Abstract
The aim of the present study was to perform comparative genomics using gene expression profile datasets of mice and humans who had been treated with anticancer drugs, to determine the similarities and differences in the antitumor mechanisms in the two mammals. This involved data mining of antitumor gene expression regulation, and screening of genetic loci from experimental mouse models of antitumor targets, to provide a theoretical basis of drug design. Subsequently, 9 overlapping genes with opposite expression patterns were identified across mouse and human cell lines that were treated with a specific cyclin-dependent kinase 4/6 inhibitor, PD0332991. These genes included LIM homeobox 2, adenomedullin, bone marrow stromal cell antigen 1, caveolin 1, histone cluster 1 (HIST1) H2B family member C, HIST1 H3 family member F, low density lipoprotein-receptor related protein 11, prolyl 4-hydroxylase subunit α1 and torsin family 3 member A. In addition, the janus kinase-signal transducer and activator of transcription signaling pathway, Toll-like receptor signaling pathway, T cell receptor signaling pathway and the nucleotide-binding oligomerization domain-like receptor signaling pathway were identified as candidate pathways for explaining antitumor mechanisms.
Keywords: anticancer, transcriptomic, human, mouse
Introduction
Disorders of the cell cycle are closely associated with the occurrence and development of tumors and are regulated by complex regulatory networks, particularly those involving cyclin-dependent kinases (CDKs), including CDK1, CDK2, CDK3, CDK4, CDK5, CDK6 and CDK7 (1,2). Of these, CDK4/6 has been previously reported to be expressed in various types of cancer, including lung, oral and breast cancer, and hepatocellular carcinoma (3–6). The role of CDK4/6 in tumor cell proliferation is more important than other CDKs and has been established to be one of the essential signaling molecules involved in cell differentiation and metastasis (7). Aberrant CDK activity may lead to uncontrolled proliferation in many tumors, which suggests that inhibiting the activity of CDKs may have a therapeutic benefit. The CDK4/6 inhibitor, PD0332991, has been demonstrated to inhibit tumor cell cycle replication and proliferation in mice and rat animal models in addition to human cancer cell lines including MCF7 (8–11). It has become the prospect for the development of new antitumor drugs. Although PD0332991 is particularly effective in specific antitumor models that induced Rb-dependent cytostasis, identifying the underlying mechanistic pathways of CDK inhibitors is required to fully understand therapeutic responses and this has been investigated in a panel of breast cancer cell lines (12).
Microarray technology is widely used to investigate the biological mechanisms in response to treatment with PD0332991 in humans and mice (8–11). In the present study, a comparative genomics approach was used with relevant gene expression profile datasets of mice and humans that had undergone anticancer drug treatments. The similarities and differences in the antitumor mechanisms in the two mammals at the transcriptomic level were determined. This was achieved by data mining the process of antitumor gene expression regulation, and by screening of genetic loci in a mouse model of an antitumor target, to provide the theoretical basis of drug design.
Materials and methods
Microarray data collection and preprocessing
The microarray dataset of the anticancer efficiency of PD0332991 was obtained from the National Centre for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo). Datasets were reanalyzed if they met the following conditions: i) The data was genome-wide; ii) the dataset contained human and mouse samples; iii) the number of cases and controls in each dataset was ≥3; and iv) complete raw microarray data were available. Based on these criteria, the GSE40514 dataset [contributed by Choi et al (13)] was selected for reanalysis. This superseries is composed of two subseries including GSE40512 and GSE40513. In the dataset of GSE40512, human T-ALL cell line KOPTK1 cells were cultured in the presence of the CDK4/6 inhibitor PD 0332991 with 1 mM or vehicle (VO) for 48 h and the experiment was performed in triplicate. A total of 6 RNA samples (3 vehicle treated and 3 PD0332991 treated samples) were tested by using the platform of Affymetrix Human Genome U133 Plus 2.0 Array (HG-U133_Plus_2, GPL570). In the dataset of GSE40513, mouse breast cancer V720 cells were cultured in the presence of the CDK4/6 inhibitor PD 0332991 with 1 mM or vehicle (VO) for 24 h and the experiment was performed in triplicate. A total of 6 RNA samples (3 vehicle treated and 3 PD 0332991 treated samples) were tested by using the platform of Affymetrix Mouse Genome 430 2.0 Array (Mouse430_2, GPL1261).
R version 3.2.0 (www.r-project.org) and Bioconductor version 3.5 (www.bioconductor.org) were used for data preprocessing (14). The Robust Multichip Average (RMA) algorithm was used in the oligo package to normalize the raw expression data and to generate normalized gene expression intensity in human and mouse cell lines (15). Gene annotation of the probes of human and mouse cell lines was performed using custom written Python code version 2.7 (www.python.org). Probes with no gene annotation or ones that matched multiple gene symbols were removed. Then, the average expression value of multiple probe identities that matched to an official gene symbol was calculated, and this value represented the expression intensity of the corresponding gene symbol. Finally, 20,307 human gene symbols and 20,968 mouse gene symbols were identified. There were 13,976 homologous genes in both human and mouse cell lines. These 13,976 genes were selected for further analysis.
Differential expression gene analysis
Differential expression gene analysis was performed using R version 3.2.2 and the Bioconductor library. The empirical Bayes algorithm (eBayes) in the limma package was used to detect differentially expressed genes between PD0332991-treated and vehicle-treated samples in human and mouse cell lines (16). Fold-change (FC) was calculated as the mean gene expression value in PD0332991-treated samples divided by the mean gene expression value in the vehicle-treated samples. Upregulated genes were considered as a logarithmic transformed fold-changes (log2(FC))≥log2(1.5) and a false discovery rate (FDR) was the adjusted P-value of ≤0.05. Downregulated genes were considered as log2(FC)≤-log2(1.5) and a FDR P-value of ≤0.05.
Gene set enrichment analysis
Java Gene set Enrichment Analysis (GSEA) version 2.2.2 (http://software.broadinstitute.org/gsea/index.jsp) was used for human and mouse samples. The curated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway software version 5.1 (http://www.genome.jp/kegg) was chosen as the gene sets database to perform the enrichment analysis. Gene sets with <15 or >500 genes were excluded. Phenotype label was set as breast cancer vs. control. The t-statistic mean of the genes was calculated for each KEGG pathway using a permutation test 1,000 times. The upregulated pathways were considered as a normalized enrichment score (NES) >0 and the downregulated pathways were considered as a NES <0. Pathways with a FDR P-value of ≤0.05 were considered to be significantly enriched. A Venn diagram was produced using InteractiVenn (www.interactivenn.net) to demonstrate the enriched KEGG pathways in the human and mouse groups (17).
Results and Discussion
Differentially expressed genes in human T-ALL cells and mouse breast cancer cells
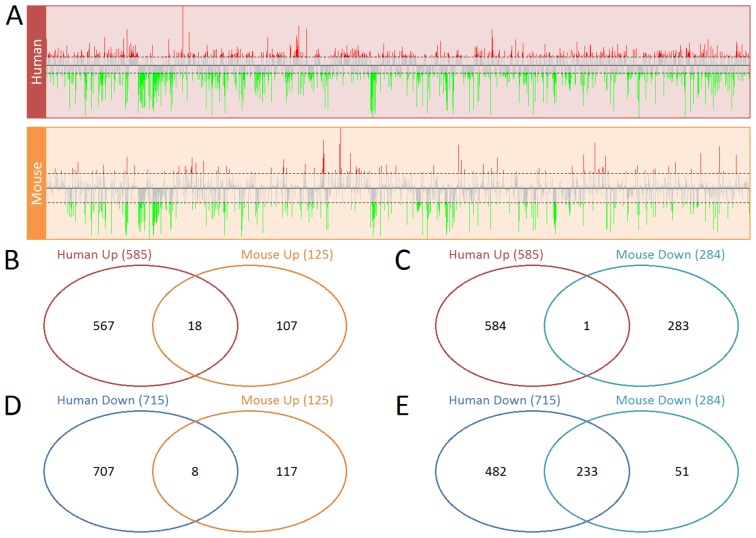
In total, there were 13,976 homologous genes expressed in human and mouse gene expression profiles. Volcano plots are presented in Fig. 1. A greater number of dysregulated genes were identified in the human group compared with the mouse group. A total of 585 upregulated genes and 715 downregulated genes of PD0332991-treated samples were identified in human KOPTK1 cells. By comparison, 125 upregulated genes and 284 downregulated genes were identified in mouse V720 cells. There were 233 common downregulated genes in both human and mouse samples, which was presented in the Venn diagram in Fig. 2. However, there were only 18 upregulated genes that overlapped in these two groups (Fig. 2). The overlapping genes which were significantly up or downregulated across human and mouse cells are presented in Tables I and II.
Figure 1.
Volcano plots of the gene expression profiles of (A) human T-lymphoblast acute lymphoblastic leukemia cells and (B) mouse breast cancer cells. The red points represent upregulated genes and the green points represent downregulated genes. The vertical dotted grey lines represents the log(FC) cutoff and the horizontal grey line represents the logarithmic transformed FDR P-value cutoff. FC, fold-change; FDR, false discovery rate.
Figure 2.
Differentially expressed genes in human T-lymphoblast acute lymphoblastic leukemia cells and mouse breast cancer cells. (A) The log (fold-change) of the dysregulated genes in human and mouse cells. Red lines represent the upregulated genes and the green lines represent the downregulated genes. All genes were sorted by name. Venn diagram of (B) upregulated genes in human and mouse cells, (C) upregulated genes in human cells and downregulated genes in mouse cells, (D) downregulated genes in human cells and the upregulated genes in mouse cells and (E) downregulated genes in human and mouse cells.
Table I.
Enriched Kyoto Encyclopedia of Genes and Genomes pathway analysis in human T-lymphoblast acute lymphoblastic leukemia cells.
| A, Upregulated | ||
|---|---|---|
| Pathway | NES | FDR |
| Allograft rejection | 2.22 | <0.001 |
| Cytokine-cytokine receptor interaction | 2.18 | <0.001 |
| JAK-STAT signaling pathway | 2.10 | <0.001 |
| Toll-like receptor signaling pathway | 2.06 | <0.001 |
| Type I diabetes mellitus | 2.04 | 0.001 |
| Lysosome | 2.00 | 0.001 |
| Autoimmune thyroid disease | 2.00 | 0.001 |
| Intestinal immune network for IgA production | 1.96 | 0.001 |
| Hematopoietic cell lineage | 1.92 | 0.003 |
| T cell receptor signaling pathway | 1.85 | 0.007 |
| Asthma | 1.82 | 0.009 |
| Natural killer cell mediated cytotoxicity | 1.78 | 0.014 |
| O-glycan biosynthesis | 1.69 | 0.032 |
| Leishmania infection | 1.70 | 0.033 |
| SNARE interactions in vesicular transport | 1.65 | 0.050 |
| B, Downregulated | ||
| Pathway | NES | FDR |
| Cell cycle | −2.49 | <0.001 |
| DNA replication | −2.46 | <0.001 |
| Oocyte meiosis | −2.26 | <0.001 |
| Mismatch repair | −2.14 | <0.001 |
| Homologous recombination | −2.12 | <0.001 |
| Pyrimidine metabolism | −1.98 | 0.001 |
| Base excision repair | −1.98 | 0.001 |
| Nucleotide excision repair | −1.94 | 0.001 |
| Progesterone-mediated oocyte maturation | −1.88 | 0.003 |
| One carbon pool by folate | −1.84 | 0.005 |
| Glycolysis/gluconeogenesis | −1.68 | 0.034 |
JAK-STAT, janus kinase-signal transducer and activator of transcription; SNARE, soluble N-ethylmaleimide-sensitive factor activating protein receptor; FDR, false discovery rate; NES, normalized enrichment score.
Table II.
Enriched Kyoto Encyclopedia of Genes and Genomes pathway analysis in mouse breast cancer cells.
| A, Upregulated | ||
|---|---|---|
| Pathway | NES | FDR |
| Cytosolic DNA-sensing pathway | 2.27 | <0.001 |
| NOD-like receptor signaling pathway | 2.17 | <0.001 |
| Hematopoietic cell lineage | 1.98 | 0.003 |
| Focal adhesion | 1.99 | 0.004 |
| Toll-like receptor signaling pathway | 1.83 | 0.018 |
| Leishmania infection | 1.85 | 0.019 |
| Prion diseases | 1.84 | 0.020 |
| Cytokine-cytokine receptor interaction | 1.79 | 0.024 |
| JAK-STAT signaling pathway | 1.77 | 0.026 |
| ECM-receptor interaction | 1.77 | 0.028 |
| Glycosaminoglycan degradation | 1.73 | 0.037 |
| Type I diabetes mellitus | 1.72 | 0.040 |
| Complement and coagulation cascades | 1.70 | 0.045 |
| Lysosome | 1.69 | 0.046 |
| B, Downregulated | ||
| Pathway | NES | FDR |
| Cell cycle | −2.71 | <0.001 |
| DNA replication | −2.67 | <0.001 |
| Homologous recombination | −2.47 | <0.001 |
| Oocyte meiosis | −2.41 | <0.001 |
| Spliceosome | −2.37 | <0.001 |
| Nucleotide excision repair | −2.32 | <0.001 |
| Mismatch repair | −2.27 | <0.001 |
| Base excision repair | −2.20 | <0.001 |
| Progesterone-mediated oocyte maturation | −2.11 | <0.001 |
| RNA degradation | −2.10 | <0.001 |
| Pyrimidine metabolism | −1.99 | 0.001 |
| RNA polymerase | −1.71 | 0.019 |
| Cysteine and methionine metabolism | −1.62 | 0.043 |
JAK-STAT, janus kinase-signal transducer and activator of transcription; NOD, nucleotide-binding oligomerization domain; ECM, extracellular matrix; FDR, false discovery rate; NES, normalized enrichment score.
To investigate the anticancer efficiency of PD0332991, 9 genes with diverse regulatory patterns between mouse and human were identified (Fig. 3). In the PD0332991 group, LIM homeobox 2 (LHX2) was overexpressed in humans compared with the vehicle group, whereas in mice, LHX2 expression was reduced (Fig. 3). Dysregulated expression of LHX2 in human cancers has been previously reported (18). In addition, cancer-associated epigenetic alterations were significantly associated with LHX2 regulation, including DNA methylation and microRNA regulation (19–22). In the present study, adrenomedullin (ADM), bone marrow stromal cell antigen 1 (BST1), caveolin 1 (CAV1), histone cluster 1 H2B family member C (HIST1H2BC), histone cluster 1 H3 family member F (HIST1H3F), low density lipoprotein-receptor related protein 11 (LRP11), prolyl 4-hydroxylase subunit α1 (P4HA1) and torsin family 3 member A (TOR3A) were downregulated in human KOPTK1 cells, and were upregulated in mouse V720 cells. HIST1H3F demonstrated the largest difference between PD0332991- and vehicle-treated samples in human and mouse cell lines (log2(FC)=−1.77, FDR=5.99E-6 in human; log2(FC)=1.21, FDR=8.20E-4 in mice) from the 8 genes investigated. The histone HIST1H3F has been previously identified as a biomarker for predicting cancer risk (23). In addition, the mean log2 gene expression value of HIST1H2BC in humans was ~4. However, the mean value in mice was ~12. This suggested that there was a large difference between human and mouse cell lines. HIST1H2BC is one of the most significant susceptible locus in the transcriptome study of schizophrenia, which has been also reported to be a suitable reference gene in non-small cell lung cancer (24,25). The association between ADM and epithelial-mesenchymal transition, and the underlying signaling pathways in intrahepatic cholangiocellular carcinoma has been previously elucidated (26). The overexpression of ADM in tumor cells may accelerate breast cancer bone metastasis, which suggests that it may be a potential target for therapeutic intervention against bone metastases (27). The association between movement disorders and genetic polymorphisms of BST1 has been confirmed in different populations, including Parkinson's and Alzheimer's disease (28–30). BST1 has been also investigated in a previous study on the drug resistance of tumors, including anticancer drugs for artesunate resistance (31). CAV1 may have a tumor suppression role in breast cancer and its expression is regulated by CpG island shore methylation (32). LRP11 is a member of the low-density lipoprotein receptor family, and has a potential role in mediating cellular drug uptake (33). One of the isoforms of P4HA, P4HA1, has an important role in prostate cancer progression and is associated with microRNA-124 (34). TOR3A functions with the gene encoding the Ras-related protein RAB1B, which regulates molecular transport, protein trafficking and developmental disorders (35).
Figure 3.
Expression profiles of nine target genes in human T-lymphoblast acute lymphoblastic leukemia cells and mouse breast cancer cells. The dark color bars represent the gene expression value in the PD0332991-treatment group and the light color bars represent the gene expression values in vehicle-treatment group. Error bars represent standard deviation of the mean expression values of each gene of three replicates. LHX2, LIM homeobox 2; ADM, adenomedullin; BST1, bone marrow stromal cell antigen 1; CAV1, caveolin 1; HIST1H2BC, histone cluster 1 H2B family member C; HIST1H3F, histone cluster 1 H3 family member F; LRP11, low density lipoprotein-receptor related protein 11; P4HA1, prolyl 4-hydroxylase subunit α1; TOR3A, torsin family 3 member A.
Anticancer efficiency of PD0332991 on multiple pathways levels
GSEA analysis was performed to identify the signaling pathways that were significantly associated with anticancer effect of PD0332991. The findings of the present study revealed that there were 15 upregulated pathways and 11 downregulated pathways in PD0332991-treated human KOPTK1 cells. In addition, there were 14 upregulated pathways and 13 downregulated pathways in PD0332991-treated mouse V720 cells (Fig. 4A and B). There were 7 common upregulated pathways and 9 common downregulated pathways in the human and mouse samples. However, oppositely dysregulated pathways in the two groups were not identified. The details of these pathways are presented in Tables I and II. The downregulated pathways in human and mouse cell lines were primarily associated with cell cycle, DNA replication and nucleotide metabolism. The upregulated pathways in the two groups treated with PD0332991 were involved in various intracellular signaling pathways, including the JAK-STAT, Toll-like receptor, T cell receptor and nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathways.
Figure 4.
The comparison of enriched Kyoto Encyclopedia of Genes and Genomes pathways analysis in response to PD0332991 treatment between human and mouse. (A) Upregulated pathways in human T-lymphoblast acute lymphoblastic leukemia cells (red) and mouse breast cancer cells (orange). (B) Downregulated pathways in human (blue) and mouse (cyan) cells.
It has been previously reported that the JAK/STAT pathway may be aberrantly activated in solid tumors including breast cancer, whose activation in malignant and nonmalignant cells contributes to the cancer pathogenesis and therapeutic response (36,37). Toll-like receptor (TLR)3 signaling is an integral component of solid tumors, and inhibition of this pathway may increase the effectiveness of current treatments of breast cancer in combination with anticancer drugs (38). The TLR8 signaling pathway has been associated with the functional regulation of tumor-specific ∆γ T cells, which are important contributors to innate immunity against cancer (39). Gene polymorphisms of NOD1/caspase recruitment domain (CARD)4 and NOD2/CARD15 may be associated with altered risk of diverse malignancies, including breast cancer (40).
In conclusion, the present study identified 9 overlapping genes with opposite expression patterns across PD0332991-treated human T-ALL cells and mouse breast cancer cells. In addition, various cellular signaling pathways were identified to be closely associated with the efficacy of anticancer drugs. Therefore, this data may aid future breast cancer drug screening and design.
References
- 1.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 2.Björklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenperä J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 3.Acevedo M, Vernier M, Mignacca L, Lessard F, Huot G, Moiseeva O, Bourdeau V, Ferbeyre G. A CDK4/6-dependent epigenetic mechanism protects cancer cells from PML-induced senescence. Cancer Res. 2016;76:3252–3264. doi: 10.1158/0008-5472.CAN-15-2347. [DOI] [PubMed] [Google Scholar]
- 4.O'Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 5.Yu BD, Becker-Hapak M, Snyder EL, Vooijs M, Denicourt C, Dowdy SF. Distinct and nonoverlapping roles for pRB and cyclin D: Cyclin-dependent kinases 4/6 activity in melanocyte survival. Proc Natl Acad Sci USA. 2003;100:14881–14886. doi: 10.1073/pnas.2431391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baughn LB, Di Liberto M, Wu K, Toogood PL, Louie T, Gottschalk R, Niesvizky R, Cho H, Ely S, Moore MA, Chen-Kiang S. A novel orally active small molecule potently induces G1 arrest in primary myeloma cells and prevents tumor growth by specific inhibition of cyclin-dependent kinase 4/6. Cancer Res. 2006;66:7661–7667. doi: 10.1158/0008-5472.CAN-06-1098. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YX, Sicinska E, Czaplinski JT, Remillard SP, Moss S, Wang Y, Brain C, Loo A, Snyder EL, Demetri GD, et al. Antiproliferative effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo. Mol Cancer Ther. 2014;13:2184–2193. doi: 10.1158/1535-7163.MCT-14-0387. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Boyson CA, Di Liberto M, Huang X, Hannah J, Dorn DC, Moore MA, Chen-Kiang S, Zhou P. CDK4/6 inhibitor PD 0332991 sensitizes acute myeloid leukemia to cytarabine-mediated cytotoxicity. Cancer Res. 2015;75:1838–1845. doi: 10.1158/0008-5472.CAN-14-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, Toogood PL. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 10.Menu E, Garcia J, Huang X, Di Liberto M, Toogood PL, Chen I, Vanderkerken K, Chen-Kiang S. A novel therapeutic combination using PD 0332991 and bortezomib: Study in the 5T33MM myeloma model. Cancer Res. 2008;68:5519–5523. doi: 10.1158/0008-5472.CAN-07-6404. [DOI] [PubMed] [Google Scholar]
- 11.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 13.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H, Sicinski P. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. 3.0. 1. [Google Scholar]
- 15.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou F, Gou S, Xiong J, Wu H, Wang C, Liu T. Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma. Mol Biol Rep. 2014;41:8163–8167. doi: 10.1007/s11033-014-3716-2. [DOI] [PubMed] [Google Scholar]
- 19.de Groot JS, Pan X, Meeldijk J, van der Wall E, van Diest PJ, Moelans CB. Validation of DNA promoter hypermethylation biomarkers in breast cancer-a short report. Cell Oncol (Dordr) 2014;37:297–303. doi: 10.1007/s13402-014-0189-1. [DOI] [PubMed] [Google Scholar]
- 20.Shi X, Zhan L, Xiao C, Lei Z, Yang H, Wang L, Zhao J, Zhang HT. miR-1238 inhibits cell proliferation by targeting LHX2 in non-small cell lung cancer. Oncotarget. 2015;6:19043–19054. doi: 10.18632/oncotarget.4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MS, Lee J, Oh T, Moon Y, Chang E, Seo KS, Hoehn BD, An S, Lee JH. Genome-wide identification of OTP gene as a novel methylation marker of breast cancer. Oncol Rep. 2012;27:1681–1688. doi: 10.3892/or.2012.1691. [DOI] [PubMed] [Google Scholar]
- 22.Rauscher GH, Kresovich JK, Poulin M, Yan L, Macias V, Mahmoud AM, Al-Alem U, Kajdacsy-Balla A, Wiley EL, Tonetti D, Ehrlich M. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer. 2015;15:816. doi: 10.1186/s12885-015-1777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirisola V, Mora R, Esposito AI, Guastini L, Tabacchiera F, Paleari L, Amaro A, Angelini G, Dellepiane M, Pfeffer U, Salami A. A prognostic multigene classifier for squamous cell carcinomas of the larynx. Cancer Lett. 2011;307:37–46. doi: 10.1016/j.canlet.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Sanders AR, Göring HH, Duan J, Drigalenko EI, Moy W, Freda J, He D, Shi J. MGS and Gejman PV: Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22:5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saviozzi S, Cordero F, Lo Iacono M, Novello S, Scagliotti GV, Calogero RA. Selection of suitable reference genes for accurate normalization of gene expression profile studies in non-small cell lung cancer. BMC Cancer. 2006;6:200. doi: 10.1186/1471-2407-6-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou C, Zheng Y, Li L, Zhai W, Li R, Liang Z, Zhao L. Adrenomedullin promotes intrahepatic cholangiocellular carcinoma metastasis and invasion by inducing epithelial-mesenchymal transition. Oncol Rep. 2015;34:610–616. doi: 10.3892/or.2015.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siclari VA, Mohammad KS, Tompkins DR, Davis H, McKenna CR, Peng X, Wessner LL, Niewolna M, Guise TA, Suvannasankha A, Chirgwin JM. Tumor-expressed adrenomedullin accelerates breast cancer bone metastasis. Breast Cancer Res. 2014;16:458. doi: 10.1186/s13058-014-0458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung SJ, Jung Y, Hong M, Kim MJ, You S, Kim YJ, Kim J, Song K. Alzheimer's disease and Parkinson's disease genome-wide association study top hits and risk of Parkinson's disease in Korean population. Neurobiol Aging. 2013;34(2695):e1–e7. doi: 10.1016/j.neurobiolaging.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Saad M, Lesage S, Saint-Pierre A, Corvol JC, Zelenika D, Lambert JC, Vidailhet M, Mellick GD, Lohmann E, Durif F, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population. Hum Mol Genet. 2011;20:615–627. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- 30.UK Parkinson's Disease Consortium1; Wellcome Trust Case Control Consortium II, corp-author. Spencer CC, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, Barker RA, Bellenguez C, et al. Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sertel S, Eichhorn T, Sieber S, Sauer A, Weiss J, Plinkert PK, Efferth T. Factors determining sensitivity or resistance of tumor cell lines towards artesunate. Chem Biol Interact. 2010;185:42–52. doi: 10.1016/j.cbi.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Rao X, Evans J, Chae H, Pilrose J, Kim S, Yan P, Huang RL, Lai HC, Lin H, Liu Y, et al. CpG island shore methylation regulates caveolin-1 expression in breast cancer. Oncogene. 2013;32:4519–4528. doi: 10.1038/onc.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung NS, Wasan KM. Potential role of the low-density lipoprotein receptor family as mediators of cellular drug uptake. Adv Drug Deliv Rev. 2004;56:1315–1334. doi: 10.1016/j.addr.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Chakravarthi BV, Pathi SS, Goswami MT, Cieślik M, Zheng H, Nallasivam S, Arekapudi SR, Jing X, Siddiqui J, Athanikar J, et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5:6654–6669. doi: 10.18632/oncotarget.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Úbeda R, García-Vázquez FA, Romar R, Gadea J, Muñoz M, Hunter RH, Coy P. Oviductal transcriptome is modified after insemination during spontaneous ovulation in the sow. PLoS One. 2015;10:e0130128. doi: 10.1371/journal.pone.0130128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, McKenney AS, Papalexi E, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov. 2015;5:316–331. doi: 10.1158/2159-8290.CD-14-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintás-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: Mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933–1940. doi: 10.1158/1078-0432.CCR-12-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz AL, Dickerson E, Dagia N, Malgor R, McCall KD. TLR signaling inhibitor, phenylmethimazole, in combination with tamoxifen inhibits human breast cancer cell viability and migration. Oncotarget. 2016 doi: 10.18632/oncotarget.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng G, Wang HY, Peng W, Kiniwa Y, Seo KH, Wang RF. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity. 2007;27:334–348. doi: 10.1016/j.immuni.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Kutikhin AG. Role of NOD1/CARD4 and NOD2/CARD15 gene polymorphisms in cancer etiology. Hum Immunol. 2011;72:955–968. doi: 10.1016/j.humimm.2011.06.003. [DOI] [PubMed] [Google Scholar]