Abstract
The development of targeted tyrosine kinase inhibitors (TKIs) has succeeded in altering the course of chronic myeloid leukemia (CML). However, a number of patients have failed to respond or experienced disease relapse following TKI treatment. Proviral integration site for moloney murine leukemia virus-1 (PIM-1) is a serine/threonine kinase that participates in regulating apoptosis, cell cycle, signal transduction and transcriptional pathways, which are associated with tumor progression, and poor prognosis. SMI-4a is a selective PIM-1 kinase inhibitor that inhibits PIM-1 kinase activity in vivo and in vitro. The present study aimed to explore the mechanism underlying the antitumor effect of SMI-4a in K562 and imatinib-resistant K562 (K562/G) cell lines. It was demonstrated that SMI-4a inhibited the proliferation of K562 and K562/G cells using a WST-8 assay. The Annexin V-propidium iodide assay demonstrated that SMI-4a induced apoptosis of K562 and K562/G cells in a dose-, and time-dependent manner. Furthermore, Hoechst 33342 staining was used to verify the apoptosis rate. The clone formation assay revealed that SMI-4a significantly inhibited the colony formation capacity of K562 and K562/G cells. Western blot analysis demonstrated that SMI-4a decreased phosphorylated (p)-Ser9-glycogen synthase kinase (GSK) 3β/pGSK3β and inhibited the translocation of β-catenin. In addition, the downstream gene expression of apoptosis regulator Bax and poly(ADP-ribose) polymerase-1 was upregulated, and apoptosis regulator Bcl-2 and Myc proto-oncogene protein expression levels were downregulated. Immunofluorescence results demonstrated changes in the expression level of β-catenin in the plasma and nucleus. The results of the present study suggest that SMI-4a is an effective drug to use in combination with current chemotherapeutics for the treatment of imatinib-resistant CML.
Keywords: SMI-4a, chronic myeloid leukemia, cell apoptosis, cell cycle, GSK 3β
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell-derived disorder, which is characterized by the presence of the fusion gene BCR-ABL 1 encoding a tyrosine kinase whose constitutive activity is essential for CML emergence, maintenance, and progression (1,2). Imatinib mesylate, a target tyrosine kinase inhibitor (TKI), has been shown to exert a significant and lasting response in chronic-phase CML patients. However, 20–30% of patients fail to respond suboptimally or experience disease relapse after an initial response with imatinib (3). Despite the development of second-generation TKIs, such as dasatinib and nilotinib, fraction of CML patients in chronic phase and a percentage of patients in advanced phase are either primary refractory to TKIs or eventually develop resistance (4).
Proviral integration site for moloney murine leukemia virus (PIM) kinases belongs to the active serine/threonine kinase family (5). PIM-1 participates in regulating apoptosis, cell cycle, signal transduction, and transcriptional pathways, which are linked to cell survival (6). Accumulating preclinical studies have shown that overexpression of PIM-1 in cancer cells can substantially contribute to malignant transformation, tumor progression, and poor prognosis (7,8). Targeting PIM-1 sensitized prostate and colon cancer cells (9,10) and improving the efficacy of Akt inhibitors and epidermal growth factor receptor (EGFR) are the primary objectives of targeted therapies in prostate cancer (9).
In hematological malignancies, PIM-1 is overexpressed in acute leukemia (5), T-cell lymphoma (11), mantle cell lymphoma (MCL) (12), and chronic lymphocytic leukemia (CLL) (13). Being a small molecular selective PIM kinase inhibitor, SMI-4a has been found to exhibit cytotoxicity activity in 25 leukemic cell-lines, including human and murine pre-T-LBL cells, myeloid leukemia cells expressing BCR/ABL, PML/RARA, AML/ETO, and mutant FLT3, and human pre-B-LBL cells (11). Knockdown of PIM-1 kinases impairs the survival of resistant forms of FLT3- and BCR-ABL-transformed leukemia cells (5). The exact anti-leukemia role of PIM-1 kinase inhibitor as a therapeutic target in CML is not clearly understood. In this study, we investigated the cytotoxicity and mechanism of action of SMI-4a on human CML cell-line K562 and imatinib-resistant K562 cell line (K562/G). Our investigation establishes the utility of PIM-1 kinase inhibitiors in the treatment of imatinib-resistant CML, which may provide a new therapeutic approach.
Materials and methods
Cell isolation and cell culture
Bone marrow mononuclear cells (BMMCs) from patients with CML and peripheral blood mononuclear cells (PBMCs) from healthy individuals were enriched by Ficoll-Hypaque density gradient centrifugation. K562 and K562/G (imatinib induced resistant) cell lines were provided by Quentin Liu lab, the Third Affiliated Hospital, Sun Yat-sen University. All primary cells, K562 and K562/G cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Hyclone) at 37°C in 5% CO2. SMI-4a was obtained from Sigma (St. Louis, MO, USA). CML patient and healthy individual samples were obtained with the written informed consent in accordance with the Declaration of Helsinki and the Approval by the Medical Ethical Committee of the Third Affiliated Hospital of Sun Yat-sen University.
Cell proliferation and viability
Cell proliferation was evaluated by WST-8 assay using a cell counting kit (Dojindo Laboratories, Gaithersburg, MD) according to the manufacturer's protocols. Briefly, 1×104 cells were seeded into 96-well plates, respectively. The cells were treated with increasing concentrations of SMI-4a and incubated at 37°C in 5% CO2 for 24 and 48 h. Subsequently, 10 µl WST-8 solutions were added to each well and incubated at 37°C for 4 h. The plates were read at 450 nm using a microplate reader.
Cell cycle analysis
Cells were collected, fixed, and resuspended in phosphate-buffered saline containing 100 µg/ml RNaseA, 0.2% Triton X-100, and 50 µg/ml propidium iodide (PI; all Sigma). Cell cycle was analyzed by flow cytometry and data was analyzed using Modifit LT for Mac v2.0 software (BD Biosciences, Franklin Lakes, NJ, USA).
Colony formation assay
K562 and K562/G cells were incubated in methylcellulose culture in triplicates. Briefly, 1 ml culture mixture containing 2×103 cells, 0.9% methylcellulose (R&D Systems, Inc., Minnneapolis, MN, USA), and various concentrations of SMI-4a was plated in each plate and incubated at 37°C in 5% CO2 for 2 weeks. The colonies (>40 cells) were evaluated by direct counting using an inverted microscope.
Apoptosis analysis using flow cytometry and microscopy
Apoptosis was evaluated using Annexin V-PI binding assay (KeyGENBioTECH, Nanjing, Jiangsu, China) according to the manufacturer's protocols. Cells treated with 0, 20, 40, and 80 µM SMI-4a for 24 and 48 h were collected and stained with Annexin V-PI for 15 min at 37°C in the dark. Apoptosis analysis was conducted using flow cytometry and the data was analyzed using FlowJo software version 7.6 (FlowJo, LLC., Ashland, OR, USA).
Hoechst 33342 (Sigma) was used to examine nuclear fragmentation of apoptosis cells. Cells were harvested and stained with Hoechst 33342 (10 µg/ml) for 15 min, and the slides were viewed by a fluorescence microscope (IX71S1F3; Olympus Corp., Tokyo, Japan). Nuclei of apoptosis and normal cells were counted in ten random areas per coverslip (at least 100 cells were counted). Data was obtained from the three independent experiments.
Western blot analysis
K562 and K562/G cellular proteins were isolated using lysis buffer. The cell nucleus and plasma proteins were extracted by The Nuclear and Cytoplasmic Protein Extraction kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China). Protein concentrations were determined using Bradford's method. 40 µg proteins were separated on 8 to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% bovine serum albumin (BSA; MP Biomedicals, Santa Ana, California, USA) for 1 h at room temperature and then incubated PIM-1 (1:1,000; cat. no. 3247), Bcl-2 (1:1,000; cat. no. 4223), Bax (1:1,000; cat. no. 5023), cleaved poly (ADP-ribose) polymerase (PARP) (1:1,000; cat. no. 9532), c-Myc (1:1,000; cat. no. 5605), glycogen synthase kinase 3β (GSK-3β) (1:1,000; cat. no. 9315), p-GSK-3β (Ser9) (1:1,000; cat. no. 9322), β-catenin (1:1,000; cat. no. 8480), H3 (1:1,000; cat. no. 4499) and β-actin (1:2,000; cat. no. 12262) antibody (Cell Signaling Technology Corp, Beverly, MA). Subsequently, the membranes were incubated with HRP conjugate goat anti rabbit IgG (H + L) (1:5,000; cat. no. SA00001-2) and HRP conjugate goat anti mouse IgG(H + L) (1:5,000; cat. no. SA00001-1, Proteintech Group, Rosemont, Pennsylvania, USA) and detected using enhanced chemiluminescence reagents (Sigma) according to the manufacturer's protocols.
Immunofluorescence analysis
1×105 cells were plated onto glass slides and fixed with 4% paraformaldehyde (Sigma) and permeabilized in 0.1% Triton X-100. After blocking with phosphate buffer saline (PBS) containing 10% FBS, the cells were incubated with monoclonal antibodies against β-catenin (1:50; cat. no. 8480; Cell Signaling Technology Corp, Beverly, MA). Slides were then stained with Alexa Fluor 594 secary antibody (1:100; cat. no. R37119; Invitrogen, Carlsbad, California, USA). Images were obtained using a fluorescence microscope (IX71S1F3; Olympus Corp.) at magnification, ×400.
Statistical analysis
We used SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA) to analysis experiment data which presented as mean ± standard deviation.
One-way analysisof variance was performed to compare groups, followed by Fisher's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
SMI-4a induces cell cycle arrest in S phase
In order to determine whether SMI-4a could induce cell cycle arrest, the effect of SMI-4a on the CML cell cycle distribution was analyzed using PI staining. After the cells were treated with different concentrations of SMI-4a for 24 and 48 h, the portion of cells in G2/M phase decreased, while the portion of cells in S phase increased (Fig. 2A). Treatment of K562 cells with 80 µM SMI-4a for 48 h resulted in a decrease in the portion of cells in G2/M phase from 5.57±0.31 to 0.43±0.002%, and increase in the portion of cells in S phase from 51.66±2.37 to 62.57±3.54%. Compared to the control group, the S phase cells of 80 µM group had increased significantly (P<0.05) (Fig. 2B). The similar effect was showed on K562/G cells. Treatment of K562/G cells with 80 µM WSMI-4a for 48 h resulted in a decrease in the portion of cells in G2/M phase from 8.12±0.43 to 0.1±0.006%, and increase in the portion of cells in S phase from 42.78±2.38 to 60.72±3.41% (P<0.05) (Fig. 2C).
Figure 2.
SMI-4a induces S phase arrest in cell cycle of CML cells. (A) K562 and K562/G cells were stained with PI and subjected to flow cytometric analysis. Data shown is representative of three independent experiments. (B) K562 and (C) K562G cells data analysis revealed the proportion of cells in each phase of the cell cycle. Data is presented as mean ± SEM; *P<0.05 vs. 0 µM group. PI, propidium iodide; SEM, standard error of mean.
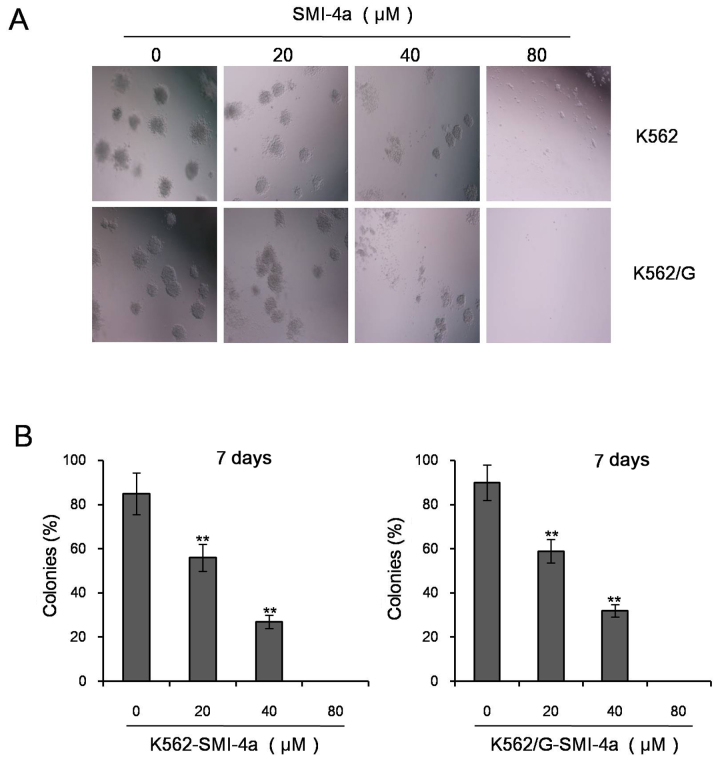
SMI-4a reduces colony formation capacity in CML cells
In order to identify the effect of SMI-4a on colony formation in CML cells, K562 and K562/G cells treated with various concentrations of SMI-4a were incubated in methylcellulose culture for 1 week. The results have demonstrated that SMI-4a suppressed colonies in both the cells in a dose-dependent manner. Compared to the control group, colonies were significantly decreased at the concentration of 20 µM in K562 and K562/G cells (P<0.05). It suggesting that SMI-4a significantly inhibited the colony formation capacity in CML cells (Fig. 3).
Figure 3.
SMI-4a reduces colony formation capacity of CML cells. (A) K562 and K562/G cells, treated with various concentrations of SMI-4a, were incubated in methylcellulose culture for 1 week and observed under microscope (magnification, ×200). (B) Analysis of K562 and K562/G cells in three independent experiments. Data is presented as mean ± SEM, **P<0.01 vs. 0 µM group. SEM: standard error of mean.
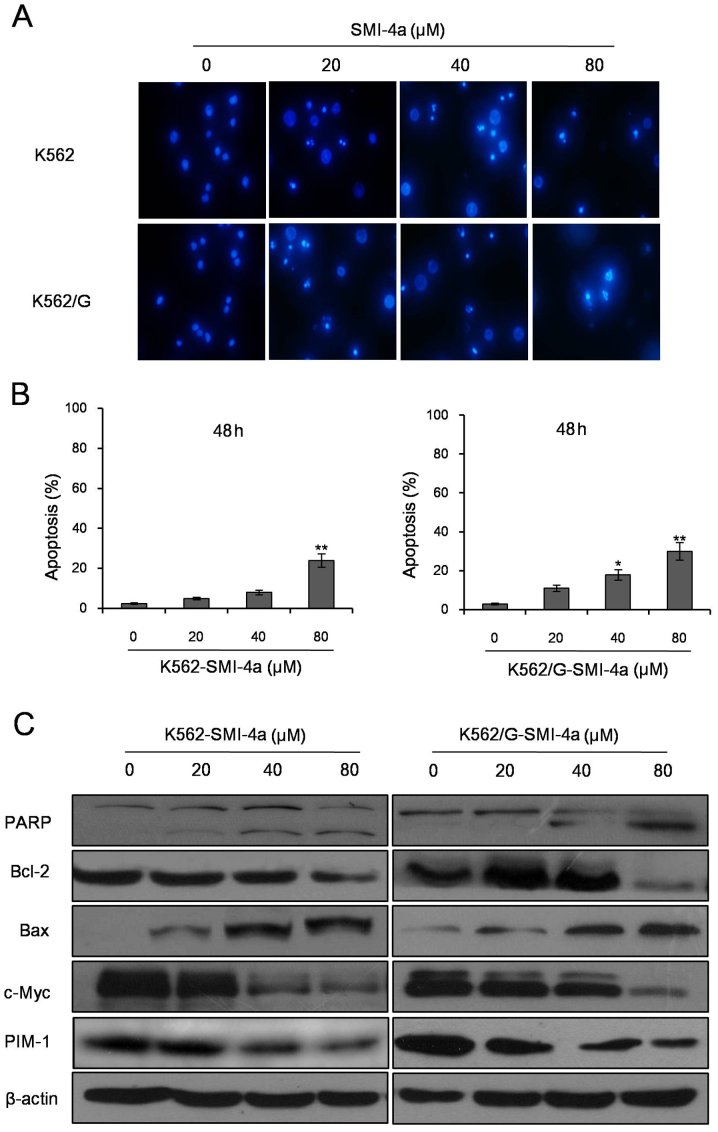
SMI-4a induces apoptosis in CML cells
Annexin V-PI double staining assay showed that SMI-4a induced apoptosis of CML cells in both dose- and time-dependent manners (Fig. 4A). Flow cytometry analysis showed that the total apoptosis rates were 15.34±1.74 and 28.59±2.84% in K562 cells treated with SMI-4a at 80 µM for 24 and 48 h (Fig. 4B). However, in K562/G cells treated with SMI-4a, the total apoptosis rates were 19.12±2.03 and 32.59±3.49% at the concentrations of 80 µM for 24 and 48 h (Fig. 4B). Next, K562 and K562/G cells were stained with Hoechst 33342 dye after exposure to SMI-4a for 48 h and observed under a fluorescence microscope. The results showed that the nuclei of untreated cells were circular, while the nuclei of cells treated with SMI-4a were condensation or ruptured (Fig. 5A and B). Compared to the control group, when the concentration of SMI-4a were 80 µM in K562 cells and 40 µM in K562/G cells, the cells apoptosis rates were significantly increased (P<0.05). In order to verify the apoptosis on protein level, the expression levels of apoptosis-related proteins were measured using western blot (Fig. 5C). The results showed that SMI-4a inhibit the expression of PIM-1 in dose-dependent manner in K562 and K562/G cells. As the concentration of SMI-4a increased, the expression of cleaved PARP and Bax increased as well as the expression of Bcl-2 and c-Myc decreased.
Figure 4.
SMI-4a induces apoptosis of CML cells by Annexin V-PI double staining. (A) Apoptotic cells were measured by Annexin V-PI double staining assay. Data shown is a representative of three independent experiments. (B) Analysis of K562 and K562/G cells in three independent experiments. Data is presented as mean ± SEM, *P<0.05 vs. 0 µM group, **P<0.01 vs. 0 µM group. SEM, standard error of mean.
Figure 5.
SMI-4a induces apoptosis of CML cells by Hoechst 333342 dye staining. (A) K562 and K562/G cells were stained with Hoechst 333342 dye after exposure to SMI-4a for 48 h and examined under a fluorescence microscope (magnification, ×400). (B) Statistical presentation of the apoptosis findings in three independent experiments. (C) K562 and K562/G cells were treated with different concentrations of SMI-4a for 48 h and the expression of apoptosis-related proteins was determined using western blot analysis. Data is presented as mean ± SEM, *P<0.05 vs. 0 µM group, **P<0.01 vs. 0 µM group. SEM, standard error of mean.
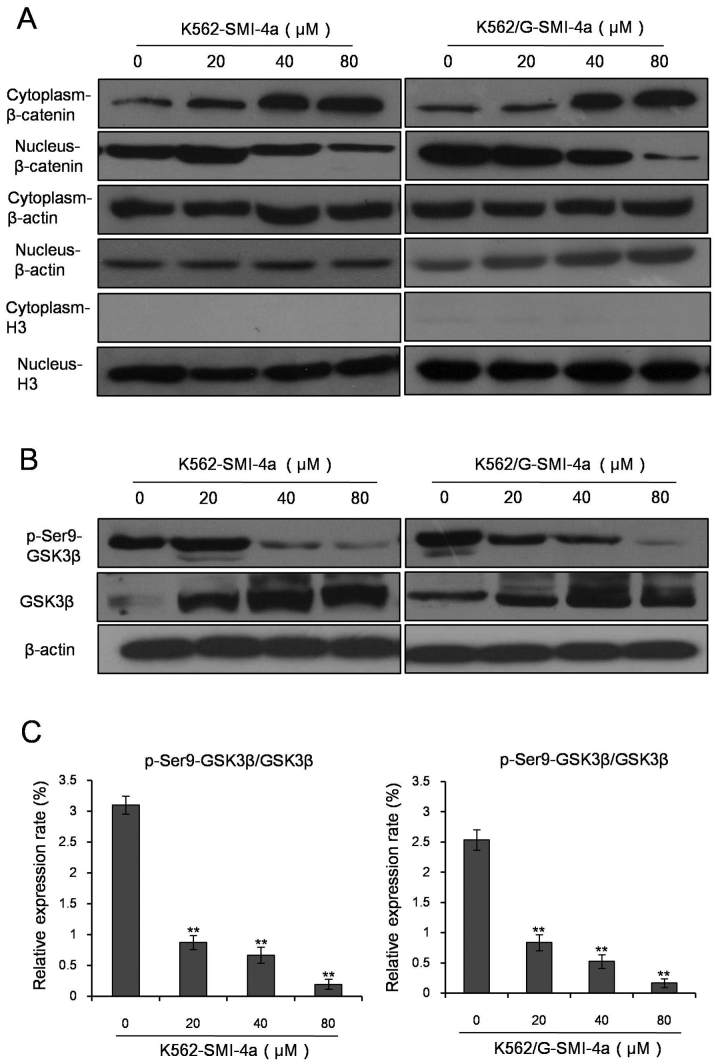
SMI-4a enhances the activity of GSK-3β kinase and inhibits the translocation of β-catenin
Translocation of β-catenin is critical to the activation of Wnt/β-catenin signal transduction pathway. In our study, western blot analysis was used to test the effects of SMI-4a with different concentrations on the variation of expression level of β-catenin inplasma and nucleus of K562 cells and K562/G cells (Fig. 6A). The resuts showed that as the concentration of SMI-4a increased, the expression level of the β-catenin increased in plasma, whereas decreased in nucleus.
Figure 6.
SMI-4a inhibites the phosphorylation of GSK-3β and decreases the expression of β-catenin in nucleus of CML cells. (A) Western blot analysis was used to test the effects of different concentrations of SMI-4a on the variation of expression level of β-catenin in plasma and nucleus of K562 and K562/G cells. (B) K562 and K562/G cells were treated with different concentrations of SMI-4a for 48 h and the expression of GSK-3β and p-Ser9-GSK-3β in K562 and K562/G cells was determined using western blot analysis. (C) ImageJ (NIH, Bethesda, MA, USA) was used to analyze the relative expression of pSer9-GSK-3β and GSK-3β. Data is presented as mean ± SEM, **P<0.01 vs. 0 µM group. GSK-3β, Glycogen synthase kinase 3β; p-Ser9-GSK-3β, phosphorylated glycogen synthase kinase 3β (Ser9); SEM, standard error of mean.
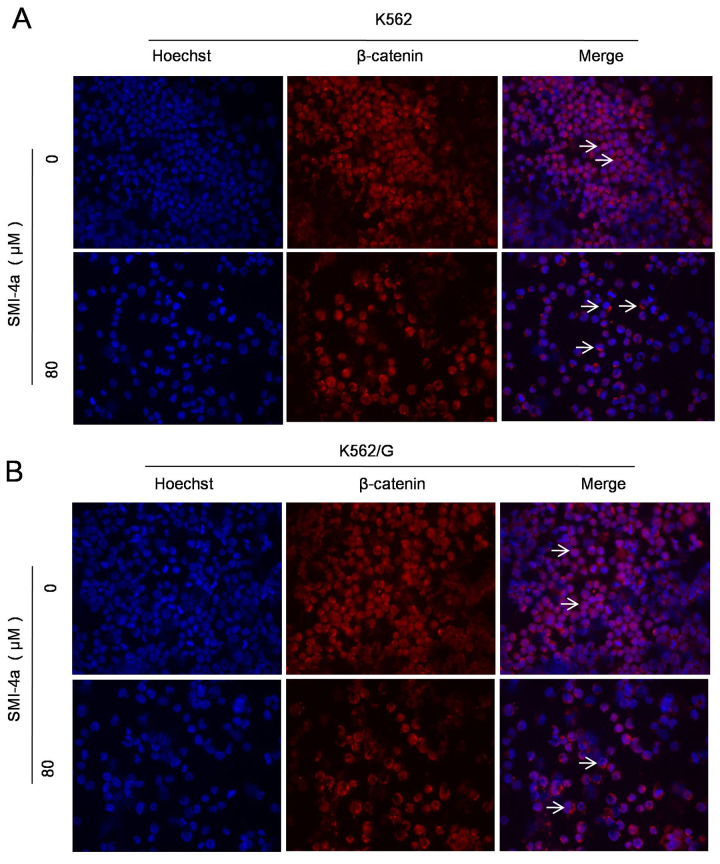
GSK-3β is a key enzyme of the translocation of β-catenin and plays a crucial role in the cell proliferation and apoptosis. So we tested the phosphorylation level of GSK-3β (Ser9), which is closely related to the translocation of β-catenin. The results showed that as the concentration of SMI-4a increased, the expression level of GSK-3β increased, while the expression level of p-Ser9-GSK-3β kinase decreased (Fig. 6B). p-Ser9-GSK-3β/GSK-3β was significantly decreased indicating that the activity of GSK-3β is enhanced (Fig. 6C). Immunofluorescence study results demonstrated the changes in the expression level of β-catenin in plasma and nucleus after SMI-4a treatment in K562 (Fig. 7A) and K562/G (Fig. 7B) cells. It is showed that when the concentration of SMI-4a were 80 µM, the expression of β-catenin in the nucleus was signicantly decreased.
Figure 7.
SMI-4a descresaes the expression of β-catenin in CML cells. K562 (A) and K562/G (B) cells were treated with 80 µM SMI-4a for 48 h and the variation of expression level of β-cateninin plasma and nucleus was examined by immunofluorescence (magnification, ×400).
Discussion
CML is a myeloproliferative neoplasm, which is associated with chromosomal translocation that gives rise to the Philadelphia chromosome and BCR-ABL gene. BCR-ABL encodes a constitutively active tyrosine kinase, which is causative for the disease. Before the introduction of TKI therapy, the disease was inevitably life-threatening (14,15). The development of targeted agents: TKIs, such as imatinib, dasatinib, and nilotinib have succeeded in altering the course of the disease. Imatinib is recommended as a first-line treatment in chronic-phase CML patients. However, few patients have failed to respond or experienced disease relapse after imatinib treatment. Even with a change in dasatinib or nilotinib, fraction of CML patients in chronic phase and a percentage of patients in advanced phase are either primary refractory or eventually develop resistance (14).
Imatinib resistance involves BCR-ABL-independent and dependent resistance mechanisms. BCR-ABL-independent mechanisms include non-adherence or intolerance to imatinib, pharmacokinetic factors (16), intracellular imatinib influx and efflux abnormality (17,18), activation of alternative signaling pathways (19), development of multidrug resistance (MDR) (15), and stem cells persistence (20). BCR-ABL-dependent mechanisms include BCR-ABL duplication, amplification, and mutations (4). So the challenge of overcoming resistance to imatinib therapy persists in the management of CML.
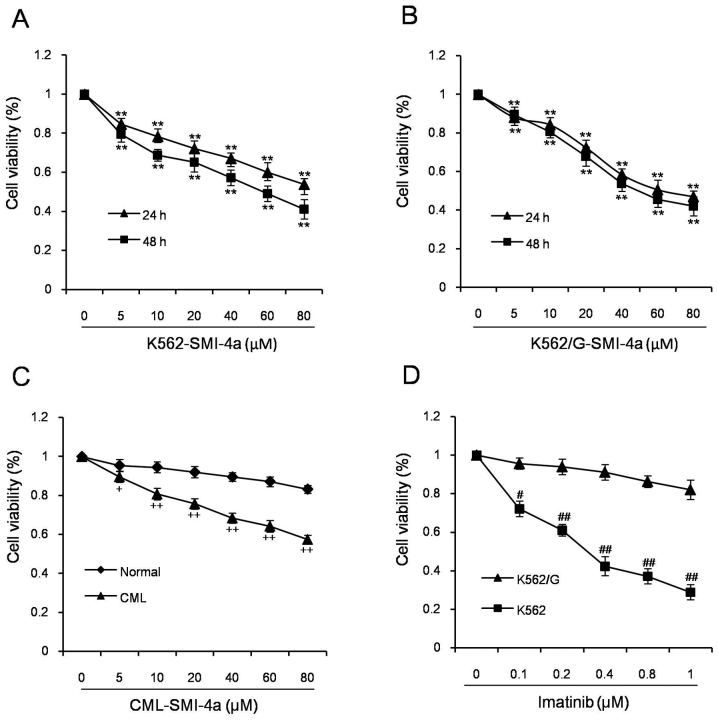
SMI-4a is a small selective PIM kinase inhibitor that inhibits PIM-1 kinase activity in vivo and in vitro. Our study showed that SMI-4a inhibited the proliferation of K562 cells in both dose- and time-dependent manner, which were consistent to the previous studies (11). To investigate the anti-proliferative efficacy on cell-lines refractory to imatinib, we evaluated its ability to suppress the growth of K562/G cells. The results demonstrated that the anti-proliferation effect was similar to K562. From this finding, it was speculated that SMI-4a can be an alternate treatment for the CML patients with imatinib resistant.
Cell cycle analysis showed that with the increase of SMI-4a concentration and duration of exposure time, cell cycle was arrested in S phase, which indicated that the anti-proliferative effect of SMI-4a achieved by the induction of S-phase cell cycle was arrest. However, it was not consistent to previous studies. Lin indicated that SMI-4a induced pre-T-LBL cells a G1 cell-cycle arrest by increasing the levels of p27Kip1 (11). If there were other cell cycle regulatory factors participating in the process need further studies.
Annexin V-PI assay and Hoechst analysis showed that SMI-4a had effect on programed cell death of K562 and K562/G cells. Consistent results were obtained on protein levels. Bax and cleaved PARP increased and Bcl-2 decreased after SMI-4a treatment. It can be seen the antileukemia effect of SMI-4a is similar with other PIM kinase inhibitors (13).
In BCR-ABL+ CML, Wnt/β-catenin signaling is aberrantly activated. Deletion of β-catenin in BCR-ABL+CML mouse model led to impaired leukemogenesis and delay of disease (21). β-catenin activation in CML cells depend on the BCR-ABL-mediated phosphorylation, which promotes the stabilization of β catenin by blocking its binding to APC/Axin/GSK3 complex. Subsequently, β-catenin translocates into the nucleus to form a complex with TCF/LEF factors and activates the expression of downstream gene, such as c-Myc (22). GSK-3β is a key enzyme of the translocation of β-catenin and plays crucial role in the cell proliferation and apoptosis. In our study, as the concentration of SMI-4a increased, the expression level of GSK 3β kinase increased, while the expression level of p-Ser9-GSK 3β kinase decreased. p-Ser9-GSK 3β/GSK 3β was decreased indicating that the activity of GSK 3β kinase increase and phosphorylation and deactivation of β-catenin take place in plasma. So β-catenin could not translocate to nucleus to bind to TCF/LEF operator and initiate the transcription of downstream proliferation related gene c-Myc, and the cell growth was inhibited.
Conclusion
SMI-4a can inhibit the proliferation and trigger programmed cell death of K562 cells and imatinib-resistent K562/G cells by enhancing the activity of GSK 3β. Active GSK 3β inhibited the translocation of β-catenin, and decreased the proliferation related gene This antitumor effect of SMI-4a is similar on K562 cells and imatinib-resistent K562/G cells, so we proposed that SMI-4a can be used as a complementary therapy for imatinib-resistent CML patients.
Figure 1.
SMI-4a inhibits cell proliferation of CML cells. K562 cells (A) and K562/G cells (B) were treated with various concentration of SMI-4a for 24 and 48 h. Mononuclear cells in bone marrow from patients with CML and normal individuals (C) were exposed to different concentrations of SMI-4a for 24 h. (D) K562 and K562/G cells were treated with various concentration of imatinib for 24 h. Data is presented as mean ± SEM; *P<0.05 vs. 0 µM group, **P<0.01 vs. 0 µM group, +P<0.05 vs. Normal group, ++P<0.01 vs. Normal group, #P<0.05 vs. K562 group, ##P<0.01 vs. K562 group. CML, chronic myeloid leukemia; N, normal individual; SEM, standard error of mean.
Acknowledgements
The present study was supported by the Science and Technology Project of Guangdong (grant no. 2011B080701008 to X.-F. Liu), Natural Science Fund of Guangdong Province (grant no. 2014A030310292 to L.-L. Liu). Major Project of Guangzhou Healthcare Innovation (grant no. 201400000003–4 to D.-J. Lin). We thank the members of Quentin Liu lab and Medical Research Center, the Third Affiliated Hospital, Sun Yat-sen University for presenting K562 and K562/G cell lines and technical support.
Glossary
Abbreviations
- CML
chronic myeloid leukemia
- TKIs
tyrosine kinase inhibitors
- PIM-1
proviral integration site for moloney murine leukemia virus-1
- GSK 3β
glycogen synthase kinase 3β
- PARP
poly (ADP-ribose) polymerase-1
- EGFR
epidermal growth factor receptor
- MCL
mantle cell lymphoma
- CLL
chronic lymphocytic leukemia
- IC50
50% inhibitory concentration
- MDR
multidrug resistance
References
- 1.Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2012 update on diagnosis, monitoring, and management. Am J Hematol. 2012;87:1037–1045. doi: 10.1002/ajh.23282. [DOI] [PubMed] [Google Scholar]
- 2.Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, Lim AQ, Chang CT, Fruman DA, Rizzieri DA, et al. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci USA. 2013;110:E2298–E2307. doi: 10.1073/pnas.1301838110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintás-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16:122–131. doi: 10.1177/107327480901600204. [DOI] [PubMed] [Google Scholar]
- 4.Jabbour EJ, Cortes JE, Kantarjian HM. Resistance to tyrosine kinase inhibition therapy for chronic myelogenous leukemia: A clinical perspective and emerging treatment options. Clin Lymphoma Myeloma Leuk. 2013;13:515–529. doi: 10.1016/j.clml.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118:693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beharrya Z, Mahajanb S, Zemskovac M, Lin YW, Tholanikunnel BG, Xia Z, Smith CD, Kraft AS. The Pim protein kinases regulate energy metabolism and cell growth. Proc Natl Acad Sci USA. 2011;108:528–533. doi: 10.1073/pnas.1013214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foulks JM, Carpenter KJ, Luo B, Xu Y, Senina A, Nix R, Chan A, Clifford A, Wilkes M, Vollmer D, et al. A small-molecule inhibitor of PIM kinases as a potential treatment for urothelial carcinomas. Neoplasia1. 2014;6:403–412. doi: 10.1016/j.neo.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YY, Taniguchi T, Baba T, Li YY, Ishibashi H, Mukaida N. Identification of a phenanthrene derivative as a potent anticancer drug with Pim kinase inhibitory activity. Cancer Sci. 2012;103:107–115. doi: 10.1111/j.1349-7006.2011.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Y, Bayakhmetov S. PIM1 kinase as a promise of targeted therapy in prostate cancer stem cells. Mol Clin Onco. 2016;4:13–17. doi: 10.3892/mco.2015.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weirauch U, Beckmann N, Thomas M, Grünweller A, Huber K, Bcher F, Hartmann RK, Aigner A. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia. 2013;15:783–794. doi: 10.1593/neo.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YW, Beharry ZM, HillA EG, Song JH, Wang W, Xia Z, Zhang Z, Aplan PD, Aster JC, Smith CD, Kraft AS. A small molecule inhibitor of Pim protein kinases blocks the growth of precursor T-cell lymphoblastic leukemia/lymphoma. Blood. 2010;115:824–833. doi: 10.1182/blood-2009-07-233445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Q, Chen LS, Neelapu SS, Miranda RN, Medeiros LJ, Gandhi V. Transcription and translation are primary targets of Pim kinase inhibitor SGI-1776 in mantle cell lymphoma. Blood. 2012;120:3491–3500. doi: 10.1182/blood-2012-02-412643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114:4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rumjanek VM, Vidal RS, Maia RC. Multidrug resistance in chronic myeloid leukaemia: How much can we learn from MDR-CML cell lines? Biosci Rep. 2013;33:e00081. doi: 10.1042/BSR20130067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamad A, Sahli Z, EI Sabban M, Mouteirik M, Nasr R. Emerging therapeutic strategies for targeting chronic myeloid leukemia stem cells. Stem Cells Int. 2013;2013:724360. doi: 10.1155/2013/724360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apperley JF. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 17.Mahon FX, Deininger MW, Schultheis B, Chabrol J, Reiffers J, Goldman JM, Melo JV. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: Diverse mechanisms of resistance. Blood. 2000;96:1070–1079. [PubMed] [Google Scholar]
- 18.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: Implications for drug resistance. Blood. 2004;104:3739–3745. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci USA. 2006;103:16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Li D. Stem cell and kinase activity-independent pathway in resistance of leukaemia to BCR-ABL kinase inhibitors. J Cell Mol Med. 2007;11:1251–1262. doi: 10.1111/j.1582-4934.2007.00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheller M, Schönheit J, Zimmermann K, Leser U, Rosenbauer F, Leutz A. Cross talk between Wnt/β-catenin and Irf8 in leukemia progression and drug resistane. J Exp Med. 2013;210:2239–2256. doi: 10.1084/jem.20130706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mancini M, Leo E, Takemaru K, Campi V, Borsi E, Castagnetti F, Gugliotta G, Santucci MA, Martinelli G. Chibby drives β catenin cytoplasmic accumulation leading to activation of the unfolded protein response in BCR-ABL1+ cells. Cell Signal. 2013;25:1820–1827. doi: 10.1016/j.cellsig.2013.05.019. [DOI] [PubMed] [Google Scholar]