Abstract
Basic fibroblast growth factor (bFGF) and fibroblast growth factor receptor 1 (FGFR1) are associated with drug resistance in lung cancer. In the present study, mouse monoclonal antibodies (mAb) against human bFGF, targeting the binding site of bFGF with FGFR1 were produced, and the antitumor activity and inhibition of metastasis was studied in Lewis lung carcinoma (LLC). A total of four hybridoma cell strains that stably secreted bFGF mAb were obtained. mAbE12 was selected as the most effective for use in the following studies, with a relative affinity constant of 5.66×108 l/mol. mAbE12 was demonstrated to inhibit cell proliferation and tumor growth in vitro and in vivo. Furthermore, mAbE12 blocked migration and metastasis of LLC cells in vitro and in vivo. This occurred due to a mAbE12-induced upregulation of E-cadherin expression through the protein kinase B-glycogen synthase kinase 3 β-Snail pathway. These results suggested that mAbE12 may be a potential antibody for the treatment of lung cancer.
Keywords: bFGF, monoclonal antibody, lung cancer, metastasis
Introduction
Metastasis is responsible for 90% of cases of cancer-associated mortality, including lung cancer. Tumor metastasis includes two processes: An initial invasion of tumor cells, and a secondary colonization process (1,2). The former process is well known while the latter requires further study to elucidate multiple issues, including how altered proteins and intracellular signaling molecules induce epithelial mesenchymal transition (EMT).
Multiple novel drugs have been identified, including molecular targeted drugs, however the lung cancer mortality rate remains high (3,4). The current clinical drugs for lung cancer are primarily epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors. However, multiple EGFR mutations during lung cancer cause drug resistance. Following screening of gene expression in lung cancer tumor tissue, basic fibroblast growth factor (bFGF), fibroblast growth factor receptor (FGFR)1 and FGFR 2 have been demonstrated to exhibit high expression levels, and be associated with drug resistance in lung cancer (5–7). There are fewer FGFR mutations in lung cancer, thus bFGF-FGFR is a reliable potential target.
Previous studies have suggested that FGFRs, in particular FGFR1, demonstrate high expression levels in lung cancer (8,9). This effect is achieved by enhancing the affinity of FGFR to its ligand through increasing the number of gene copies and gene mutations. Furthermore, bFGF collaborates with other factors to strengthen the involvement of the bFGF-FGFR signaling pathway (10–13). For example, during the EMT process, transforming growth factor-β induced isoform switching of FGFRs, causing the cells to become sensitive to bFGF (14). Similarly, the FGF-phosphoinositide 3-kinase-protein kinase B (AKT)-glycogen synthase kinase 3β (GSK3β) signaling pathway regulates EMT and increases the invasiveness of tumor cells (2).
In the initial stages of tumorigenesis, there are junctions between tumor cells and normal epithelial tissues, including tight junctions, adhesion junctions, gap junctions and desmosomes. These junctions limit the movement of tumor cells to form a complete tissue. With the development of cancer and deterioration, certain surface adhesion molecules of tumor cells are altered and the normal junctions between cells are lost. E-cadherin is involved in this process (15). The adherent junctions mediated by E-cadherin are not a static structure, however are a dynamic one. It has previously been suggested that FGF signaling promotes EMT through downregulating the expression of E-cadherin (16).
In the present study, a mouse monoclonal antibody against human bFGF was produced, targeting the binding site of bFGF with FGFR1. The monoclonal antibody (mAb) E12 inhibited Lewis lung carcinoma (LLC) metastasis through increasing the expression of E-cadherin via the AKT-GSK3-β-snail family transcriptional repressor 1 (Snail) pathway, suggesting that mAbE12 may be a promising anticancer antibody.
Materials and methods
Mouse Lewis lung carcinoma cells (LLC) and mouse myeloma cells (SP2/0) were maintained in the Guangdong Province Key Laboratory of Molecular Immunology and Antibody Engineering (Guangzhou, China). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). TRIzol reagent, and the M-MLV First-Strand cDNA Synthesis kit were purchased from Thermo Fisher Scientific, Inc. Blend Taq-Plus and Cell Counting Kit-8 (CCK-8) were purchased from Toyobo Co., Ltd. (Osaka, Japan). AKT rabbit mAb (catalog no. 4685S), phosphorylated (p) -AKT rabbit mAb (# 13038S), GSK3-β rabbit mAb (catalog no. 5676S), p-GSK3-β rabbit mAb (catalog no. 9322S), Snail mAb (catalog no. 3879S), E-cadherin mAb (catalog no. 3195S) and goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase (HRP) antibodies (catalog no. 7074S) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Balb/c mice and C57BL/6 mice were purchased from the Southern Medical University Animal Centre (Guangzhou, China). Male Balb/c mice and C57BL/6 mice (2-month old, 18–25 g, n=5) were housed in a 12 h dark and light cycle with free access to water and food, at a temperature of 26°C and humidity of 40–60%. All efforts were made to minimize suffering. The pMD18-T plasmid vector was purchased from Takara Bio, Inc. (Otsu, Japan).
Animal immunization and cell fusion
A total of 5 Balb/c mice were immunized with recombinant human bFGF, and then the mice were sacrificed after anesthesia (2% pelltobarbitalum natricum, 50 mg/kg, intraperitoneal injection) and B cells were harvested from the spleen of mice and fused with SP2/0 mouse myeloma cells. Hybridoma cell lines secreting mAb against the binding site of bFGF with FGFR1IIIC were screened out by indirect and competitive enzyme linked immunosorbent assay (provided in house by the Antibody Engineering laboratory (Guangzhou, China). The present study was approved by the Laboratory Animal Ethics Committee of Jinan University (Guangzhou, China) and animal experiments were performed in compliance with the guidelines for the Welfare of Experimental Animals in Jinan University.
Indirect ELISA
Recombinant human bFGF (50 ng/well; Department of Anesthesia and Perioperative Care, University of California, USA) was used to coat microtiter ELISA wells. The ELISA wells were blocked with 5% non-fat dry milk in PBS buffer containing 0.5% Tween-20 for 1 h at 37°C, bFGF incubated with purified antibody (purified with Protein G Sepharose (GE Healthcare, Chicago, IL, USA; catalog no. 17-0405-03, 1:400,000) at 37°C for 1 h, then HRP-conjugated goat anti-mouse antibody (1:8,000) at 37°C for 40 min. The absorbance values were measured at 450 µm on an automatic plate reader.
Competitive ELISA
The conditions used for competitive ELISA were maintained according to the method of the Indirect ELISA. ELISA wells containing bFGF were incubated with purified bFGF antibody (1,400,000) and biotin-FGFR1IIIC (1 µg/ml, Department of Anesthesia and Perioperative Care, University of California) at 37°C for 1 h. HRP-conjugated streptavidin (catalog no. N100; 1:1,000; Thermo Fisher Scientific, Inc.) was incubated at 37°C for 40 min. The absorbance values were measured at 450 µm on an automatic plate reader.
bFGF mAb production and identification
The hybridoma cell lines secreting mAb against bFGF were cloned, and ascites were produced by injecting 1×105 hybridoma cells into 5 Balb/c mice respectively. The mAb in the ascites was purified using affinity chromatography with Protein G Sepharose (GE Healthcare), and the elution buffer (pH=2.5) contained 150 mM glycine-PBS buffer. And then identified by 12% SDS-PAGE.
Total RNA of the hybridoma cells secreting mAb against bFGF was extracted using TRIzol reagent, according to the manufacturer's protocol. The concentration of the extracted RNA was detected using a Nanodrop 2000 (Thermo Fisher Scientific, Inc.) at an absorbance of 260 nm (A260), the quality was detected by a ratio of A260/A280 and agarose gel electrophoresis (1 µg per lane). cDNA was synthesized using the M-MLV First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc., catalog no. 28025013), according to the manufacturer's protocol. The genes of the mAb heavy chain and light chain were amplified by Blend Taq-Plus (Toyobo Co., Ltd., Osaka, Japan; catalog no. BTQ-201), according to the manufacturer's protocol. The primers of the DNA sequences were as follows: Heavy chain forward, 5′-TGAGGAGACGGTGACCGTGGTCCC-3′ and reverse, 5′-GAGGTGCAGCTTCAGGAGTCAGG-3′; light chain forward, 5′-TTTGATTTCCAGCTTGGTGCCTCC-3′ and reverse, 5′-GACATTGTGATGWCACAGTCTCC-3′. The reagents in the reaction were as follows: 10X buffer (5 µl), 2 mM dNTP (5 µl), forward primer (10 pmol/µl; 1 µl), reverse primer (10 pmol/µl; 1 µl), cDNA (1 µl), Blend Taq-Plus (2.5 U/µl; 0.5 µl) ddH2O (36.5 µl). The cycling conditions were as follows: Denaturation at 94°C for 5 mins on the initial cycle and for 30 sec for the rest, annealing at 56°C for 30 sec and extension at 72°C for 40 sec; with an additional 10 min extension on the last cycle. The number of cycles was 35. The genes of the mAb heavy and light chains were inserted and ligated into the pMD18-T vector (Takara Bio, Inc., catalog no. 6011). The mAb variable region sequence was analyzed by the Beijing Genomics Institute (Beijing, China).
Analysis of the binding sites of antibody fragment of variable region (Fv), FGFR1 and bFGF
Discovery Studio 4.0 software (Accelrys Ltd., Cambridge, UK) was used in homology modeling, molecular docking and three-dimensional structure building of the antibody Fv. This three-dimensional structure was used to analyze the binding site with bFGF. The binding sites of FGFR1 (ID: 4ZSA) and bFGF (ID: 4OEG) were then downloaded from the Protein Data Bank database to compare with this result.
Effect of mAb on proliferation and migration of LLC cells
Cell suspension (100 µl; 2,000 cells/well) was dispensed in a 96-well plate and pre-incubated overnight in a CO2 incubator at 37°C. Following this, the suspension was decanted into DMEM complete medium and 1, 10, 50, 200 and 300 µg/ml mAb was added into the low serum DMEM medium (0.5% FBS) in the plate. The plate was incubated at 37°C and 5% CO2. CCK-8 solution (10 µl) was added to each well following 72 h incubation. The plate was incubated for a further 2 h and the absorbance was measured at 450 nm.
LLC cells (100 µl; 5×105 cells/ml with 200 µg/ml bFGF mAb) and 600 µl DMEM complete medium (10% FBS, 20 ng/ml bFGF and 200 µg/ml bFGF mAb) were placed in the upper and lower chamber of Transwell plates, respectively, as the experimental group. LLC cells (100 µl; 5×105 cells/ml) LLC cells and 600 µl DMEM complete medium (10% FBS, 20 ng/ml bFGF) were placed in the upper and lower chamber, respectively, as the medium group. LLC cells (100 µl; 5×105 cells/ml) and 600 µl DMEM complete medium (10% FBS, 20 ng/ml bFGF and 200 µg/ml control IgG) were placed in the upper and lower chamber, respectively, as the control group. The chamber was incubated for 24 h, following which the cells were washed twice with PBS to remove unattached cells. The remaining cells were fixed with 10% formaldehyde for 30 min. Following this, the cells were stained with crystal violet for 20 min and the migrating cells were counted using a light microscope manually by 4 researchers in 5 fields of view in each chamber.
Western blot analysis of signaling pathway
LLC cells were seeded in 6-well plates containing 0.5% FBS in DMEM medium (3×105 cells/well). Following being starved and treated with 1, 10, 50 or 200 µg/ml bFGF mAb or 1, 10, 50 or 200 µg/ml control IgG for 24 h, protein was harvested from 3×105 LLC cells using RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China; catalog no. P0013C). The concentration of the protein was measured with the BCA protein assay kit (Thermo Fisher Scientific, Inc., catalog no. 23225). Protein (30 µg) was separated by 10% SDS-PAGE (100 V; 70 min) and transferred to a polyvinyldene fluoride membrane. The membrane was blocked with 5% non-fat dry milk in PBS buffer containing 0.5% Tween-20 for 1 h at 37°C, and incubated with primary antibody (E-cadherin rabbit mAb, p-Akt, AKT rabbit mAb, p-GSK3-β, GSK-3β rabbit mAb, Snail rabbit mAb; 1:1,000) at 37°C for 2 h, and HRP-conjugated secondary antibody (1:8,000) at 37°C for 1 h. The blots were also probed with β-actin (1:1,000; catalog no. 4970) at 37°C for 2 h as a control.
Xenograft studies of LLC cells
A single-cell suspension (1×106 cells/ml) of LLC cells was prepared and 50 µl subcutaneously injected into the back of the C57BL/6 mice. Once the tumors reached ~3 mm3 volume, mice were distributed randomly into two groups (n=5). The experimental group was injected with 1 mg bFGF mAb and the control group was injected with 1 mg control IgG every 3 days, around the tumor. All mice were anesthetized (2% pelltobarbitalum natricum; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany; catatlog no. P3761; 50 mg/kg; intraperitoneal injection) and sacrificed following 6 injections. The lungs were removed and fixed with Bouin solution (Huayueyang Biotechnology Co., Ltd., Beijing, China; catalog no. WX0180) at 4°C for 24 h, then metastatic nodules were identified. The tumors from the mAbE12-treated and control groups were harvested and the volume and weight of the tumors was measured.
Statistical analysis
All numerical data are presented as the mean ± standard deviation. Statistical differences were evaluated with the Mann-Whitney U test for comparison between the groups. Analysis was conducted using SPSS software, version 20.0 (IBM Corp., Armonk, NY, USA). GraphPad Prism version 6 software (GraphPad Software, Inc, La Jolla, CA, USA) was used to create the graphs from the statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Hybridoma cell screening and identification of purified antibodies
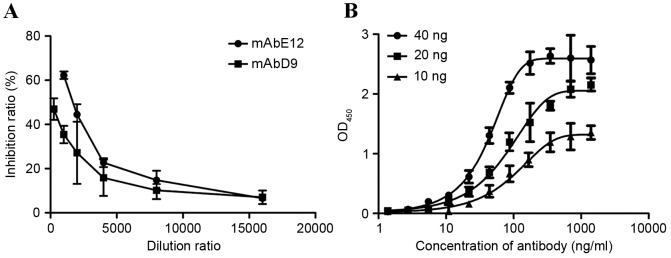
4 hybridoma cells were screened out by indirect and competitive ELISA. The competitive ELISA used cell supernatants, containing bFGF antibody, to compete with FGFR1. Of the antibodies, mAbD9 and mAbE12 demonstrated increased competitive ability compared with mAbA6 and mAbE10. The titer and the isotype of the antibodies are summarized in Table I. In order to determine the most competitive antibody, mAbD9 and mAbE12, purified mAbD9 and mAbE12 were used to compete with FGFR1. The IC50 of mAbE12 and mAbD9 were 1.96 and 1.564 µg/ml, respectively (Fig. 1A). mAbE12 was therefore more competitive than mAbD9, and mAbE12 was selected as the experimental antibody. The relative affinity of the mAbE12 antibody was measured using indirect ELISA. The relative affinity of mAbE12 antibody was Kaff=5.66×108 l/mol (Fig. 1B).
Table I.
Hybridoma screening by ELISA.
| Name | Cell supernatant dilution ratio | Ascite dilution ratio | Competitive rate (%) | Antibody isotype |
|---|---|---|---|---|
| 1A6 (mAbA6) | 1:600 | 1:700,000 | 22.77±1.8 | IgG1 |
| 2D9 (mAbD9) | 1:1,000 | 1:800,000 | 31.56±0.7 | IgG1 |
| 5F10 (mAbF10) | 1:400 | 1:500,000 | 23.77±0.7 | IgG1 |
| 2E12 (mAbE12) | 1:800 | 1:400,000 | 34.51±0.6 | IgG1 |
mAb, monoclonal antibody; IgG, immunoglobulin G.
Figure 1.
Identification of purified antibody. (A) Inhibition ratio of bFGF binding activity between bFGF antibody and FGF receptor 1βIIIC. (B) Measurement of relative affinity constant of mAbE12 antibody. Results are presented as the mean ± standard deviation. bFGF, basic fibroblast growth factor; mAb, monoclonal antibody.
Fv of mAbE12 antibody
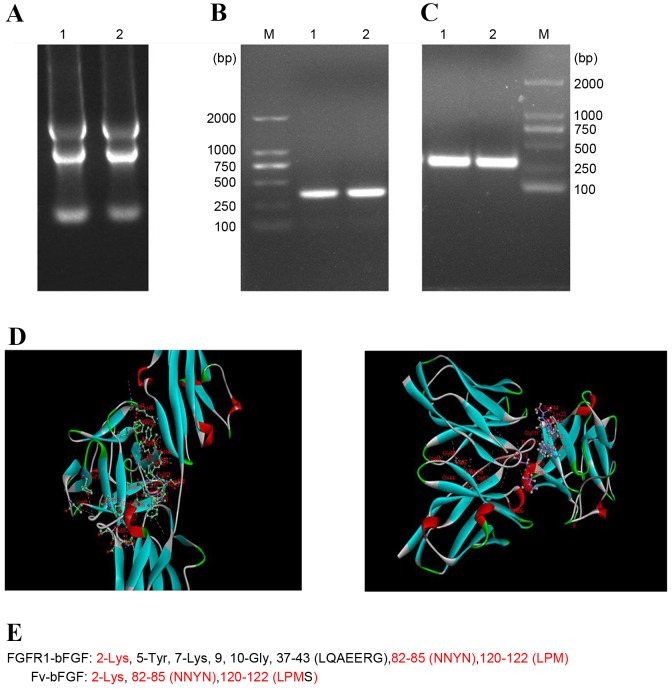
The total RNA of mAbE12 hybridoma cells was extracted (Fig. 2A) and reverse transcribed into cDNA. The heavy chain variable region and light chain variable region were ~300 bp long (Fig. 2B and C, respectively). In order to study the structure of mAbE12, Discovery Studio 4.0 software was used to model the homology, molecular docking and build three-dimensional structures of the Fv region. The binding sites of the Fv fragment of mAbE12-bFGF and bFGF-FGFR1 were also analyzed by Discovery Studio 4.0 software (Fig. 2D). The red part of the three-dimensional structures was the common binding site of Fv-bFGF and FGFR1-bFGF. Therefore, it was concluded Fv-bFGF had the same binding site as FGFR1-bFGF (Fig. 2E).
Figure 2.
Fv of mAbE12 antibody (A) Identification of isolated RNA. Reverse transcription polymerase chain reaction amplification of mAbE12 (B) heavy chain and (C) light chain Fv. (D) Three dimensional images of Fv fragment of mAbE12-bFGF and FGFR1-bFGF. The red part of the three-dimensional structures was the common binding site. (E) Comparison of binding sites between Fv-bFGF and FGFR1-bFGF. Fv, fragment of variable region; mAb, monoclonal antibody; bFGF, basic fibroblast growth factor; FGFR1, fibroblast growth factor receptor 1.
Effect of mAb on LLC cell proliferation and migration
The results of the CCK-8 assay indicated that LLC cell proliferation is inhibited by mAbE12, and the most efficient mAb concentration was 300 µg/ml (Fig. 3A) The IC50 of proliferation inhibition was 119.97±40.96 µg/ml (Fig. 3A). A stronger reduction of migrating cells was observed in the mAbE12 group compared with the IgG control group and the medium group. The migration rates were 45.73±7.98, 121.67±24.52 and 100% for the mAbE12, IgG control and medium groups, respectively (Fig. 3B).
Figure 3.
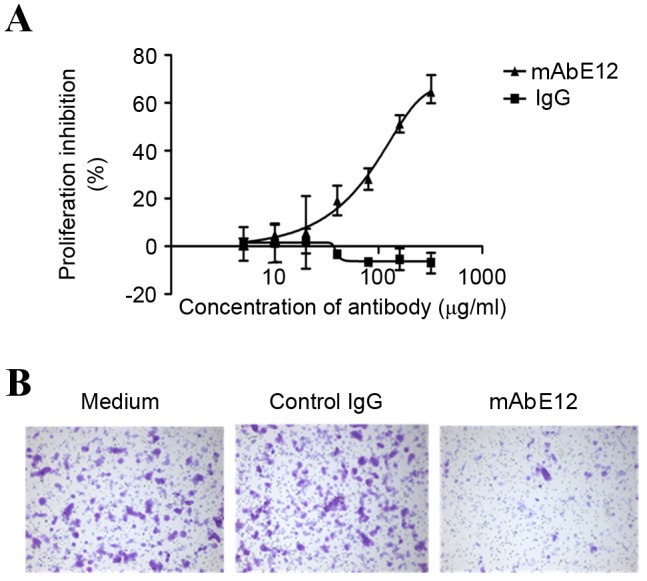
Effect of mAbE12 on (A) Lewis lung carcinoma cell proliferation and (B) migration. Results are presented as the mean ± standard deviation. mAb, monoclonal antibody; IgG, immunoglobulin G.
Effect of mAb on the LLC cell signaling pathway
Snail expression and phosphorylation of AKT(T-308) and GSK3-β (Ser-9) were visibly suppressed by treatment with 50 or 200 µg/ml mAbE12. No visible difference was observed in the IgG control group. E-cadherin expression was upregulated in the 50 and 200 µg/ml mAbE12 experimental group but remained unchanged in the IgG control group. This suggested that mAbE12 downregulated the phosphorylation of AKT and GSK3-β, downregulated Snail and upregulated E-cadherin protein levels. The suppression of the AKT/GSK3-β/Snail pathway signaling and the reduction in the number of migrating cells indicated that mAbE12 inhibited tumor metastasis through suppressing the AKT/GSK3-β/Snail pathway and upregulating the expression of E-cadherin, enhancing cell-cell junctions.
Effect of mAb on LLC cell metastasis and growth in vivo
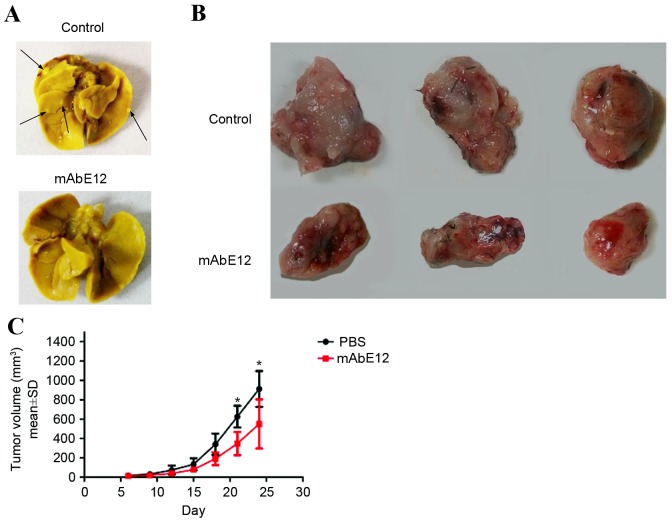
The lung colonization assay and xenograft tumor formation assay indicated that treatment of mice with mAbE12 led to a decrease in lung metastasis (Fig. 5A) and a reduction of tumor growth (Fig. 5B and C). These results demonstrated that mAbE12 inhibited the metastasis of LLC cells and tumor growth in vivo.
Figure 5.
Effect of mAb on LLC metastasis and growth in vivo. (A) Lung metastatic colonies of LLC cells with different treatments. Arrows indicate the metastatic foci in the lungs. (B) Inhibition of LLC tumor growth induced by mAbE12. (C) Tumor growth curve of mAbE12 and PBS control group. Data are presented as the mean ± SD. *P<0.05 vs. PBS control. mAb, monoclonal antibody; LLC, Lewis lung carcinoma; SD, standard deviation; PBS, phosphate-buffered saline.
Discussion
At present, small molecular compounds are used during drug treatment for patients with cancer. These have demonstrated efficacy, however also present with serious side effects. Targeted therapy, in particular antibody therapy, has provided a novel method to address this issue. Hybridoma and phage display technology are the main methods for preparing monoclonal antibodies at present (17). The present study adopted classical hybridoma technology and competitive ELISA with an avidin-biotin amplification system to screen antibodies. mAbE12, mAbD9, mAbA6 and mAbF10 cells, which are capable of secreting bFGF monoclonal antibodies, were screened. The results of the competitive ELISA demonstrated that the competitive abilities of these antibodies were different, potentially due to differences in affinity or epitope of these antibodies. mAbE12 was selected as the most competitive antibody. Initially, the reverse transcription polymerase chain reaction was used to amplify the Fv region gene. Subsequently, Discovery Studio 4.0 software was used for homology modeling, molecular docking and three-dimensional structure building. Following this, the binding site with bFgF was analyzed. The results indicated that the binding site of Fv-bFGF was similar to the binding site of FGFR1-bFGF. This further confirmed that mAbE12 screened in the present study is the target antibody for the binding site of FGFR1-bFGF, and its weak competitive ability is potentially due to its weak affinity. It also suggested that the competitive ELISA screening method established in the present study was accurate. Discovery Studio software also predicts the amino acid mutation site of the antibody, which makes it possible to then improve the affinity by site-directed mutagenesis. In future studies, Discovery Studio software should be used to predict the amino acid mutation sites and improve the affinity of the antibody.
Changes to tumor cell morphology and a variety of surface molecules is the basis of tumor metastasis. bFGF promotes the process of tumor metastasis (18). Therefore, the inhibition of the biological functions of bFGF should have an inhibitory effect on tumor metastasis. LLCs with high expression levels of bFGF and FGFR1 were selected as the object of the present study. Furthermore, LLC cell migration was decreased following treatment with mAbE12 compared with the IgG control group, which may be due to the upregulation of E-cadherin expression. E-cadherin is involved in cell interaction and signal transduction. Tumor development and progression are associated with loss or downregulation of E-cadherin. This phenomenon is often associated with changes of tumor cell morphology, metastasis and invasion (19,20). The results of in vivo and in vitro experiments demonstrated that bFGF antibody upregulates the expression level of E-cadherin, which may be one mechanism underlying the inhibition of tumor metastasis by bFGF mAb.
E-cadherin-mediated cell adhesion is associated with epithelial morphology. The loss of E-cadherin results in the acquisition of metastatic potential in tumor cells. The loss or downregulation of E-cadherin has been observed in variety of malignancies (21). In the present study, the phosphorylation level of AKT and GSK3-β were decreased when the antibody concentration was increased. The nuclear transcription factor Snail was also downregulated when treated with mAbE12 antibody. Therefore, it is possible to hypothesize that bFGF antibody upregulates E-cadherin expression in LLC, primarily by inactivating AKT, resulting in GSK3-β activation and degradation of Snail. When the nuclear transcription factor Snail is degraded by activated GSK3-β, the expression of E-cadherin will increase.
In the present study, mAbE12 antibodies were demonstrated to bind naturally expressed bFGF in LLC cells, specifically, and to inhibit the growth of LLC cells. Furthermore, mAbE12 blocked signaling pathways activated by bFGF, inhibited proliferation, and resulted in a reduction in tumor growth and lung metastasis. The primary mechanism by which this occurred was the upregulation of E-cadherin expression through regulation of the AKT-GSK3-β-Snail pathway. Understanding the mechanism of E12 may aid the identification of novel therapies for cancer.
Figure 4.
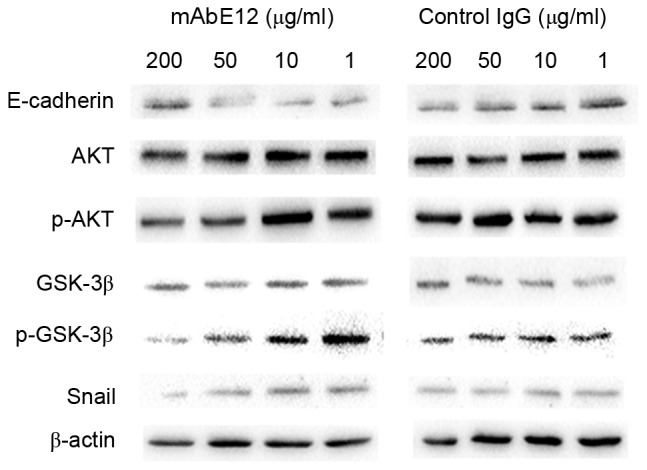
The AKT/GSK3-β/Snail pathway was suppressed by mAbE12. A dose-dependent decrease of p-AKT was observed in the mAbE12 group. The expression of Snail was downregulated and the expression of E-cadherin was upregulated in the mAbE12 group. β-actin served as a control. AKT, protein kinase B; p-, phosphorylated; Snail, snail family transcriptional repressor 1; GSK3-β, glycogen synthase kinase 3 β; mAb, monoclonal antibody.
Acknowledgements
The authors would like to thank Professor James D Marks (University of California, San Francisco, CA, USA) for providing the empirical method used to clone the bFGF mAb Fv fragment and the humanized bFGF mAb genes. The present study was supported by the State Natural Science Foundation Youth Science Foundation of China (grant no. 81202449).
Glossary
Abbreviations
- bFGF
basic fibroblast growth factor
- mAb
monoclonal antibody
- FGFR
fibroblast growth factor receptor
- EGFR
epidermal growth factor receptor
- ELISA
enzyme linked immunosorbent assay
- EMT
epithelial mesenchymal transition
- CCK-8
Cell Counting Kit-8
- LLC
Lewis lung carcinoma
- Fv
fragment of variable region
References
- 1.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Eastern Cooperative Oncology Group: Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Rades D, Setter C, Dahl O, Schild SE, Noack F. Fibroblast growth factor 2-a predictor of outcome for patients irradiated for stage II–III non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2012;82:442–447. doi: 10.1016/j.ijrobp.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 4.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 5.Azuma K, Kawahara A, Sonoda K, Nakashima K, Tashiro K, Watari K, Izumi H, Kage M, Kuwano M, Ono M, Hoshino T. FGFR1 activation is an escape mechanism in human lung cancer cells resistant to afatinib, a pan-EGFR family kinase inhibitor. Oncotarget. 2014;5:5908–5919. doi: 10.18632/oncotarget.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strutz F, Zeisberg M, Ziyadeh FN, Yang CQ, Kalluri R, Müller GA, Neilson EG. Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int. 2002;61:1714–1728. doi: 10.1046/j.1523-1755.2002.00333.x. [DOI] [PubMed] [Google Scholar]
- 7.Ribatti D, Vacca A, Rusnati M, Presta M. The discovery of basic fibroblast growth factor/fibroblast growth factor-2 and its role in haematological malignancies. Cytokine Growth Factor Rev. 2007;18:327–334. doi: 10.1016/j.cytogfr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Tiong KH, Mah LY, Leong CO. Functional roles of fibroblast growth factor receptors (FGFRs) signaling in human cancers. Apoptosis. 2013;18:1447–1468. doi: 10.1007/s10495-013-0886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomlinson DC, Knowles MA. Altered splicing of FGFR1 Is associated with high tumor grade and stage and leads to increased sensitivity to FGF1 in bladder cancer. Am J Pathol. 2010;177:2379–2386. doi: 10.2353/ajpath.2010.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int J Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- 11.Preusser M, Berghoff AS, Berger W, Ilhan-Mutlu A, Dinhof C, Widhalm G, Dieckmann K, Wöhrer A, Hackl M, von Deimling A, et al. High rate of FGFR1 amplifications in brain metastases of squamous and non-squamous lung cancer. Lung Cancer. 2014;83:83–89. doi: 10.1016/j.lungcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Billottet C, Elkhatib N, Thiery JP, Jouanneau J. Targets of fibroblast growth factor 1 (FGF-1) and FGF-2 signaling involved in the invasive and tumorigenic behavior of carcinoma cells. Mol Biol Cell. 2004;15:4725–4734. doi: 10.1091/mbc.E04-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey JH, Bianchi F, Voshol J, Bonenfant D, Oakeley EJ, Hynes NE. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 14.Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, Miyazono K, Saitoh M. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. Embo J. 2011;30:783–795. doi: 10.1038/emboj.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/S1534-5807(01)00017-X. [DOI] [PubMed] [Google Scholar]
- 17.Weisser NE, Hall JC. Applicantions of single-chain variable fragment antibodies in therapeutics and diagnostis. Biotechnol Adv. 2009;27:502–520. doi: 10.1016/j.biotechadv.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Zohn IE, Li YQ, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 19.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Bio. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi YD, Felley-Bosco E, Marti TM, Orlowski K, Pruschy M, Stahel RA. Starvation-induced activation of ATM/Chk2/p53 signaling sensitizes cancer cells to cisplatin. BMC Cancer. 2012;12:571. doi: 10.1186/1471-2407-12-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong S, Khoo A, Wei J, Bowser RK, Weathington NM, Xiao S, Zhang L, Ma H, Zhao Y, Zhao J. Serum starvation regulates E-cadherin upregulation via activation of c-Src in non-small-cell lung cancer A549 cells. Am J Physiol Cell Physiol. 2014;307:C893–C899. doi: 10.1152/ajpcell.00132.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]