Abstract
Long non-coding RNAs (lncRNAs) have been identified as key regulatory factors in various biological processes. Their dysregulation has been observed in several types of human cancer, including gastric cancer (GC). The aim of the present study was to investigate the putative roles of the lncRNA DQ786243 in the progression of GC, as well as evaluate its diagnostic and therapeutic potential in GC. The expression of DQ786243 in 82 pairs of GC tissues and adjacent healthy tissues, as well as in three GC cell lines and a human normal gastric epithelial cell line, was assessed using reverse transcription-quantitative polymerase chain reaction. In addition, the putative correlation between the expression of DQ786243 and various clinicopathological features of GC was investigated. Furthermore, the effects of silencing DQ786243 in GC cells were examined using RNA interference. Colony formation and Cell Counting kit-8 assays were used to evaluate the effects of DQ786243 on GC cell proliferation, and Transwell and wound healing assays were used to examine GC cell migration and invasion. The results of the present study demonstrated that the expression of DQ786243 was significantly upregulated in GC tissues and cell lines compared with healthy control tissues and cells. Correlation analysis revealed a significant association between the expression of DQ786243 and the TNM stage, tumor size, depth of invasion and presence of lymph node metastasis in patients with GC. Furthermore, silencing the DQ786243 was demonstrated to inhibit proliferation, and impair the migration and invasion of GC cells. The present results suggested that DQ786243 may function as an oncogenic regulator via promoting the proliferation and metastasis of GC cells. Therefore, DQ786243 may have potential as a novel biomarker for the diagnosis of GC, and may also be a promising candidate for the development of novel therapeutic approaches for the treatment of patients with GC.
Keywords: long non-coding RNA, DQ786243, gastric cancer, diagnosis
Introduction
Gastric cancer (GC) is one of the most common types of malignant tumors and has been identified as the second leading cause of cancer-associated mortality worldwide. Although a steady decline in the incidence and mortality rates of GC has been observed over the past few decades, the prognosis for patients diagnosed with advanced GC remains poor (1,2). The dysregulation of several tumor-associated genes has been implicated in gastric carcinogenesis. Therefore, the identification of reliable biomarkers and novel therapeutic targets is imperative in order to improve the quality of life and survival of patients with GC.
The development of whole genome and transcriptome sequencing technologies led to the discovery that most of the human genome is transcribed, whereas <2% of the transcripts encode proteins, suggesting that non-coding (nc)RNAs represent the majority of the human genome (3,4). Long non-coding (lnc)RNAs have been identified as a subclass of ncRNAs with a length of >200 nucleotides, and have been demonstrated to participate in the regulation of numerous biological processes, including cellular development and differentiation (5–8). Aberrant lncRNA expression has been reported in various human disorders, including cancer (9–11). Hox transcript antisense intergenic RNA has been identified as a tumor-promoting lncRNA, which has been revealed to regulate polycomb-dependent chromatin modifications, and has been associated with poor prognosis in several types of cancer (9,12,13). However, lncRNAs have also been reported to possess tumor-suppressive abilities, including maternally expressed gene 3 and growth arrest-specific transcript 5, which have been revealed to be downregulated in cancer (14,15). Therefore, it may be hypothesized that lncRNAs have potential as sensitive biomarkers and alternative therapeutic targets, and may provide effective strategies aimed at the early detection and targeted therapy of human cancer.
DQ786243 is an lncRNA, which has been reported to be upregulated in hepatocellular carcinoma (HCC) (16). DQ786243 has recently been identified as an oncogene that promotes the progression of colorectal cancer (CRC) in vitro and in vivo (17). However, the roles of DQ786243 in GC remain to be elucidated. In the present study, DQ786243 expression was revealed to be significantly increased in GC tissues and cell lines. In addition, a correlation analysis demonstrated a close association between DQ786243 levels and principal clinicopathological characteristics of patients with GC, thus suggesting a potential for DQ786243 as a novel biomarker for the effective diagnosis of GC. Furthermore, knockdown of DQ786243 was revealed to suppress proliferation, and impair the migratory and invasive capabilities of GC cells, thus suggesting that DQ786243 may have potential as an alternative therapeutic target for the treatment of patients with GC.
Materials and methods
Patients and tissue specimens
The present study was approved by the Ethics Committee of Nanjing Medical University (Nanjing, China). Written informed consent was obtained from all patients prior to enrollment in the present study.
Paired tumor tissue and adjacent healthy tissue samples were collected from 82 patients diagnosed with GC between October 2012 and May 2015 in the Department of General Surgery of the Nanjing Medical University Affiliated Wuxi Second Hospital (Wuxi, China). None of the patients enrolled in the study had received chemotherapy or radiotherapy prior to surgery. Gastric tissue specimens were collected during the resection of gastric carcinomas and were immediately frozen in liquid nitrogen for 5 min and stored at −80°C until further analysis. GC clinical stage was determined according to the 7th edition of the Cancer Staging Manual of the American Joint Committee on Cancer (18). The clinicopathological characteristics of the patients are summarized in Table I.
Table I.
Correlation between long non-coding RNA DQ786243 expression and clinicopathological features of patients with gastric cancer.
| DQ786243 expression | ||||||
|---|---|---|---|---|---|---|
| High | Low | |||||
| Variable | No. | n | % | n | % | P-value |
| Sex | 0.657 | |||||
| Male | 45 | 21 | 46.7 | 24 | 53.3 | |
| Female | 37 | 20 | 54.1 | 17 | 45.9 | |
| Age (years) | 0.659 | |||||
| >60 | 43 | 23 | 53.5 | 20 | 46.5 | |
| ≤60 | 39 | 18 | 46.2 | 21 | 53.8 | |
| Tumor size (cm) | 0.043 | |||||
| ≤5 | 48 | 19 | 39.6 | 29 | 60.4 | |
| >5 | 34 | 22 | 64.7 | 12 | 35.3 | |
| Differentiation status | 0.347 | |||||
| Well/moderate | 55 | 25 | 45.5 | 30 | 54.5 | |
| Poor/undifferentiated | 27 | 16 | 59.3 | 11 | 40.7 | |
| Depth of invasion | 0.008 | |||||
| T1-T3 | 43 | 15 | 34.9 | 28 | 65.1 | |
| T4 | 39 | 26 | 66.7 | 13 | 33.3 | |
| Lymph node metastasis | 0.002 | |||||
| Absent (N0) | 35 | 10 | 28.6 | 25 | 71.4 | |
| Present (N1-N3) | 47 | 31 | 66.0 | 16 | 34.0 | |
| Distant metastasis | 0.359 | |||||
| Absent (M0) | 77 | 37 | 48.1 | 40 | 51.9 | |
| Present (M1) | 5 | 4 | 80.0 | 1 | 20.0 | |
| TNM stage | 0.004 | |||||
| I–II | 40 | 13 | 32.5 | 27 | 67.5 | |
| III–IV | 42 | 28 | 66.7 | 14 | 33.3 | |
Correlation analysis was performed using Pearson's χ2 or Fisher's exact (for distant metastasis) test.
Cell culture
The SGC-7901, BGC-823 and MGC-803 human GC cell lines were purchased from the Cell Resource Center of the Institute of Basic Medical Sciences of the Chinese Academy of Medical Sciences (Beijing, China). The GES-1 human gastric epithelial cell line was preserved in Nanjing Medical University Affiliated Wuxi Second Hospital (Wuxi, China). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were maintained at 37°C in a humidified 5% CO2 atmosphere and the culture medium was replaced every 2 days.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd., Dalian, China). RT reaction mixtures contained 1 µg total RNA, 1 µl RT Enzyme Mix 1, 1 µl RT Primer Mix, 4 µl 5X PrimeScript Buffer 2 and 4 µl ddH2O. The temperature protocol was as follows: At 37°C for 15 min and at 85°C for 5 sec. qPCR was performed on cDNA using the QuantiFast SYBR-Green PCR kit (Qiagen GmbH, Hilden, Germany) on a StepOnePlus™ Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR reaction mixtures contained 10 µl 2X PCR Master Mix, 4 µl sense and antisense primers, 2 µl cDNA and 4 µl ddH2O. Thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 10 sec and combined annealing/extension at 60°C for 30 sec. The specificity of each PCR reaction was confirmed using a melting curve analysis. PCR primers were synthesized by Invitrogen; Thermo Fisher Scientific, Inc., and the sequences were as follows: DQ786243, forward 5′-TAGGCGGACATTGTGGTGAGT-3′ and reverse 5′-CTTCTGCTGGGCTGTTGAGTG-3′; and GAPDH, forward 5′-ACGGATTTGGTCGTATTGGGC-3′ and reverse 5′-TTGACGGTGCCATGGAATTTG-3′. Relative RNA expression was calculated using the 2−ΔΔCq method (19), and the results were normalized to the expression of GAPDH. All samples were assessed in triplicate.
Small interfering (si)RNA transfection
To avoid off-target effects, siRNA against Q786243 (si-DQ786243) consisted of three target-specific siRNAs, which were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences of individual siRNA components were as follows: si-DQ786243-1, 5′-CATGGGTACCCGGATGATGTTATAT-3′; si-DQ786243-2, 5′-CCATGGGTACCCGGATGATGTTATA-3′; si-DQ786243-3, 5′-GCCATGGGTACCCGGATGATGTTAT-3′, as previously described (17). Negative control siRNA (si-NC; 5′-TTCTCCGAACGTGTCACGT-3′) was also purchased from Shanghai GenePharma Co., Ltd. SGC-7901 and MGC-803 cells at a density of 3×105 cells/ml were seeded into 6-well plates and transfected with siRNAs using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) as the transfection reagent, according to the manufacturer's protocol. After 48 h of transfection, cells were harvested for RT-qPCR to verify the silencing of DQ786243 expression. A total of 3 groups of GC cells were used: Untransfected cells (blank), cells transfected with negative control siRNA (si-NC) and cells transfected with si-DQ786243 (si-DQ786243).
Cellular proliferation assays
A total of 100 µl SGC-7901 and MGC-803 cells from the 3 groups (blank, si-NC and si-DQ786243) were seeded into 96-well plates at a density of 1×103 cells/well. After 24, 48, 72 and 96 h of incubation at 37°C in DMEM or RPMI-1640 medium, cellular viability was evaluated using a Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. The absorbance was measured at a wavelength of 450 nm using a Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.).
A colony formation assay was also performed. GC cells (blank, si-NC and si-DQ786243 groups; 500 cells/well) were seeded into 6-well plates with DMEM or RPMI-1640 medium supplemented with 10% FBS and cultured at 37°C with 5% CO2 for 14 days. Colonies were fixed with methanol at room temperature for 20 min and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The total number of visible colonies was determined under an optical microscope (Olympus Corporation, Tokyo, Japan; magnification, ×100). All experiments were repeated three times.
Cellular migration and invasion assays
Cellular migration was assessed using 6.5 mm Transwell inserts with 8.0-µm pore polycarbonate membranes (Costar; Corning Incorporated, Corning, NY, USA). A cell invasion assay was performed using Transwell inserts coated with Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, ~2×105 transfected and non-transfected SGC-7901 and MGC-803 cells were suspended in DMEM or RPMI-1640 medium supplemented with 1% FBS and were seeded into the upper chambers of the inserts. Medium supplemented with 10% FBS was added into the lower chambers as a chemoattractant. Following 24 h of incubation at 37°C, cells on the upper surface of the membrane were removed; cells that had migrated to the lower membrane were fixed with 100% methanol at room temperature for 20 min and stained with crystal violet. Cells were observed using an optical microscope (Olympus Corporation, Tokyo, Japan; magnification, ×200). Cells were counted in 5 random fields from each well, and the average number of migrated or invaded cells was calculated. The assays were performed in triplicate.
Wound healing assay
Transfected and non-transfected SGC-7901 and MGC-803 cells were cultured in DMEM or RPMI-1640 medium supplemented with 10% FBS and maintained at 37°C with 5% CO2 for 24–48 h, until 90–100% confluent. Linear scratches were created on the cell layer using a pipette tip, and cells were maintained in serum-free DMEM or RPMI-1640 medium. The wound healing process was observed under an optical microscope (Olympus Corporation; magnification, ×100) for 48 h and the images were analyzed using ImageJ software version 1.48 u (National Institutes of Health, Bethesda, MD, USA). Experiments were performed in triplicate.
Statistical analysis
The statistical significance of the differences between groups was assessed using Student's t-test for pair-wise comparisons or one-way analysis of variance followed by Fisher's least significant difference post hoc test for multiple comparisons. Correlation analysis was performed using Pearson's χ2 test or Fisher's exact test. P<0.05 was considered to indicate a statistically significant difference. Data are expressed as the mean ± standard deviation of three independent experiments. Statistical analysis was performed using the SPSS software version 20.0 (IBM Corp., Armonk, NY, USA).
Results
DQ786243 is upregulated in GC
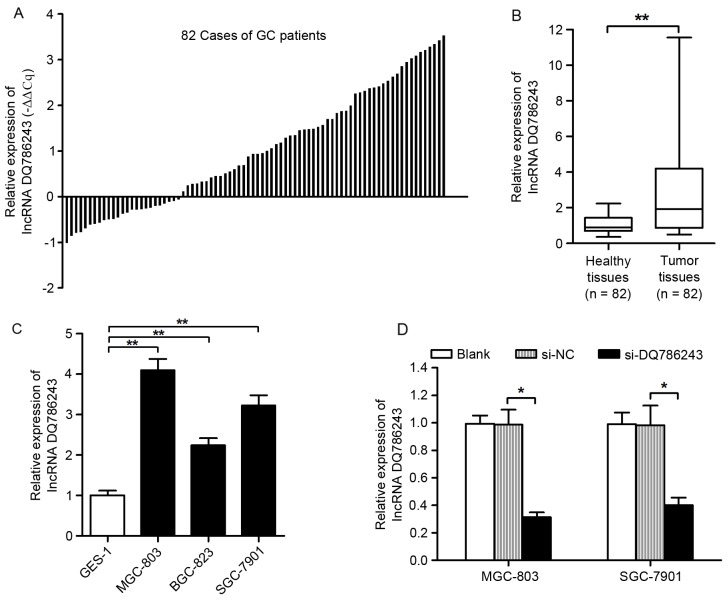
The expression levels of DQ786243 were assessed in 82 paired samples from GC and adjacent healthy tissues using RT-qPCR. The present results demonstrated that DQ786243 expression was upregulated in 69.5% (57/82) of GC tissue samples compared with in matched non-cancerous tissue samples (Fig. 1A). As presented in Fig. 1B, the relative expression levels of DQ786243 were significantly increased in tumor tissue compared with adjacent healthy tissue (P<0.01). In addition, DQ786243 expression levels in the MGC-803, SGC-7901 and BGC-823 GC cell lines were significantly increased compared with in GES-1 healthy gastric epithelial cells (P<0.01; Fig. 1C). These results suggested that DQ786243 may be aberrantly overexpressed in GC, and its dysregulation may be associated with disease progression.
Figure 1.
Expression of long non-coding RNA DQ786243 in GC tissue samples and cell lines. (A) Relative expression levels of DQ786243 in paired samples form GC and adjacent non-cancerous tissues. Data are presented as -ΔΔCq = −(ΔCqtumor - ΔCqnormal). DQ786243 was revealed to be upregulated in 69.5% (57/82) of patients with GC. (B) DQ786243 expression levels in tumor tissue were significantly increased compared with in adjacent healthy tissue samples. Data are expressed as fold-changes in tumor tissue relative to healthy tissue. (C) Relative expression levels of DQ786243 in the human MGC-803, BGC-823 and SGC-7901 GC cell lines, and in the GES-1 healthy gastric epithelial cell line. DQ786243 expression was significantly higher in GC cells compared with healthy cells. (D) Relative expression levels of DQ786243 in MGC-803 and SGC-7901 cells following transfection with si-DQ786243 or si-NC. Data are expressed as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01. GC, gastric cancer; si, small interfering; NC, negative control; lncRNA, long non-coding RNA.
Correlation of DQ786243 expression with clinicopathological features of GC
In order to investigate the clinical relevance of DQ786243 in GC progression, the relationship between the expression of DQ786243 in GC tissue and the clinicopathological characteristics of patients with GC was investigated. The median value of DQ786243 expression levels in GC tissue samples was used as the cut-off value to divide the 82 GC cases into the DQ786243-high group (n=41 patients) and the DQ786243-low group (n=41 patients), as previously described (20). As presented in Table I, higher DQ786243 expression was revealed to be associated with larger tumor sizes (P=0.043), deeper invasion (P=0.008), lymphatic metastasis (P=0.002) and advanced TNM stage (P=0.004) in patients with GC. However, no significant correlation was detected between DQ786243 expression and other clinicopathological factors, such as sex (P=0.657), age (P=0.659), differentiation status (P=0.347) and the presence of distant metastases (P=0.359).
Effect of DQ786243 knockdown on GC cell proliferation
Since DQ786243 was revealed to be upregulated in GC cell lines, its roles in GC progression were investigated using RNA interference. MGC-803 and SGC-7901 cells were transfected with siRNAs specifically targeting DQ786243. Silencing of DQ786243 expression following siRNA transfection was confirmed using RT-qPCR (P<0.05; Fig. 1D).
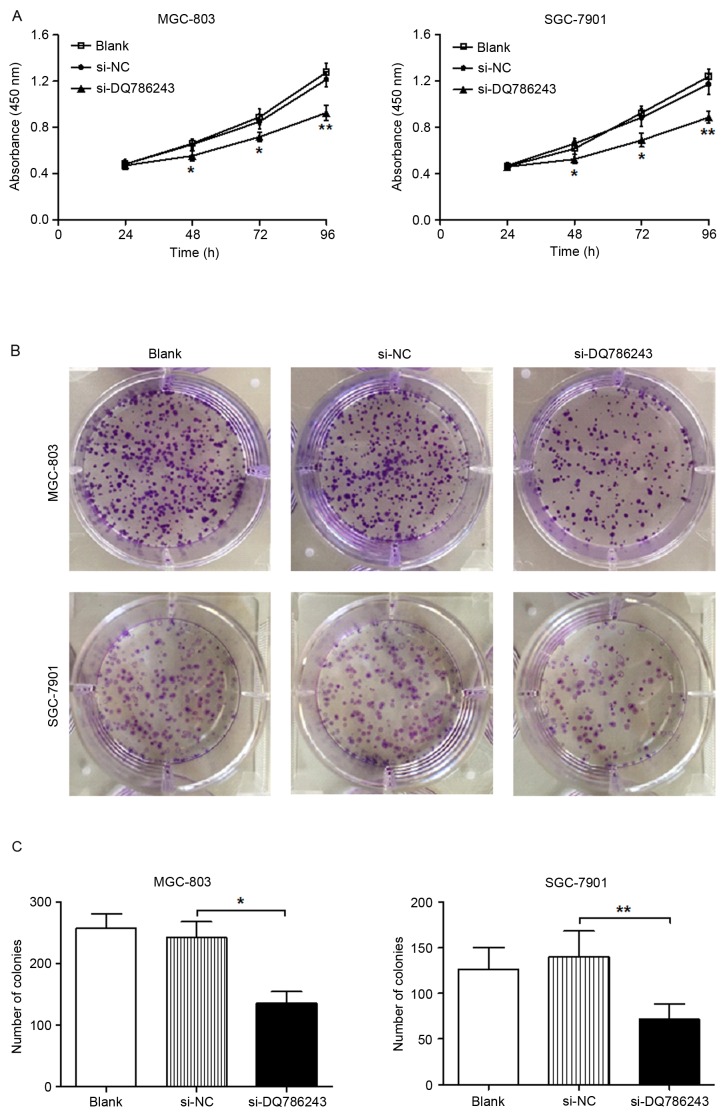
A CCK-8 assay was used to investigate cellular proliferation. The present results demonstrated that DQ786243 knockdown significantly suppressed the proliferation of MGC-803 and SGC-7901 cells (P<0.01; Fig. 2A). In addition, a colony formation assay revealed that the number of colonies generated by MGC-803 (P<0.05) and SGC-7901 (P<0.01) cells was significantly reduced following transfection with si-DQ786243 (Fig. 2B and C). These results indicated that silencing the expression of DQ786243 inhibited the proliferation of GC cells in vitro.
Figure 2.
Long non-coding RNA DQ786243 enhances the proliferation of GC cells in vitro. (A) A Cell Counting kit-8 assay was performed on MGC-803 and SGC-7901 cells following transfection with si-DQ786243 or si-NC to investigate the effects of DQ786243 on cellular proliferation. (B) Quantification of a colony formation assay in MGC-803 and SGC-7901 cells following transfection with si-DQ786243 or si-NC. (C) Knockdown of DQ786243 expression significantly impaired the colony formation capabilities of GC cells. Data are expressed as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01. GC, gastric cancer; si, small interfering; NC, negative control; lncRNA, long non-coding RNA.
Effect of DQ786243 knockdown on GC cell migration and invasion
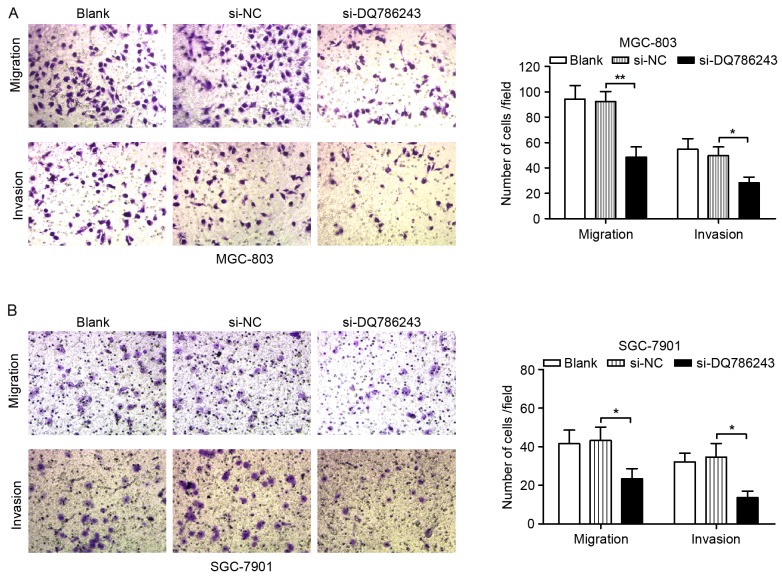
To investigate whether DQ786243 may participate in GC cell metastasis, the migratory and invasive capabilities of GC cells were evaluated. As demonstrated using Transwell migration and invasion assays, silencing of DQ786243 expression significantly suppressed migration (P<0.01) and invasion (P<0.05) in MGC-803 (Fig. 3A) and SGC-7901 (P<0.05; Fig. 3B) cells.
Figure 3.
Long non-coding RNA DQ786243 promotes the migration and invasion of GC cells in vitro. Knockdown of DQ786243 expression significantly suppressed migration and invasion of (A) MGC-803 and (B) SGC-7901 cells. Magnification, ×200. Data are expressed as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01. GC, gastric cancer; si, small interfering; NC, negative control.
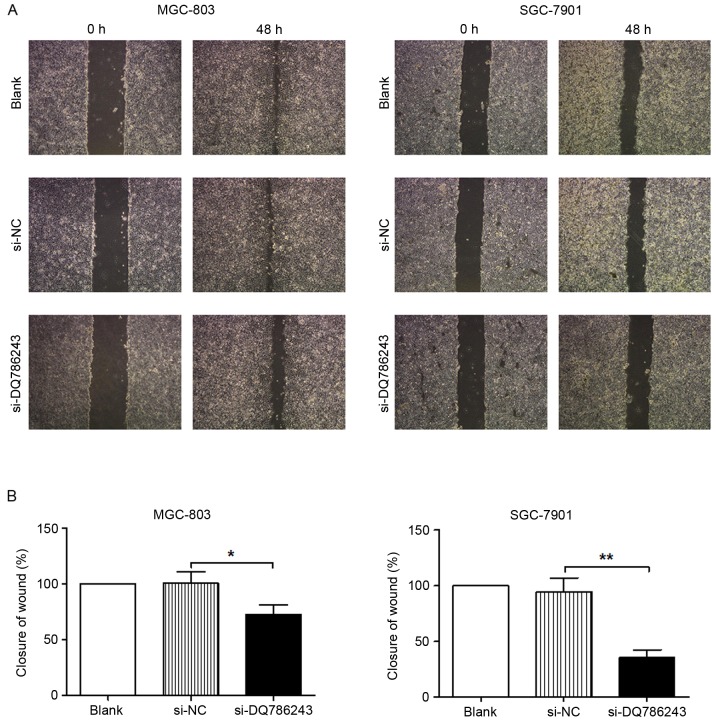
Similar results were obtained following a wound healing assay. Following knockdown of DQ786243 expression, wound healing in MGC-803 (P<0.05) and SGC-7901 (P<0.01) cells was significantly impaired (Fig. 4A and B). The present results indicated that suppression of DQ786243 expression impaired the migration and invasion of GC cells in vitro.
Figure 4.
In vitro wound healing assay of GC cells. (A) Representative photomicrographs of MGC-803 and SGC-7901 cells following transfection with si-DQ786243 or si-NC. Blank cells were not transfected. Magnification, ×100. (B) Knockdown of DQ786243 expression significantly impaired wound closure in MGC-803 and SGC-7901 cells. Data are expressed as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01. GC, gastric cancer; si, small interfering; NC, negative control.
Discussion
GC constitutes a major public health concern and a significant burden on society worldwide, with ~951,600 new cases and ~723,100 cases of mortality reported in 2012 (1,2). Therefore, the identification of novel biomarkers for the reliable early diagnosis, as well as alternative therapeutic strategies for the treatment of patients with GC is of critical importance. In the present study, lncRNA DQ786243 was revealed to be overexpressed in GC tissue samples and cell lines compared with in healthy tissue samples and cells. A positive correlation was detected between increased DQ786243 expression and TNM stage, tumor size, depth of invasion and lymph node metastasis in patients with GC. Furthermore, following the downregulation of DQ786243 expression, the proliferative, migratory and invasive capabilities of GC cells were significantly suppressed in vitro. The present results suggested that DQ786243 may function as an oncogene in GC and may be involved in the progression of the disease.
Accumulating evidence has identified lncRNAs as critical players in the regulation of several biological processes, including cell cycle progression, chromatin remodeling, and the transcriptional and post-transcriptional regulation of genes (21,22). In addition, aberrant lncRNA expression has been reported in various types of cancer. The oncogenic lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) has been associated with the metastasis and poor prognosis of non-small cell lung cancer (23). MALAT-1 overexpression has also been demonstrated to promote the growth and metastasis of gallbladder cancer cells, through the activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase signaling pathway (24). lncRNA is highly upregulated in liver cancer, is upregulated in hepatocellular carcinoma mediated by Hepatitis B virus X protein, and has been demonstrated to enhance cancer cell proliferation through the inhibition of p18 (25). In addition, overexpression of the lncRNA colon cancer-associated transcripts 1 (CCAT1) has been demonstrated in CRC tissue samples, thus suggesting its potential as a highly specific marker for the reliable diagnosis of CRC and tumor-associated diseases (26). The transcription factor c-Myc has been reported to activate CCAT1, thus promoting the proliferation and migration of gastric carcinoma cells (27). Other lncRNAs, including H19, gastric carcinoma high expressed transcript 1 and ZXF1, have also been revealed to be dysregulated in various types of cancer (28–30). However, the molecular mechanisms underlying the involvement of lncRNAs in gastric carcinogenesis, and the clinical implications for the treatment of patients with GC, remain to be fully elucidated.
DQ786243 is an lncRNA which was initially reported to be upregulated in hepatocellular carcinoma (16). Recently, DQ786243 has been identified as an oncogene in CRC that promotes disease progression, whereas its knockdown was reported to inhibit the proliferation, migration and invasion of CRC cells, through the regulation of cell cycle- and apoptosis-associated proteins, and of the epithelial-mesenchymal transition process (17). Therefore, the present study investigated the roles of DQ786243 in GC, and suggested that DQ786243 may function as an oncogene during the development and progression of GC. However, further experiments using larger sample sizes are required to elucidate the molecular mechanisms and the exact biological functions of DQ786243 in GC.
In conclusion, the present study demonstrated that DQ786243 was significantly upregulated in human GC cell lines and in tissue samples isolated from patients with GC. Correlation analysis revealed that increased DQ786243 expression was associated with larger tumor size, deeper invasion, lymph node metastasis and advanced TNM stage in patients with GC. Furthermore, following knockdown of DQ786243 expression, proliferation, migration and invasion of GC cells were significantly impaired. These results suggested that DQ786243 may have potential as a diagnostic biomarker for the early detection of GC, and may be used as a potential target for the development of novel therapeutic strategies for the treatment of patients with GC.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 6.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 11.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 12.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 15.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 17.Sun L, Xue H, Jiang C, Zhou H, Gu L, Liu Y, Xu C, Xu Q. LncRNA DQ786243 contributes to proliferation and metastasis of colorectal cancer both in vitro and in vivo. Biosci Rep. 2016;36 doi: 10.1042/BSR20160048. pii: e00328. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, Zhou B, Chen JJ, Chen P. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17:640–646. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 21.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 24.Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A, Dayanc BE, Ritter G, Gomceli I, et al. Colon cancer associated transcript-1: A novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598–1606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 28.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Zhou XF, Pan GF, Zhao JP. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68:401–407. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]