colon cancer tissues and cells. (A) Reverse transcription-quantitative polymerase chain reaction was performed to detect mRNA expression levels of GALNT4 in (A) 20 colon cancer/non-tumor tissues and (B) colon cancer and normal colon cell lines. Experiments were conducted in triplicate. *P<0.05 vs. normal cells. GALNT4, polypeptide N-acetylgalactosaminyltransferase 4.
Discussion
The present study investigated the biological role of miR-4262 in the progression of colon cancer. A significant downregulation of miR-4262 in colon cancer tissues and cells was observed, compared with matched non-tumor tissues. Additionally, functional experiments demonstrated that miR-4262 suppressed cell growth in vitro. The investigation regarding the cell cycle demonstrated that miR-4262 inhibited the proliferation of colon cancer cells through induction of cell cycle arrest at the G1/S phase. The present study, to the best of our knowledge, is the first to elucidate the role of miR-4262 in colon cancer.
GALNT4 was identified as a direct functional target of miR-4262 using a bioinformatics prediction program. Computational analysis revealed that there were binding sites for the miR-4262 seed sequence at the 3′UTR of GALNT4. Furthermore, restoration of miR-4262 expression led to decreased luciferase activity of wild-type GALNT4 3′UTR whereas the site-directed mutation abrogated miR-4262 regulation. In addition, results from the RT-qPCR and protein expression analysis indicated that the ectopic expression of miR-4262 suppressed mRNA and protein levels of GALNT4 simultaneously. These results therefore suggest that miR-4262 regulates the expression of GALNT4 by directly targeting the 3′UTR of GALNT4 in colon cancer. The function of GALNT4 in the regulation of colon cancer cell lines was then examined and an increase in GALNT4 was observed in colon cancer tissues and cells.
GALNT4 has previously been demonstrated to preferentially O-glycosylate the threonine residues at positions 44 and 57 of the P-selecting lycoprotein ligand (PSGL-1) (13). P-selectin (CD62P) has been demonstrated to be a key factor in the hematogenous dissemination of tumor cells, in particular, facilitating growth and metastasis of colon carcinomas (14). Liu et al (15) suggested that the Thr57 residue of PSGL-1 is an O-glycan carrier site required for P-selectin binding, which suggests that O-glycosylation of PSGL-1 by GALNT4 may be important for numerous P-selectin mediated cell-cell interactions. Therefore, f urther studies may be required to investigate the role of miR-4262 in the regulation of GALNT4-mediated O-glycosylation of PSGL-1.
In conclusion, the results of the present study demonstrated that miR-4262 inhibited the proliferation of colon cancer cells through induction of cell cycle arrest at the G1/S phase, mediated by GALNT4. These findings may greatly expand current understanding of the molecular basis of miR-4262/GALNT4 regulation in colon cancer, and may aid future developments of novel therapies. However, further experiments to examine the functional effect of GALNT4 in the pathogenesis of colon cancer will be required in the future.
References
- 1.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancerin in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancerstatistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Niang B, Jin L, Chen X, Guo X, Zhang H, Wu Q, Padhiar AA, Xiao M, Fang D, Zhang J. GalNAc-T4 putatively modulates the estrogen regulatory network through FOXA1 glycosylation in human breast cancer cells. Mol Cell Biochem. 2016;411:393–402. doi: 10.1007/s11010-015-2601-1. [DOI] [PubMed] [Google Scholar]
- 5.Kufe DW. Mucins in cancer: Function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira DM, RodriguesP M, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Mei Q, Li F, Quan H, Liu Y, Xu H. Busulfan inhibits growth of human osteosarcoma through miR-200 family microRNAs in vitro and in vivo. Cancer Sci. 2014;105:755–762. doi: 10.1111/cas.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Xiao W, Sun J, Han D, Zhu Y. MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via suppressing Akt phosphorylation in glioblastoma. Tumour Biol. 2014;35:8653–8658. doi: 10.1007/s13277-014-2131-6. [DOI] [PubMed] [Google Scholar]
- 10.Bao H, Gao F, Xie G, Liu Z. Angiotensin-converting enzyme 2 inhibits apoptosis of pulmonary endothelial cells during acute lung injury through suppressing Mir-4262. Cell Physiol Biochem. 2015;37:759–767. doi: 10.1159/000430393. [DOI] [PubMed] [Google Scholar]
- 11.Shapovalov Y, Hoffman D, Zuch D, de Mesy Bentley KL, Eliseev RA. Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J Biol Chem. 2011;286:22331–22338. doi: 10.1074/jbc.M111.250092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, et al. Cloning of a human UDP-N-acetyl-alpha-D-Galactosamine: Polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 1998;273:30472–30481. doi: 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Short HJ, Qian KX, Elhammer AP, Geng JG. Characterization of glycoprotein ligands for P-selectin on a human small cell lung cancer cell line NCI-H345. Biochem Biophys Res Commun. 2001;288:637–644. doi: 10.1006/bbrc.2001.5806. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Ramachandran V, Kang J, Kishimoto TK, Cummings RD, McEver RP. Identification of N-terminal residues on P-selectin glycoprotein ligand-1 required for binding to P-selectin. J Biol Chem. 1998;273:7078–7087. doi: 10.1074/jbc.273.12.7078. [DOI] [PubMed] [Google Scholar]