Supplemental Digital Content is available in the text.
Keywords: alleles, genetics, genome-wide association study, odds ratio, pharmacogenetics
Abstract
Background—
Intravenous immunoglobulin (IVIG) is the treatment of choice in Kawasaki disease (KD). IVIG is used to prevent cardiovascular complications related to KD. However, a proportion of KD patients have persistent fever after IVIG treatment and are defined as IVIG resistant.
Methods and Results—
To develop a risk scoring system based on genetic markers to predict IVIG responsiveness in KD patients, a total of 150 KD patients (126 IVIG responders and 24 IVIG nonresponders) were recruited for this study. A genome-wide association analysis was performed to compare the 2 groups and identified risk alleles for IVIG resistance. A weighted genetic risk score was calculated by the natural log of the odds ratio multiplied by the number of risk alleles. Eleven single-nucleotide polymorphisms were identified by genome-wide association study. The KD patients were categorized into 3 groups based on their calculated weighted genetic risk score. Results indicated a significant association between weighted genetic risk score (groups 3 and 4 versus group 1) and the response to IVIG (Fisher’s exact P value 4.518×10−03 and 8.224×10−10, respectively).
Conclusions—
This is the first weighted genetic risk score study based on a genome-wide association study in KD. The predictive model integrated the additive effects of all 11 single-nucleotide polymorphisms to provide a prediction of the responsiveness to IVIG.
Kawasaki disease (KD) is an acute systemic vasculitis syndrome. This disease mainly affects children who are younger than 5 years of age.1 Since KD was first reported by Dr Kawasaki in 1967 in Japan,2 it has now been recognized as a worldwide pediatric disease. Epidemiological studies indicate that the prevalence of KD is higher in East Asia than North America and Europe.3 Although the cause remains unclear, a genetic contribution to disease susceptibility is well established.4 Several genome-wide association studies (GWASs)5–7 and candidate gene association studies8–12 have identified susceptibility loci for KD.
See Editorial by Portman and Shrestha
KD is thought to be the leading cause of acquired heart disease in children in the developed countries.13–15 High-dose intravenous immunoglobulin (IVIG) is currently acknowledged to be the standard treatment for KD patients. Receiving a single high dose of IVIG within 10 days after disease onset can significantly reduce the risk of coronary artery lesions (CALs).16 However, 10% to 20% of KD patients have recrudescent fever after the initial IVIG treatment, and these patients are at higher risk of CAL and acquired cardiac complications in adulthood.12,17,18
The molecular mechanisms of IVIG for anti-inflammation in KD remain unclear. Potential mechanisms include the blockade of the Fc receptor,19–21 neutralization of the pathogenic or the toxic products of unknown infectious agents, immune-modulatory effects,22 stimulation of suppressor activity, and modulation of the cytokines.23 Several genome-wide association studies have revealed common variants that strongly associated with KD, CAL formation, and IVIG treatment.24–26 Here, we applied a GWAS-based weighted genetic risk score (wGRS) to develop a genetic model to predict the IVIG resistance in KD.
Methods
Patients Studied
The DNA of children who have been diagnosed as KD and received IVIG treatment at Chang Gung Memorial Hospital, Kaohsiung Medical Center from the year 2001 to 2007 were collected. The informed consent was obtained from all patients and guardians. The acquisition and subsequent use of samples were approved by the Institutional Review Board of Chang Gung Memorial Hospital (IRB No. 98-1438B), and the methods were performed in accordance with the approved guidelines. We adopted the same diagnostic criteria of KD as reported previously27–29 and further excluded the samples with acute fever for <5 days. A diagnosis of KD was made by a clinician using the suggested universal criteria for KD proposed by the American Heart Association. In any child with fever lasting for >5 days who fulfilled four of the following criteria: diffuse mucosal inflammation with strawberry tongue and fissure lips, bilateral nonpurulent conjunctivitis, indurative angioedema over the hands and feet, dysmorphic skin rashes, and unilateral cervical lymphadenopathy.30 Every patient diagnosed as KD was treated with a single dose of IVIG infusion (2 g/kg) within a 12-hour period. Aspirin (3 to 5 mg per kg per day) was given until all signs of inflammation were resolved or regression of CAL under 2-dimensional echocardiography test by visualizing coronary arteries (including left and right) diameter on the parasternal short-axis view of the aorta. Following the guidelines of Japanese Ministry of Health guidelines, we defined CAL as an increment of the internal diameter of 3 mm (≤5 years old) or 4 mm (>5 years old), or 1.5× larger of internal diameter than the adjacent segment. We defined IVIG responsiveness as defervescence within 48 hours after the first administration of IVIG and no recurrence (temperature >38°C) for at least 7 days, combined with remission of inflammatory signs. KD patients who had persistence of fever beyond 48 hours were defined as IVIG resistant.
DNA Extraction and Genome-Wide Scanning
The blood cells were subject to DNA extraction for oligonucleotide arrays. Samples were first treated with 0.5% sodium dodecylsulfate lysis buffer and then protease K (1 mg/mL) for 4 hours at 60°C. DNA was extracted using Gentra (Qiagen, Valencia, CA) extraction kit followed by 70% alcohol precipitation and Gentra Puregene Blood Kit (Qiagen). Single-nucleotide polymorphisms (SNPs) were detected by using Affymetrix Genome-Wide Human SNP Array 6.0 platform (Affymetrix, Inc).
Statistical Analysis
Quality Control
PLINK31 was applied for quality control of genome-wide scan data. We excluded the SNPs with missing call rates exceeding 1.0%, a P value of Hardy–Weinberg equilibrium <1×10−05 and minor allele frequency <5.0%. The SNPs reference of Affymetrix Genome-Wide Human SNP Array 6.0 platform was NCBI36 (hg18). CrossMap (Version 0.1.5) was used to lift over data to NCBI37 (hg19). SHAPEIT32 and IMPUTE233 were applied for the haplotype phasing and genotype imputation. HapMap 3 genotype data were incorporated34 with our Taiwanese data to perform principal component analysis (PCA). PCA was performed by using Genome-wide Complex Trait Analysis,35 which performed PCA by the same algorithm implemented in EIGENSTRAT and output corresponding eigenvalues and eigenvectors, to identify sample substructure on autosomal genotype data.
SNP Association Analysis
To satisfy the 603698 SNPs to filtering criteria, we performed association analysis by using the mixed linear model algorithm implemented in Genome-wide Complex Trait Analysis that accounts for the polygenic effect of all SNPs during association test. Then, we calculated the fixed effect of all SNPs by excluding candidate markers (mixed linear model with candidate marker excluded), which prevented loss of power because of double fitting of the candidate markers. Manhattan plot was plotted by Haploview36 software. Then we evaluated the residual population stratification by calculating genomic inflation λ value and visualized corresponding test statistics using quantile-quantile plot in R (http://www.r-project.org/).
Weighted Genetic Risk Scoring System
wGRS system proposed by De Jager et al37 was applied to calculate the cumulative effects of candidate SNPs. In this study, the allelic odds ratios were natural logarithm transformed to become the weight of each SNP. The wGRS was calculated by multiplying the weight by the risk allele number (0, 1, or 2) and taking the sum across 11 SNPs, as shown in the following equation:
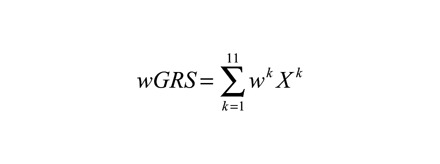 |
(1) |
where k is SNP, wk is the corresponding weight of SNP (ln(OR)) and Xk is the number of the risk allele (0, 1, or 2). wGRS of IVIG responders and nonresponders has been compared by Wilcoxon rank-sum test with continuity correction. KD patients were then categorized by corresponding wGRS into 4 groups: group 1 (wGRS<|mean−SD|), group 2 (|mean−SD| ≤wGRS<median), group 3 (median≤wGRS<|mean+SD|), and group 4 (wGRS≥|mean+SD|). wGRS was also compared between groups, and relevant statistical parameters were calculated by using group 1 as a reference. The subgroup analysis was further performed to confirm the intragroup difference of wGRS between IVIG responders and nonresponders.
To further access the performance of wGRS in the prediction of IVIG responsiveness, we conducted a receiver operating characteristic (ROC) curve analysis.38 For every discrimination threshold, 95% confidence intervals (CIs) were calculated by 2000 stratified bootstrapping, which contained the same number of both groups (24 cases and 126 controls) as the original sample. The area under the ROC curve was also calculated to evaluate the accuracy of wGRS predictors. Finally, the difference between area under the ROC curves was tested by DeLong method, an asymptotically exact method to calculate the uncertainty of an AUC as described by DeLong et al.39
Results
Sample Substructure Evaluation of GWAS
KD patients (n=150) treated at Kaohsiung Chang Gung Memorial Hospital were included in this study. In total, 867 877 SNPs were genotyped in 24 IVIG nonresponders (male=58.33%, age=2.24 [mean]±2.82 [SD] years old) and 126 IVIG responders (male=60.32%, age=2.16±2.41 years old). After marker-level quality control, 264 179 of 867 877 (30.44%) markers were filtered. SNPs were defined to pass quality control with following criteria: minor allele frequency >5.0%, genotyping call rate >99%, and passed Hardy–Weinberg equilibrium test in controls (P<1×10−05). SNPs of 603 698 of 867 877 (69.56%) remained for further analysis. In sample-level quality control step, all subjects passed the criteria of sample call rate >99% and were retained for further analysis. PCA was performed to assess sample structure and identify sample outliers. Inspecting the first 3 PCA axes, no significant population stratification (Figure 1A and 1B) was identified. P values from an allelic association test showed no inflation and revealed a low possibility of sample substructure. (Genomic inflation factor λ value λGC=1.015; Figure 1C)
Figure 1.

Evaluation of population stratification in Kawasaki disease (KD) genome-wide association study (GWAS) subjects (24 cases and 126 controls). A, Principal components analysis showed similar distributions of the top 3 principal components (PC1, PC2, and PC3) in cases and controls. B, Subjects from Taiwanese KD GWAS study and HapMap 3 database were analyzed by using Genome-wide Complex Trait Analysis v1.24 software. The top 2 principal components (PC1 and PC2) were plotted. PCA results indicated that Taiwanese KD subjects were clustered together. C, Allelic association using mixed linear model (MLM) was evaluated by quantile-quantile (Q-Q) plot. The genomic inflation λ value (λGC) was 1.015, revealed a low possibility of population stratification.
Risk Loci Affecting IVIG Responsiveness
To identify the risk loci affecting IVIG responsiveness in Taiwanese KD patients, we conducted a mixed linear model association test. Considering the modest sample size in our study, genetic relation matrix was estimated and incorporated into the mixed linear model as a polygenic effect to prevent false-positive association and increase statistical power.40 Eight SNPs that reached genome-wide suggestive significance (5×10−06<P<5×10−08) were identified (Figure 2; Table 1). Among all identified possible candidate loci, SNP rs9380548A/T that showed the strongest association signal (P=1.03×10−06) was located in an intergenic region (6p24.1) of chromosome 6 and the 3′ upstream from ADTRP. The risk allele (A allele) showed an increasing risk (odds ratio, 1.6305) of IVIG unresponsiveness when compared with the nonrisk allele (T allele). Another SNP rs742490G/C (P=2.67×10−06) was located at intron of EXOC2 (6p25.3), the odds ratio by which risk allele (G allele) compared with nonrisk allele (C allele) was 1.4895. rs7634A/T (P=1.51×10−06; odds ratio, 1.2676), rs1250301G/A (P=3.41×10−06; odds ratio, 1.4635) and rs2797915T/C (P=3.41×10−06; odds ratio, 1.4635) located on chromosome 10 were also found to be associated with IVIG nonresponsiveness. rs7634A/T was at 3′ untranslated region of KLF6 (10p15.2), whereas rs1250301G/A and rs2797915T/C were in an intergenic region between ZNF438 and ZEB1 (10p11.22). Moreover, rs4130857A/G, rs4585205T/C, and rs4602399G/T at an intergenic region between MIR548A3 and ZPLD1 (3q13.11) were also found to be associated with IVIG nonresponsiveness (P=4.87×10−06; odds ratio, 1.2660).
Figure 2.
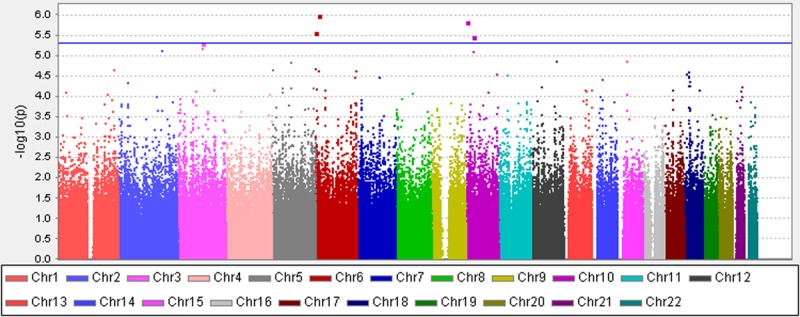
Manhattan plot for the Kawasaki disease intravenous immunoglobulin (IVIG) responsiveness analysis. Association values (−log10 p) obtained using the mixed linear model association test were plotted against chromosome (based on genomic build hg19). The blue line indicated the genome-wide suggestive significant threshold (P<5×10−6, −log10 P<5.3). CEU indicates Northern Europeans from Utah; CHB, Han Chinese in Bejing, China; JPT, Japanese in Tokyo; and YRI, Yoruba in Ibadan.
Table 1.
Genome-Wide Association Results

wGRS Analysis for Prediction of IVIG Responsiveness
To predict the responsiveness of KD patients to IVIG, we applied a scoring system based on wGRS proposed by De Jager et al.37 In total, 11 SNPs with P<1×10−05 were included in the wGRS algorithm. The wGRS of patients was calculated and further analyzed (Table I in the Data Supplement). The wGRS showed a significant difference (Wilcoxon rank-sum test P=1.071×10−10) between IVIG responsive and nonresponsive groups (Figure IA and IB in the Data Supplement). Patients were categorized into 4 strata: group 1 (wGRS<0.042), group 2 (0.042≤wGRS<0.636), group 3 (0.636≤wGRS<1.681), and group 4 (wGRS≥1.681). The cumulative effects of SNPs were evaluated (Table 2; Table I in the Data Supplement). As shown in Table 2, all patients (42 of 42) in group 1 showed good responsiveness to IVIG treatment, whereas 14 of 21 IVIG nonresponders were in the group 4. The results indicated that the patient categorized in group 4 would be estimated to have 164.33 fold higher risk for IVIG resistance compared with those in group 1 (P=8.224×10−10; 95% CI, 8.83–3059.50). The sensitivity and specificity of group 4 versus group 1 were 100% and 85.7%, respectively.
Table 2.
wGRS Analysis

To assess the effect of boundaries selected on comparison groups, we repeated the association test between IVIG responsiveness and groups by different thresholds. First, we categorized the samples into 3 strata, that is, group 1 (wGRS<|mean−SD|), group 2 (|mean−SD|≤wGRS<|mean+SD|), and group 3 (wGRS≥|mean+SD|). The results were also significant, revealing a higher ratio of IVIG resistance in the group with the higher wRGS score (Table II in the Data Supplement). In addition, we recategorized the samples into 3 strata, including group 1 (wGRS<lowest tertile), group 2 (lowest tertile≤wGRS<highest tertile), and group 3 (wGRS≥highest tertile). Again, the association tests revealed that individuals with IVIG resistance had higher wGRS scores than individuals who were IVIG responsive (Table III in the Data Supplement). Together, we confirmed the correlation between IVIG responsiveness and stratification according to the wGRS.
Furthermore, we performed the analysis based on 7 SNPs after removing 3 SNPs that were in high linkage disequilibrium. As rs4130857, rs4585205, and rs4602399 on chromosome 3 and rs1250301 and rs2797915 on chromosome 10 are in highly linkage disequilibrium, we selected rs4130857 and rs1250301 (in total 7 SNPs) for the wGRS analysis. As expected, we observed a similar association compared with the results using 11 SNPs (Table IV in the Data Supplement).
ROC Curve Analysis of wGRS
Further analysis between groups 4 and 1 indicated a positive predictive value of 0.667, negative predictive value of 1.000, positive likelihood ratio of 6.993 (moderate evidence to rule in disease), and negative likelihood ratio of 0 (strong evidence to rule out disease). Besides, the odds ratio of IVIG resistant is estimated to be 16.65 in group 3 and 4.32 in group 2 (P=4.518×10−03; 95% CI, 0.94–294.67, sensitivity = 100%, specificity = 46.7%; P=1.000; 95% CI, 0.17–109.79, sensitivity = 100%, specificity = 59.2%, respectively), by using group 1 as the reference. Subcategory case–control analysis was further performed to confirm the difference of intragroup wGRS in each group (Figure IC in the Data Supplement). Results revealed a significant difference between case and control in group 4 (P=5.742×10−03), whereas no difference was found between groups 2 and 3 (P=0.2398 and P=0.0688, respectively).
Finally, ROC curves were constructed to evaluate the performance of wGRS in categorizing KD patients into the IVIG-responsive or nonresponsive group. The results indicated that high area under the ROC curve (91.1%) revealed a good separating ability of wGRS predictors (Figure 3A). The best predictive value of wGRS was 1.3, and the corresponding specificity and sensitivity were 88.9% and 79.2%, respectively. After adjusted for sex effects, the performance of wGRS remains well, with the best value of 2.8. The corresponding specificity and sensitivity were 81.7% and 79.2%, respectively (Figure 3B). Comparison between 2 ROCs showed a P value of 0.10502 (Z statistic of 1.621) under the null hypothesis of a true difference in AUC was equal to 0.
Figure 3.
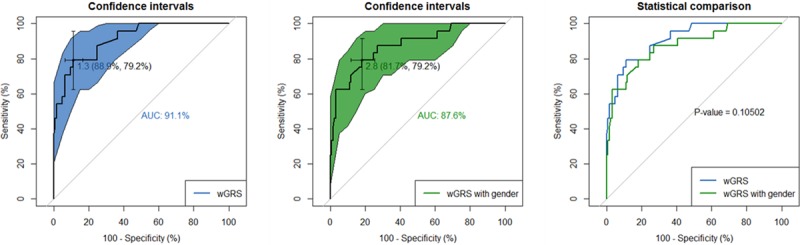
Receiver operating characteristic (ROC) curve analysis of weighted genetic risk score (wGRS). A, ROC of wGRS with 1-Specificity as x axis and sensitivity with y axis. The confidence intervals of ROC were calculated by DeLong method. The most predictive wGRS value (1.3) and the corresponding specificity and sensitivity (88.9% and 79.2%, respectively) were shown. B, ROC of wGRS in considered the effect of sex was shown. C, Comparison between ROC of wGRS and wGRS with sex.
To infer the generalizability of our model, we compared the frequencies of 11 SNPs across different populations (Table V in the Data Supplement) according to the HAPMAP database. The frequencies of the 11 SNPs from the Taiwanese cohort were similar to other East Asian populations, including Han Chinese in Bejing, China and Japanese in Tokyo, but differed from Northern Europeans from Utah, and Yoruba in Ibadan populations.
Discussion
KD is a systemic vasculitis that mostly affected children of <5-years old, though, increased prevalence world widely. The most severe complication is CALs that will cause lifelong sequelae for children. IVIG or intravenous γ-globulin has been administrated for the treatment of KD since 1983.41 Administration of high dose IVIG significantly reduced the CAL rate from 49.0% to 18.4%. Currently, a single high dose of IVIG (2 g/kg) is considered as a gold standard in the treatment of KD. The molecular mechanisms of IVIG for the treatment of KD remain unclear. Several studies have identified epidemiological and laboratory characteristics as predictors of IVIG resistance. The biomarkers include patients’ age, illness day, serum markers (including platelet count, erythrocyte sedimentation rate, hemoglobin concentration, C-reactive protein, eosinophil counts, lactate dehydrogenase, plasma level of albumin, alanine aminotransferase, clusterin, and granulocyte colony-stimulating factor), and sonographic gallbladder abnormalities.42–47 In addition, genetic variations were reported to associate with IVIG resistance in KD patients. The genetic markers include FcγR family, inositol 1,4,5-trisphosphate 3-kinase C (ITPKC), caspase-3 (CASP3), CD209, CCR2-CCR5, CCL3L1, interleukin 1 beta (IL-1B), and platelet-activating factor acetylhydrolase.48 However, for each KD patient, the clinical value of single identified risk genetic variants is less informative because of the modest odds ratio. This limitation causes the difficulty to translate the genomics findings into clinical applications.
Chung et al applied wGRS system to categorize breast cancer patients based on their risk of chemotherapy-induced alopecia. Their study demonstrated a potential for clinical use of wGRS.49 In this study, we also applied wGRS analysis based on the GWAS data to develop a predictive model for IVIG responsiveness. The advantage of wGRS algorithm is to identify the cumulative effects of multiple SNPs for IVIG drug responsiveness in KD patients. By using wGRS to aggregate the odds ratio of multiple genetic risk variants, the predictive accuracy of IVIG drug responsiveness can be improved largely. Indeed, our study showed the inspiring result that wGRS scoring system can significantly increase the sensitivity and specificity of prediction of IVIG responsiveness (Figure 3A and 3B).
We recognize both strengths and limitations to our study. Because of low prevalence of KD and rare cases of IVIG resistance, the sample size of cohort used in this study was small. In addition, the effect size of each SNP was modest. The wide CIs for the odds ratio suggest that replication of these findings in larger cohorts should be performed.
In short, our study combined with the wGRS algorithm and GWAS analysis (GWAS of 603 698 SNPs). We integrated the additive effects of all 11 risk SNPs from GWAS results to establish a predictive model and to identify a new algorithm model as a rational prediction information for IVIG responsiveness in KD patients.
Acknowledgments
We thank all affected individuals and their families who devoted their time and effort to participate in this study. The genome-wide association study (GWAS) data of this study were supported by Academia Sinica Genomic Medicine Multicenter Study, Taiwan. We are grateful to Jane C. Burns (University of California San Diego), Dr Chisato Shimizu (University of California San Diego), Dr Jihoon Kim (University of California San Diego), and Yusuke Nakamura (University of Chicago), for providing valuable suggestions in the revised article and data analysis.
Sources of Funding
This study was partly supported by a grant from the Ministry of Science and Technology, Taiwan (MOST 105-2628-B-038-001-MY4), a grant from Taipei Medical University (TMU 105-5807-001-400) and grant from Chang Gung Memorial Hospital (CMRPG8D1561). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Disclosures
None.
Supplementary Material
Footnotes
Dr Kuo and H.S.-C. Wong contributed equally to this work.
The Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/doi:10.1161/CIRCGENETICS.116.001625/-/DC1.
CLINICAL PERSPECTIVE
We have identified candidate single-nucleotide polymorphisms in select genetic loci to predict intravenous immunoglobulin responsiveness in Kawasaki disease patients. We also developed a scoring system that may be implemented in clinical practice to predict outcome of first-line therapy of Kawasaki disease children.
References
- 1.Newburger JW, Takahashi M, Gerber MA, Gewitz MH, Tani LY, Burns JC, et al. Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease; Council on Cardiovascular Disease in the Young; American Heart Association. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Pediatrics. 2004;114:1708–1733. doi: 10.1542/peds.2004-2182. doi: 10.1542/peds.2004-2182. [DOI] [PubMed] [Google Scholar]
- 2.Kawasaki T. Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes in children. Arerugi. 1967;16:178–222. [PubMed] [Google Scholar]
- 3.Bayers S, Shulman ST, Paller AS. Kawasaki disease: part I. Diagnosis, clinical features, and pathogenesis. J Am Acad Dermatol. 2013;69:501.e501–501.e511. doi: 10.1016/j.jaad.2013.07.002. quiz 511–512. [DOI] [PubMed] [Google Scholar]
- 4.Uehara R, Yashiro M, Nakamura Y, Yanagawa H. Kawasaki disease in parents and children. Acta Paediatr. 2003;92:694–697. doi: 10.1080/08035320310002768. [DOI] [PubMed] [Google Scholar]
- 5.Burgner D, Davila S, Breunis WB, Ng SB, Li Y, Bonnard C, et al. International Kawasaki Disease Genetics Consortium. A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet. 2009;5:e1000319. doi: 10.1371/journal.pgen.1000319. doi: 10.1371/journal.pgen.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onouchi Y, Ozaki K, Burns JC, Shimizu C, Terai M, Hamada H, et al. Japan Kawasaki Disease Genome Consortium; US Kawasaki Disease Genetics Consortium. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. 2012;44:517–521. doi: 10.1038/ng.2220. doi: 10.1038/ng.2220. [DOI] [PubMed] [Google Scholar]
- 7.Lee YC, Kuo HC, Chang JS, Chang LY, Huang LM, Chen MR, et al. Taiwan Pediatric ID Alliance. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat Genet. 2012;44:522–525. doi: 10.1038/ng.2227. doi: 10.1038/ng.2227. [DOI] [PubMed] [Google Scholar]
- 8.Kuo HC, Yang KD, Juo SH, Liang CD, Chen WC, Wang YS, et al. ITPKC single nucleotide polymorphism associated with the Kawasaki disease in a Taiwanese population. PLoS One. 2011;6:e17370. doi: 10.1371/journal.pone.0017370. doi: 10.1371/journal.pone.0017370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onouchi Y, Gunji T, Burns JC, Shimizu C, Newburger JW, Yashiro M, et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat Genet. 2008;40:35–42. doi: 10.1038/ng.2007.59. doi: 10.1038/ng.2007.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo HC, Onouchi Y, Hsu YW, Chen WC, Huang JD, Huang YH, et al. Polymorphisms of transforming growth factor-β signaling pathway and Kawasaki disease in the Taiwanese population. J Hum Genet. 2011;56:840–845. doi: 10.1038/jhg.2011.113. doi: 10.1038/jhg.2011.113. [DOI] [PubMed] [Google Scholar]
- 11.Kuo HC, Huang YH, Chien SC, Yu HR, Hsieh KS, Hsu YW, et al. Genetic variants of CD209 associated with Kawasaki disease susceptibility. PLoS One. 2014;9(8):e105236. doi: 10.1371/journal.pone.0105236. doi: 10.1371/journal.pone.0105236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo HC, Chang WC. Genetic polymorphisms in Kawasaki disease. Acta Pharmacol Sin. 2011;32:1193–1198. doi: 10.1038/aps.2011.93. doi: 10.1038/aps.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayers S, Shulman ST, Paller AS. Kawasaki disease: part II. Complications and treatment. J Am Acad Dermatol. 2013;69:513.e511–513.e518. doi: 10.1016/j.jaad.2013.06.040. quiz 521–522. [DOI] [PubMed] [Google Scholar]
- 14.Harnden A, Alves B, Sheikh A. Rising incidence of Kawasaki disease in England: analysis of hospital admission data. BMJ. 2002;324:1424–1425. doi: 10.1136/bmj.324.7351.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y, Yashiro M, Uehara R, Oki I, Watanabe M, Yanagawa H. Monthly observation of the number of patients with Kawasaki disease and its incidence rates in Japan: chronological and geographical observation from nationwide surveys. J Epidemiol. 2008;18:273–279. doi: 10.2188/jea.JE2008030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newburger JW, Takahashi M, Beiser AS, Burns JC, Bastian J, Chung KJ, et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N Engl J Med. 1991;324:1633–1639. doi: 10.1056/NEJM199106063242305. doi: 10.1056/NEJM199106063242305. [DOI] [PubMed] [Google Scholar]
- 17.Singh S, Sharma D, Bhattad S, Phillip S. Recent advances in Kawasaki disease - Proceedings of the 3rd Kawasaki Disease Summit, Chandigarh, 2014. Indian J Pediatr. 2016;83:47–52. doi: 10.1007/s12098-015-1858-4. doi: 10.1007/s12098-015-1858-4. [DOI] [PubMed] [Google Scholar]
- 18.Pośnik-Urbańska A, Szymanowska Z, Kawecka-Jaszcz K. The probability of Kawasaki diseases in young patients with cardiac disorders–retrospective studies. Przegl Lek. 2003;60:792–796. [PubMed] [Google Scholar]
- 19.Shrestha S, Wiener H, Shendre A, Kaslow RA, Wu J, Olson A, et al. Role of activating FcγR gene polymorphisms in Kawasaki disease susceptibility and intravenous immunoglobulin response. Circ Cardiovasc Genet. 2012;5:309–316. doi: 10.1161/CIRCGENETICS.111.962464. doi: 10.1161/CIRCGENETICS.111.962464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ, et al. Hong Kong–Shanghai Kawasaki Disease Genetics Consortium; Korean Kawasaki Disease Genetics Consortium; Taiwan Kawasaki Disease Genetics Consortium; International Kawasaki Disease Genetics Consortium; US Kawasaki Disease Genetics Consortium; Blue Mountains Eye Study. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. 2011;43:1241–1246. doi: 10.1038/ng.981. doi: 10.1038/ng.981. [DOI] [PubMed] [Google Scholar]
- 21.Kuo HC, Hsu YW, Wu MS, Woon PY, Wong HS, Tsai LJ, et al. FCGR2A promoter methylation and risks for intravenous immunoglobulin treatment responses in Kawasaki disease. Mediators Inflamm. 2015;2015:564625. doi: 10.1155/2015/564625. doi: 10.1155/2015/564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Wang W, Gong F, Fu S, Zhang Q, Hu J, et al. Evaluation of intravenous immunoglobulin resistance and coronary artery lesions in relation to Th1/Th2 cytokine profiles in patients with Kawasaki disease. Arthritis Rheum. 2013;65:805–814. doi: 10.1002/art.37815. doi: 10.1002/art.37815. [DOI] [PubMed] [Google Scholar]
- 23.Kuo HC, Yang YL, Chuang JH, Tiao MM, Yu HR, Huang LT, et al. Inflammation-induced hepcidin is associated with the development of anemia and coronary artery lesions in Kawasaki disease. J Clin Immunol. 2012;32:746–752. doi: 10.1007/s10875-012-9668-1. doi: 10.1007/s10875-012-9668-1. [DOI] [PubMed] [Google Scholar]
- 24.Lou J, Zhong R, Shen N, Lu XZ, Ke JT, Duan JY, et al. Systematic confirmation study of GWAS-identified genetic variants for Kawasaki disease in a Chinese population. Sci Rep. 2015;5:8194. doi: 10.1038/srep08194. doi: 10.1038/srep08194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onouchi Y. Susceptibility genes for Kawasaki disease. Nihon Rinsho. 2014;72:1554–1560. [PubMed] [Google Scholar]
- 26.Peng Q, Chen CH, Wu Q, Yang Y. Meta-analyses of the associations of genome-wide association study- linked gene loci with Kawasaki disease. Zhonghua Er Ke Za Zhi. 2013;51:571–577. [PubMed] [Google Scholar]
- 27.Liang CD, Kuo HC, Yang KD, Wang CL, Ko SF. Coronary artery fistula associated with Kawasaki disease. Am Heart J. 2009;157:584–588. doi: 10.1016/j.ahj.2008.11.020. doi: 10.1016/j.ahj.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Kuo HC, Wang CL, Liang CD, Yu HR, Chen HH, Wang L, et al. Persistent monocytosis after intravenous immunoglobulin therapy correlated with the development of coronary artery lesions in patients with Kawasaki disease. J Microbiol Immunol Infect. 2007;40:395–400. [PubMed] [Google Scholar]
- 29.Kuo HC, Wang CL, Liang CD, Yu HR, Huang CF, Wang L, et al. Association of lower eosinophil-related T helper 2 (Th2) cytokines with coronary artery lesions in Kawasaki disease. Pediatr Allergy Immunol. 2009;20:266–272. doi: 10.1111/j.1399-3038.2008.00779.x. doi: 10.1111/j.1399-3038.2008.00779.x. [DOI] [PubMed] [Google Scholar]
- 30.Newburger JW, Takahashi M, Gerber MA, Gewitz MH, Tani LY, Burns JC, et al. Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease; Council on Cardiovascular Disease in the Young; American Heart Association; American Academy of Pediatrics. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 2004;110:2747–2771. doi: 10.1161/01.CIR.0000145143.19711.78. doi: 10.1161/01.CIR.0000145143.19711.78. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.De Jager PL, Chibnik LB, Cui J, Reischl J, Lehr S, Simon KC, et al. Steering Committee of the BENEFIT Study; Steering Committee of the BEYOND Study; Steering Committee of the LTF Study; Steering Committee of the CCR1 Study. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–1119. doi: 10.1016/S1474-4422(09)70275-3. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 40.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–106. doi: 10.1038/ng.2876. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furusho K, Sato K, Soeda T, Matsumoto H, Okabe T, Hirota T, et al. High-dose intravenous gammaglobulin for Kawasaki disease. Lancet. 1983;2:1359. doi: 10.1016/s0140-6736(83)91109-1. [DOI] [PubMed] [Google Scholar]
- 42.Rigante D, Valentini P, Rizzo D, Leo A, De Rosa G, Onesimo R, et al. Responsiveness to intravenous immunoglobulins and occurrence of coronary artery abnormalities in a single-center cohort of Italian patients with Kawasaki syndrome. Rheumatol Int. 2010;30:841–846. doi: 10.1007/s00296-009-1337-1. doi: 10.1007/s00296-009-1337-1. [DOI] [PubMed] [Google Scholar]
- 43.Kuo HC, Liang CD, Wang CL, Yu HR, Hwang KP, Yang KD. Serum albumin level predicts initial intravenous immunoglobulin treatment failure in Kawasaki disease. Acta Paediatr. 2010;99:1578–1583. doi: 10.1111/j.1651-2227.2010.01875.x. doi: 10.1111/j.1651-2227.2010.01875.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuo HC, Yang KD, Liang CD, Bong CN, Yu HR, Wang L, et al. The relationship of eosinophilia to intravenous immunoglobulin treatment failure in Kawasaki disease. Pediatr Allergy Immunol. 2007;18:354–359. doi: 10.1111/j.1399-3038.2007.00516.x. doi: 10.1111/j.1399-3038.2007.00516.x. [DOI] [PubMed] [Google Scholar]
- 45.Sleeper LA, Minich LL, McCrindle BM, Li JS, Mason W, Colan SD, et al. Pediatric Heart Network Investigators. Evaluation of Kawasaki disease risk-scoring systems for intravenous immunoglobulin resistance. J Pediatr. 2011;158:831.e3–835.e3. doi: 10.1016/j.jpeds.2010.10.031. doi: 10.1016/j.jpeds.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egami K, Muta H, Ishii M, Suda K, Sugahara Y, Iemura M, et al. Prediction of resistance to intravenous immunoglobulin treatment in patients with Kawasaki disease. J Pediatr. 2006;149:237–240. doi: 10.1016/j.jpeds.2006.03.050. doi: 10.1016/j.jpeds.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 47.Ou-Yang MC, Kuo HC, Lin IC, Sheen JM, Huang FC, Chen CC, et al. Plasma clusterin concentrations may predict resistance to intravenous immunoglobulin in patients with Kawasaki disease. ScientificWorld J. 2013;2013:382523. doi: 10.1155/2013/382523. doi: 10.1155/2013/382523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo HC, Hsu YW, Wu MS, Chien SC, Liu SF, Chang WC. Intravenous immunoglobulin, pharmacogenomics, and Kawasaki disease. J Microbiol Immunol Infect. 2016;49:1–7. doi: 10.1016/j.jmii.2014.11.001. doi: 10.1016/j.jmii.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Chung S, Low SK, Zembutsu H, Takahashi A, Kubo M, Sasa M, et al. A genome-wide association study of chemotherapy-induced alopecia in breast cancer patients. Breast Cancer Res. 2013;15:R81. doi: 10.1186/bcr3475. doi: 10.1186/bcr3475. [DOI] [PMC free article] [PubMed] [Google Scholar]


