Abstract
The transmembrane protease serine 2:v-ets erythroblastosis virus E26 oncogene homolog (TMPRSS2:ERG) gene fusion is common in prostate cancer, while its functional role is not fully understood. The present study aimed to investigate the significance of the TMPRSS2:ERG gene fusion in human prostate cancers using bioinformatics tools. Comprehensive alteration analysis of TMPRSS2 and ERG in 148 different human cancer studies was performed by cBioPortal, and the mRNA expression level of the ERG gene was evaluated using Oncomine analysis. Furthermore, lentiviral short hairpin (sh)RNA-mediated knockdown of TMPRSS2:ERG was performed to study the impact of ERG silencing on cell proliferation and cell cycle distribution in prostate cancer cells. The results demonstrated that the TMPRSS2 and ERG genes were mostly altered in prostate cancer, and the most frequent alteration was gene fusion. Oncomine analysis demonstrated that the ERG gene was significantly upregulated in prostate clinical samples compared with the normal prostate gland in four independent datasets, and a positive association was observed between potassium inwardly-rectifying channel subfamily J member 15, down syndrome critical region gene 4, potassium inwardly-rectifying channel subfamily J member 6 and ERG gene expression. There were 272 mutations of the ERG gene identified in the cBioPortal database; among the mutations, 2 missense mutations (R367C and P401H) were regarded as functional mutations (functional impact score >1.938). Furthermore, the present study successfully knocked down ERG gene expression through a lentiviral-mediated gene silencing approach in VCaP prostate cancer cells. The ERG mRNA and protein expression levels were both suppressed significantly, and a cell-cycle arrest at G0/G1 phase was observed after ERG gene silencing. In conclusion, these bioinformatics analyses provide novel insights for TMPRSS2:ERG fusion gene study in prostate cancer. Target inhibition of ERG expression could significantly cause cell growth arrest in prostate cancer cells, which could be a potentially valuable target for prostate cancer treatment. However, the precise mechanism of these results remains unclear; therefore, further studies are required.
Keywords: prostate cancer, transmembrane protease serine 2, v-ets erythroblastosis virus E26 oncogene homolog, fusion gene, bioinformatics
Introduction
Prostate cancer is one of the most frequent malignancies and the most common leading cause of cancer-associated death in men all over the world, particularly in developed countries (1,2). In the past decades, prostate specific antigen (PSA) was the only widely-used serum biomarker for prostate cancer. However, due to the extensive use of serum PSA testing, the prostate cancer-specific mortality has increased significantly, which results in over-diagnosis or over-treatment (3). Multiple technologies have been applied to identify novel prostate cancer biomarkers in tissues and blood of patients. Nevertheless, no biomarker has been identified to replace the routine use of PSA at present.
Recently, gene fusion transcripts of transmembrane protease serine 2 (TMPRSS2):v-ets erythroblastosis virus E26 oncogene homolog (ERG), also termed TMPRSS2:ERG or T2E, have been identified as promising urinary novel biomarkers in prostate cancer (4,5). A study in 2005 demonstrated that up to 55% prostate cancer cases were identified to have ERG over-expression, using a novel biostatistical method called cancer outlier profile analysis (6). Furthermore, the overexpression of ERG is in the majority of tumors driven by fusion of the ERG gene with TMPRSS2, which are both located on chromosome 21 (7). TMPRSS2 is a prostate-specific and androgen-response gene that encodes a protein belonging to the serine protease family, which functions in prostate carcinogenesis and relies on gene fusion with ETS transcription factors, such as ERG and ETV1 (8). ERG is an oncogene that encodes a member of the erythroblast transformation-specific family of transcription factors (9), which is a key regulator of cell proliferation, differentiation, angiogenesis, inflammation and apoptosis. The TMPRSS2:ERG gene fusion is the most frequent genomic alteration in prostate cancer cases and results in overexpression of the transcription factor ERG (10), which is present in both early- and late-stage prostate cancer (castration-resistant prostate cancer, CRPC) (6,11).
Numerous studies have evaluated the significance of TMPRSS2-ERG in prostate cancer patients with varying results (12), some of which indicated that the fusion gene is not an important predicator of prostate cancer mortality and recurrence (13), while other studies demonstrated that TMPRSS2:ERG fusion was associated with an increased risk of prostate cancer mortality (13–16). The present study examined the expression pattern of the TMPRSS2:ERG fusion gene in human pan-cancers, including prostate cancer, by using the publically available data from cBioPortal. Based on these findings, the present study specifically analyzed the ERG alterations, mRNA expression, mutations and interaction networks in several prostate cancer datasets. Furthermore, the functional role of ERG in prostate cancer cells was examined by lentiviral-mediated knockdown approaches. The present study provides novel insights for the TMPRSS2:ERG fusion gene study in prostate cancer.
Materials and methods
Determination of TMPRSS2 and ERG alterations across different cancer types
The frequency of TMPRSS2 and ERG gene alterations (including mutations, deletions, copy number gains and amplifications) was performed across multiple cancer types using the cBioPortal for Cancer Genomics database (www.cbioportal.org), which contains 147 common cancer studies with the details of almost 23,000 patients. All searches were performed according to the online instructions of cBioPortal.
Oncomine database analysis
ERG mRNA expression levels in prostate cancer were compared with its matched normal tissues by using The Cancer Genome Atlas (TCGA) datasets in the Oncomine database (www.oncomine.org). The threshold used to obtain the most significant probes of the queried gene for each microarray data included a two-fold difference in expression between cancers and normal tissues if P<1×10−4. The mRNA expression level of ERG was analyzed in three independent datasets.
ERG gene silencing by short hairpin (sh)RNA in VCaP cells
The prostate cancer cell line VCaP which obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Three pairs of shRNAs for ERG (GenBank ID: NM_001136154.1) were designed (Table I), synthesized and packaged by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). shRNAs were cloned into a pLKO.1 puro vector (Addgene, Inc., Cambridge, MA, USA) according to the manufacturer's protocol. Lentiviral particles were generated following transfection of 80% confluent 293T cells (Type Culture Collection of the Chinese Academy of Sciences), with 15 µg pLKO.1-shRNA-ERG plasmid or empty control vector using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific) for 10 min at room temperature according to the manufacturer's instructions. VCaP cells were initially seeded at a density of 5×106 cells/100 mm dish. After 24 h incubation, cultures were supplemented with 1×108 lentiviral particles (multiplicity of infection of 8) with 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 48 h. Subsequently, total cell RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and DNaseI (New England BioLabs, Inc., Ipswich, MA, USA). Total RNA was reverse-transcribed into cDNA using a PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's instructions. The temperature protocol that was used was as follows: At 37°C for 15 min and at 85°C for 5 sec. Quantitative polymerase chain reaction (qPCR) was used to evaluate the ERG silencing effect at the mRNA level using SYBR Premix Ex Taq (Takara Bio, Inc.) according to the manufacturer's instructions. The thermocycling conditions that were used were as follows: At 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec, at 60°C for 20 sec and at 72°C for 30 sec. The following oligonucleotide primers were used: ERG, forward 5′-ATCGCATTATGGCCAGCACT-3′, reverse 5′-TGTCCATAGTCGCTGGAGGA-3′; and β-actin, forward 5′-GGACTTCGAGCAAGAGATGG-3′ and reverse 5′-AGCACTGTGTTGGCGTACAG-3′. The relative gene expression data were assayed using the comparative Cq method as described previously (17,18).
Table I.
shRNA sequences targeting the ERG gene.
| shRNA duplex | Sequence (5′-3′) |
|---|---|
| ERG-shRNA1 | |
| Forward | CCGGTGCTCATATCAAGGAAGCCTTATCAAGAGTAGGCTTCCTTGATATGAGCTTTTT |
| Reverse | AATTAAAAAGCTCATATCAAGGAAGCCTTACTCTTGAATAGGCTTCCTTGATATGAGC |
| ERG shRNA2 | |
| Forward | CCGGTCCACCCACAGAAGATGAACTTTTCAAGAGAAGTTCATCTTCTGTGGGTGGTTTTT |
| Reverse | AATTAAAAACCACCCACAGAAGATGAACTTCTCTTGAAAAGTTCATCTTCTGTGGGTGG |
| EEG-shRNA3 | |
| Forward | CCGGTGATGATGTTGATAAAGCCTTATTCAAGAGTAAGGCTTTATCACATCATCTTTTT |
| Reverse | AATTAAAAAGATGATGTTGATAAAGCCTTACTCTTGAATAAGGCTTTATCACATCATC |
ERG, v-ets erythroblastosis virus E26 oncogene homolog; shRNA, short hairpin RNA.
Total cellular proteins were extracted from 70–80% confluent cultured cells after 48 h transfection using ice cold lysis buffer (20 mM HEPES, 0.1% SDS, 1 mM EDTA, 1 mM EGTA, 10 mM monothioglycerol, 1 mM PMSF, 5 mM leupeptin, 0.25 M sucrose). Protein concentration was determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of extracted protein samples (30 µg) were separated by standard 12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Folowing blocking with 5% non-fat dry milk at room temperature for 1 h, membranes were probed with optimally diluted primary antibodies at 4°C overnight, then incubated with a horseradish peroxidase-conjugated secondary antibody (cat no. ab6721; 1:5,000; Abcam, Cambridge, MA, USA) at room temperature for 1 h. Protein bands were visualized with enhanced chemiluminescence western blot reagents (GE Healthcare Life Sciences, Little Chalfont, UK) as described previously (19). Blots were semi-quantified by densitometry using ImageJ software version 2.0 (National Institutes of Health, Bethesda, MD, USA). Primary antibodies used were as follows: Anti-ERG monoclonal antibody (cat no. ab92513; Abcam, Cambridge, MA, USA; 1:1,000) and anti-β-actin (cat no. 4970; Cell Signaling Technology, Inc., Danvers, MA, USA; 1:1,000).
Cell proliferation assay
Cell proliferation was assessed by MTT assay according to the manufacturer's protocol (Roche Diagnostics GmbH, Mannheim, Germany). The formed formazan crystals were dissolved by adding 100 µl/well acidic SDS buffer [10% SDS, 0.16% (6 mol/l) HCl and 5% isobutyl alcohol] and incubating overnight in a CO2-free incubator at 37°C. The optical density (OD)570 absorption was measured in a microplate reader (Perkin Elmer Victor3 1420 Multilabel Plate Counter, PerkinElmer, Inc., Waltham, MA, USA). Experiments were repeated three times, and data were represented as the mean of five-replicate wells ± standard error.
Cell cycle analysis
Cell cycle analysis of control and ERG-silenced VCaP cells from 3 independent biological replicates were collected. The cells were washed in PBS, and then fixed in 70% ethanol for 30 min at −20°C. The fixed cells were washed three times, resuspended in PBS containing 10 µg/ml of RNase A for 30 min, and then incubated with 10 µg/ml propidium iodide (PI) for 30 min in the dark. Subsequently, the samples were used for DNA flow cytometry (ALTRA cell sorting system, Beckman Coulter, Inc., Brea, CA, USA) analysis. For each measurement, at least 15,000 cells were acquired. Analysis of cell cycle was performed with ModFit LT2 software version 2.0 (Verity Software House, Inc., Topsham, ME, USA).
STRING analysis
STRING software (https://string-db.org/) (20) was used to generate the network of predicted associations for ERG protein. The network was set in evidence mode, in which the associations of the proteins were predicted based on up to 7 different evidences (the presence of fusion evidence, neighborhood evidence, co-occurrence evidence, experimental evidence, text-mining evidence, database evidence and co-expression evidence).
Statistical analysis
GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analyses. All analysis was performed using the unpaired Student's t-test or analysis of variance followed by a post hoc Tukey test for multiple comparisons. The data were presented as mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Determination of TMPRSS2 and ERG gene alterations across different human cancer types
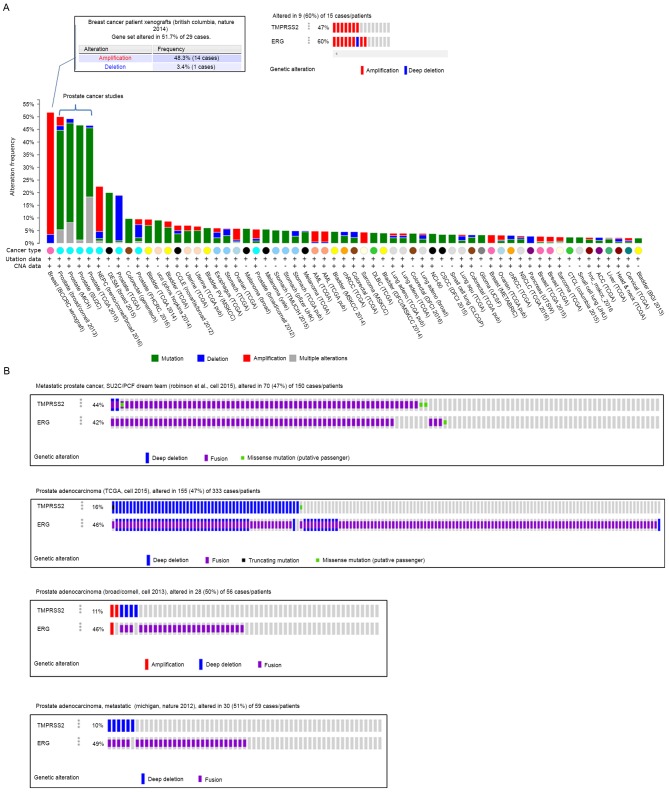
By pan-cancer analysis, it was demonstrated that the TMPRSS2 and ERG alterations (including mutations, deletions and amplifications) were mainly observed in one breast cancer study and most of the prostate cancer studies (Fig. 1A). In the breast cancer xenografts study (21) TMPRSS2 and ERG were notably altered in 51.7% of 29 cases, among which 48.3% (14 cases) were amplification. Only one case contained a deep deletion in ERG gene in the breast cancer study mentioned above.
Figure 1.
Alteration frequency analysis of TMPRSS2 and ERG gene in human cancers using cBioPortal. (A) The alteration frequencies of TMPRSS2 and ERG across 148 cancer studies (min. % altered samples: 2% are presented). Gene amplification is the highest in breast cancer while gene fusion is the most frequent in prostate cancers. The red bars indicate gene amplification, blue bars are homozygous deletions, green bars are non-synonymous mutations, and gray bars indicate multiple alterations. (B) Gene fusion frequencies of TMPRSS2 and ERG in prostate cancers in four independent studies. Purple bars represent gene fusion cases of prostate cancer. TMPRSS2, transmembrane protease serine 2; ERG, v-ets erythroblastosis virus E26 oncogene homolog.
However, in prostate cancer studies, the most frequent alteration of TMPRSS2 and ERG was gene fusion (Fig. 1B). Amplification, missense mutation and deep deletion were less frequently observed. Studies in prostate adenocarcinoma (22,23) showed that TMPRSS2 and ERG were altered in over 47% in prostate cancers, and 46% of them were gene fusion. Moreover, two metastatic prostate cancer datasets (24,25) demonstrated that 42 and 49% of the patients had ERG gene fusion (Fig. 1B).
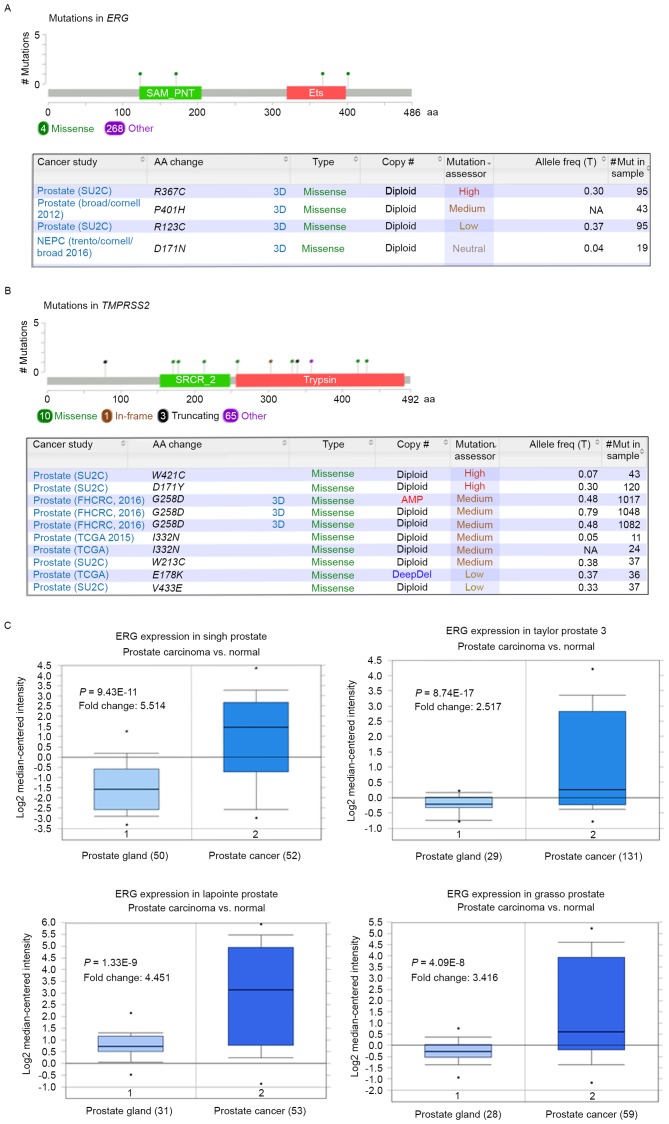
Although there was less frequent mutation than gene fusion observed in TMPRSS2 and ERG, some of them may serve important roles in prostate cancer progression. There were 272 mutations of the ERG gene identified in the cBioPortal database; among the mutations, 2 missense mutations (R367C and P401H) were regarded as functional mutations (Functional impact score >1.938). The details of missense mutations of TMPRSS2 and ERG with high mutation assessor score are presented in Fig. 2A and B.
Figure 2.
Missense mutations and mRNA expression of TMPRSS2 and ERG in prostate cancer. Missense mutations of (A) ERG and (B) TMPRSS2 in prostate cancer. (C) mRNA expression profile of ERG gene in four independent prostate cancer studies using Oncomine analysis. TMPRSS2, transmembrane protease serine 2; ERG, v-ets erythroblastosis virus E26 oncogene homolog.
ERG gene is overexpressed in prostate cancer clinical samples
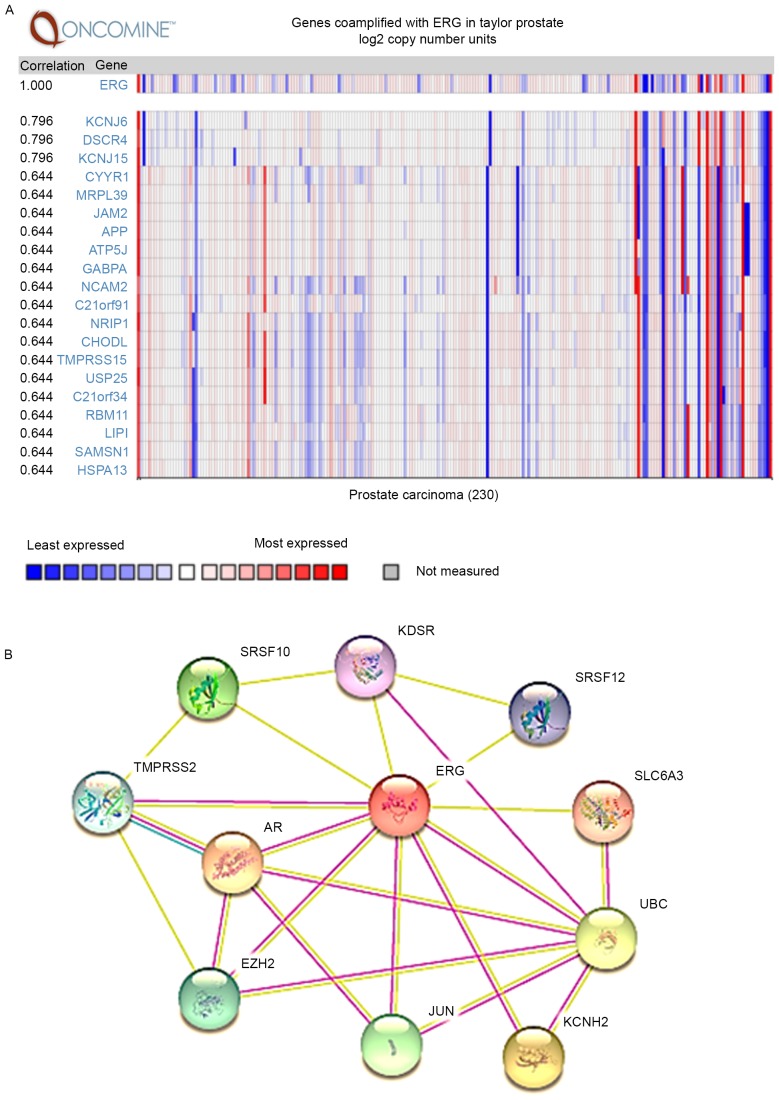
Given the high-frequent alterations of TMPRSS2 and ERG observed in prostate cancer studies, the mRNA expression profile of ERG in prostate cancer in four independent datasets were analyzed using Oncomine analysis. Notably, ERG mRNA expression levels were significantly upregulated in prostate cancer cases compared with their normal tissues in all four independent datasets (Fig. 2C). Furthermore, the co-expression gene of ERG in a cohort of 230 patients with prostate cancer was also evaluated (Taylor Prostate) (26) in the Oncomine database, as well as the interaction networks by STRING (Fig. 3B). The most correlated gene of ERG was potassium inwardly-rectifying channel subfamily J member 6 (KCNJ6), potassium inwardly-rectifying channel subfamily J member 15 (KCNJ15) and down syndrome critical region gene 4 (DSCR4; Fig. 3A).
Figure 3.
Co-expression and interaction networks of ERG in prostate cancer. (A) Co-expression analysis of ERG in prostate cancer using Oncomine database. (B) The interaction networks of ERG analyzed by STRING. TMPRSS2, transmembrane protease serine 2; ERG, v-ets erythroblastosis virus E26 oncogene homolog.
shRNA specifically reduces ERG expression in VCaP cells
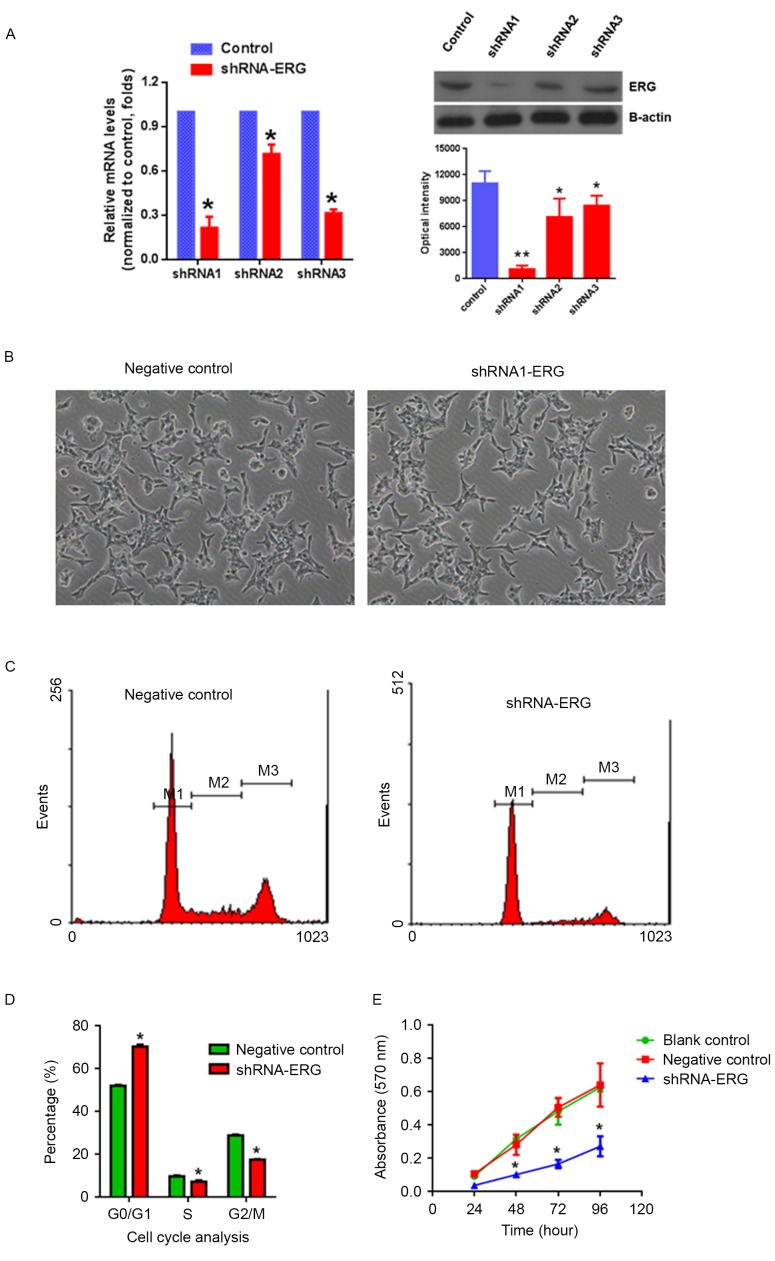
To study the role of TMPRSS2:ERG in this context of preexisting genetic alterations, lentiviral-mediated shRNA was used to knock down ERG gene expression in VCaP cells that are known to harbor the TMPRSS2:ERG gene fusion (11). Three pairs of shRNA of ERG were designed; both RT-qPCR and western blot analysis demonstrated that shRNA1-ERG exhibited the highest knockdown efficiency compared with the scramble control. The mRNA expression level of ERG was decreased by >79% and the protein expression level was reduced >93% in the shRNA-ERG1 viral-infected VCaP cells (Fig. 4A, P<0.05). In addition, there was no significant changes in cell morphology observed in the shRNA-ERG infected cells (Fig. 4B).
Figure 4.
shRNA mediate ERG gene silencing and functional studies in prostate cancer cells. (A) shRNA-ERG knockdown efficiency studied by reverse transcription-quantitative polymerase chain reaction and western blotting. (B) Phenotype characterization of VCaP cells after ERG silencing by shRNA-ERG. (C) Flow cytometric analysis of VCAP cells and (D) % of cells in each phase of the cell cycle after ERG gene silencing. (E) Cell proliferation analysis using MTT methods. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 vs. blank control or negative control. shRNA, short hairpin RNA; TMPRSS2, transmembrane protease serine 2; ERG, v-ets erythroblastosis virus E26 oncogene homolog.
Knockdown of ERG in VCaP cells inhibits cell proliferation through cell cycle arrest
Upon the shRNA-mediated knockdown of ERG in VCaP cells, cell proliferation was determined by MTT assay and the cell cycle distribution was assessed by flow cytometry. The results demonstrated that specific knockdown of the ERG gene in prostate cancer cells could cause G0/G1 cell cycle arrest (Fig. 4C and D) and significantly inhibit cell proliferation (Fig. 4E) compared with the scramble virus-infected controls in VCaP cells.
Discussion
The discovery of fusion genes involving the TMPRSS2 promoter region with ERG coding DNA sequences in >50% of prostate cancer cases has provide a significant insight for exploration of useful biomarkers for prostate cancer study and clinical treatment (11,27). However, the prognostic value of TMPRESS2:ERG gene fusion is a hotly debated topic in the current literature (13,28). The present study analyzed the TMPRSS2 and ERG gene expression and alteration in multi-cancer types by using cBioPortal, and indicated that these genes were mostly altered in prostate cancer, and the most frequent alteration was gene fusion, which was consistent with previous studies (11). Notably, some missense mutations with high mutation assessor score were identified in the TMPRSS2 and ERG gene, which may serve important roles in the gene fusion process and prostate cancer development.
Important studies in recent years clarified the significance of the TMPRSS2:ERG gene fusion in prostate cancer, and most of them indicated that the presence of the fusion gene product denotes an unfavorable outcome (7,15,29). The most direct consequence for the TMPRSS2:ERG gene fusion was the significant upregulation of the ERG gene, which is not normally expressed in prostate epithelia (30), and is likely to be involved in prostate cancer development by enhancing tumor angiogenesis (31). The high expression of ERG in prostate cancer is associated with advanced tumor stage, shorter survival time, high Gleason score and metastasis (12). Full-length ERG is a 486 amino-acid 54 kDa transcription factor, and contains an ETS DNA-binding domain and a pointed domain (32,33). Normally, ERG is highly expressed in the embryonic mesoderm and endothelium and serves a critical role in the formation of the vascular system and the urogenital tract, and in bone development (34–36). Aberrant expression of the ERG gene has a major impact on cell invasion (37) and metastasis (38), as well as the differentiation of prostate epithelium (39). The ERG gene is the first demonstration of constitutive oncogene activation in prostate cancer; however, the functional consequences and mechanisms of the TMPRSS2:ERG gene fusion are not fully understood. In particular, the co-expression genes and interaction networks have not been characterized.
Recently, interest in the TMPRSS2:ERG fusion gene in prostate cancer remains high, which is supposed to be a novel biomarker, therapy target, diagnostic and prognostic indicator in prostate cancer (7,36). Therefore, the present study also surveyed ERG gene expression by Oncomine analysis, based on RNA-Seq data, which demonstrated that the ERG gene was significantly increased in four independent prostate cancer study datasets. Based on these findings, the present study designed specific shRNA of the ERG gene for loss-of function study. It was demonstrated that ERG gene silencing could significantly inhibit prostate cancer cells proliferation, and induce G0/G1 cell cycle arrest in prostate cancer cells. These results suggested that not only the alteration of TMPRSS2 and ERG gene could be a specific marker in prostate cancer, but also could be a potential therapy target in prostate cancer. However, the exact mechanism remains unclear; therefore, further studies are required to illustrate the signaling pathways involved in this progression.
Acknowledgements
The present study was supported by Shenzhen Science and Technology Program Basic Research Project (grant no. JCYJ20150402144905865).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Velonas VM, Woo HH, dos Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Mol Sci. 2013;14:11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephan C, Rittenhouse H, Hu X, Cammann H, Jung K. Prostate-Specific Antigen (PSA) Screening and New Biomarkers for Prostate Cancer (PCa) EJIFCC. 2014;25:55–78. [PMC free article] [PubMed] [Google Scholar]
- 5.Teoh JY, Tsu JH, Yuen SK, Chan SY, Chiu PK, Lee WM, Wong KW, Ho KL, Hou SS, Ng CF, Yiu MK. Prognostic significance of time to prostate-specific antigen (PSA) nadir and its relationship to survival beyond time to PSA nadir for prostate cancer patients with bone metastases after primary androgen deprivation therapy. Ann Surg Oncol. 2014;22:1385–1391. doi: 10.1245/s10434-014-4105-8. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Palanisamy N, Siddiqui J, Chinnaiyan AM, Kunju LP. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136:935–946. doi: 10.5858/arpa.2011-0424-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hossain D, Bostwick DG. Significance of the TMPRSS2:ERG gene fusion in prostate cancer. BJU Int. 2013;111:834–835. doi: 10.1111/bju.12120. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tandefelt Gasi D, Boormans J, Hermans K, Trapman J. ETS fusion genes in prostate cancer. Endocr Relat Cancer. 2014;21:R143–R152. doi: 10.1530/ERC-13-0390. [DOI] [PubMed] [Google Scholar]
- 10.Kissick HT, On ST, Dunn LK, Sanda MG, Asara JM, Pellegrini KL, Noel JK, Arredouani MS. The transcription factor ERG increases expression of neurotransmitter receptors on prostate cancer cells. BMC Cancer. 2015;15:604. doi: 10.1186/s12885-015-1612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 12.Hagglof C, Hammarsten P, Strömvall K, Egevad L, Josefsson A, Stattin P, Granfors T, Bergh A. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS One. 2014;9:e86824. doi: 10.1371/journal.pone.0086824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: A cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–1509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demichelis F, Fall K, Perner S, Andrén O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210630. [DOI] [PubMed] [Google Scholar]
- 15.St John J, Powell K, Conley-Lacomb MK, Chinni SR. TMPRSS2-ERG Fusion gene expression in prostate tumor cells and its clinical and biological significance in prostate cancer progression. J Cancer Sci Ther. 2012;4:94–101. doi: 10.4172/1948-5956.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Wang Z, Li XF, He X, Guan LL, Tuo JL, Wang Y, Luo Y, Zhong HL, Qiu SP, Cao KY. Screening and identification of significant genes related to tumor metastasis and PSMA in prostate cancer using microarray analysis. Oncol Rep. 2013;30:1920–1928. doi: 10.3892/or.2013.2656. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Wang Z, He SY, Zhang SF, Luo HJ, Zhou K, Li XF, Qiu SP, Cao KY. Bax-interacting factor-1 inhibits cell proliferation and promotes apoptosis in prostate cancer cells. Oncol Rep. 2016;36:3513–3521. doi: 10.3892/or.2016.5172. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. Punctuated evolution of prostate cancer genomes. Cancer Res. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network: The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao JJ, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 28.Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, Kunju LP, Tomlins SA. Correlation of urine TMPRSS2:ERG and PCA3 to ERG+ and total prostate cancer burden. Am J Clin Pathol. 2012;138:685–696. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulda V, Topolcan O, Kucera R, Kripnerova M, Srbecka K, Hora M, Hes O, Klecka J, Babuska V, Rousarova M, et al. Prognostic Significance of TMPRSS2-ERG Fusion Gene in Prostate Cancer. Anticancer Res. 2016;36:4787–4793. doi: 10.21873/anticanres.11037. [DOI] [PubMed] [Google Scholar]
- 30.Deramaudt TB, Remy P, Stiegler P. Identification of interaction partners for two closely-related members of the ETS protein family, FLI and ERG. Gene. 2001;274:169–177. doi: 10.1016/S0378-1119(01)00610-2. [DOI] [PubMed] [Google Scholar]
- 31.Kissick HT, Sanda MG, Dunn LK, Arredouani MS. Development of a peptide-based vaccine targeting TMPRSS2:ERG fusion-positive prostate cancer. Cancer Immunol Immunother. 2013;62:1831–1840. doi: 10.1007/s00262-013-1482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao VN, Papas TS, Reddy ES. erg, a human ets-related gene on chromosome 21: Alternative splicing, polyadenylation, and translation. Science. 1987;237:635–639. doi: 10.1126/science.3299708. [DOI] [PubMed] [Google Scholar]
- 33.Salek-Ardakani S, Smooha G, de Boer J, Sebire NJ, Morrow M, Rainis L, Lee S, Williams O, Izraeli S, Brady HJ. ERG is a megakaryocytic oncogene. Cancer Res. 2009;69:4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 34.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayaraj P, Le Bras A, Mitchell N, Kondo M, Juliao S, Wasserman M, Beeler D, Spokes K, Aird WC, Baldwin HS, Oettgen P. Erg is a crucial regulator of endocardial-mesenchymal transformation during cardiac valve morphogenesis. Development. 2012;139:3973–3985. doi: 10.1242/dev.081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adamo P, Ladomery MR. The oncogene ERG: A key factor in prostate cancer. Oncogene. 2016;35:403–414. doi: 10.1038/onc.2015.109. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 38.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]