The unspliced form of XBP1 stabilizes MDM2 protein by inhibiting its ubiquitination and regulates the MDM2/p53 axis.
Abstract
Cell cycle progression is a tightly controlled fundamental process in living cells, with any defects being closely linked to various abnormalities. The tumor suppressor p53/p21 axis is a core pathway controlling cell cycle progression; however, its regulatory mechanism has not been fully elucidated. In an effort to unravel this crucial network, we screened a short hairpin RNA expression vector library and identified unspliced X-box binding protein 1 (XBP1-u) as a novel and critical regulator of the p53/p21 axis. Specifically, XBP1-u negatively regulates the p53/p21 axis by enhancing p53 ubiquitination, which in turn down-regulates p21 expression. We show that XBP1-u suppression induces G0-G1 phase arrest and represses cell proliferation. We further report that the carboxyl terminus of XBP1-u, which differs from that of its spliced form (XBP1-s) due to a codon shift, binds and stabilizes mouse double minute homolog 2 (MDM2) protein, a negative regulator of p53, by inhibiting its self-ubiquitination. Concomitantly, XBP-u overexpression enhances tumorigenesis by positively regulating MDM2. Together, our findings suggest that XBP1-u functions far beyond being merely a precursor of XBP1-s and, instead, is involved in fundamental biological processes. Furthermore, this study provides new insights regarding the regulation of the MDM2/p53/p21 axis.
INTRODUCTION
Cell cycle is a critical event controlling cell proliferation. It progresses in a directional manner following well-ordered events: DNA replication, spindle assembly, nuclear division, and cytokinesis. Cell cycle progression is regulated by numerous proteins, including cyclins and cyclin-dependent kinases (CDKs), whose expression oscillates throughout the cell cycle and is tightly controlled. p21 was the first reported CDK inhibitor and was identified as a tumor suppressor gene induced by p53 (1, 2). It binds to cyclins and CDKs and thus negatively regulates cell cycle progression (3). It also participates in other important physiological processes, such as DNA repair, stem cell maintenance, differentiation, and senescence; thus, loss of p21 might lead to various disorders including tumorigenesis (3–5). Mice lacking p21 display higher tumorigenesis potential, and their embryonic fibroblast cells can bypass the G1-S checkpoint upon exposure to DNA damage (6). Despite functioning as a tumor suppressor and being down-regulated in various malignancies (7, 8), p21 itself is rarely mutated in human cancers (3, 9). These findings underscore the importance of the regulatory pathways controlling p21 gene expression, which have not been fully elucidated.
Here, in an effort to unravel the regulatory mechanism of the p53/p21 axis, we screened a short hairpin RNA (shRNA) vector library and identified X-box binding protein 1 (XBP1) as a negative regulator of p21 transcriptional activity. XBP1 has been characterized as a bZIP (basic-region leucine zipper) transcription factor that interacts specifically with the conserved X2 boxes of major histocompatibility complex class II gene promoters (10). XBP1 yields two isoforms: unspliced XBP1 (XBP1-u) and spliced XBP1 (XBP1-s). Upon exposure to endoplasmic reticulum (ER) stress, XBP1-u is spliced, and the 26 nucleotides located between +541 and +566 of XBP1-u are excised, causing a codon frameshift in XBP1-s and distinct C-terminal regions between the two isoforms (11, 12). XBP1-s is considered the active form, playing a pivotal role in the unfolded protein response (UPR) network (12). However, we report herein that the regulatory effect on p21 is specific to the poorly characterized XBP1-u isoform but not to the well-known XBP1-s isoform. The C terminus of XBP1-u binds to mouse double minute 2 homolog [MDM2 (also known as HDM2)] and inhibits its homodimerization and self-ubiquitination. The accumulation of MDM2 protein, in turn, accelerates the ubiquitination and proteasomal degradation of p53, which is a positive regulator of p21 transcription. Together, our findings uncover a pivotal role of XBP1-u in regulating the cell cycle and its link to the conventional MDM2/p53 axis.
RESULTS
Screening of shRNA expression vector library leads to the identification of factors involved in p21 transcriptional regulation
We established a screening system using an shRNA expression vector library and a p21 reporter gene in HCT116WT human colon carcinoma cells. The system was validated with shRNA against positive (p53) and negative (MDM2) regulators of p21: Silencing of p53 significantly decreased p21 reporter activity, whereas silencing of MDM2 robustly increased it (fig. S1A).
Next, we screened an shRNA expression vector library containing 3354 shRNA expression vectors covering 2065 genes (Fig. 1A): 1289 genes with two vectors targeting different sites per gene and 776 genes with one shRNA expression vector per gene. This screening led to the identification of more than 300 candidates or around 10% of the overall screened genes, for which p21 reporter activity was stronger than with shMDM2, and thus, those candidates were considered potential p21 suppressors (Fig. 1B, left, and table S1). To reduce the false-positive results caused by the off-target effect of shRNA, we gave priority to the 14 genes with two shRNA expression vectors among the top 10% of potential p21 suppressors. Among them, we noticed the presence of XBP1 (Fig. 1B, right). XBP1 has been known as a critical player in ER stress (12); however, its role within the p21 network remains unknown.
Fig. 1. Screening for p21 regulators using an shRNA expression vector library.
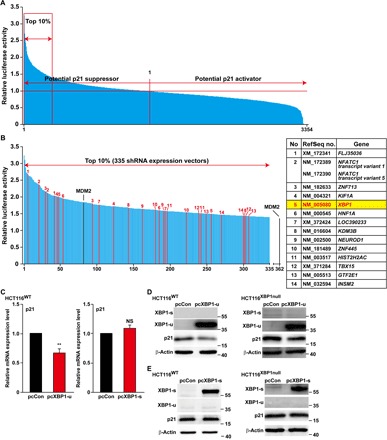
(A) Screening results of a library containing 3354 shRNA expression vectors. Relative luciferase activity was calculated as the ratio of firefly and Renilla luciferase activities. The ratios were then normalized with the average ratio of the measurement of each 96-well plate. (B) Top 10% potential p21 suppressors. Genes with both shRNA expression vectors included in the top 10% are shown in red and listed above in the right panel; MDM2 is shown in black. (C) p21 mRNA expression level in HCT116WT cells transfected with either pcXBP1-u or pcXBP1-s, as determined by quantitative reverse transcription polymerase chain reaction (qPCR). pcCon, pcDNA3.1(+); NS, not significant. (D and E) p21 protein expression level in HCT116WT and HCT116XBP1null cells transfected with either pcXBP1-u (D) or pcXBP1-s (E), as determined by Western blotting. Cells transfected with pcCon were used as control. β-Actin was used for qPCR normalization and as Western blotting loading control. qPCR data were shown relative to control and expressed as the mean ± SEM of three independent experiments. **P < 0.01 [analysis of variance (ANOVA)].
XBP1-u negatively regulates p21 transcriptional activity
We further confirmed the regulatory effect of XBP1 on the p21 promoter using two more shRNA expression vectors against XBP1 (shXBP1-3 and shXBP1-4) and by establishing HCT116XBP1null cells using the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 method. The p21 reporter activity was enhanced in both XBP1-silenced cells and HCT116XBP1null cells (fig. S1, B and C). Consistently, both XBP1 silencing and knockout robustly increased mRNA and protein expression levels of p21 (fig. S1, D to F). Together, these results indicate that XBP1 might be a novel p21 transcriptional regulator.
XBP1 is expressed as XBP1-u, which is spliced into XBP1-s upon ER stress (fig. S2, A and B). We then investigated the effect of thapsigargin, which induced XBP1 splicing and increased XBP1-s levels (fig. S2C), on p21 expression. Surprisingly, instead of suppressing it, thapsigargin promoted p21 expression (fig. S2, D and E). It should be noted that in contrast to the condition with thapsigargin addition, both the protein level and copy number of XBP1-u were significantly higher than those of XBP1-s under basal condition (that is, without thapsigargin addition), and thus, under basal condition, the shXBP1 vectors described above mainly affected the levels of XBP1-u (fig. S2, E to G). Hence, we assumed that the effect of XBP1 silencing and knockout described in this work could be attributed to the absence of XBP1-u. Next, we selectively overexpressed XBP1-u and XBP1-s in HCT116WT cells (fig. S3, A and B). Only overexpression of XBP1-u could significantly suppress p21 mRNA and protein expression, whereas XBP1-s overexpression failed to produce any significant changes (Fig. 1, C to E, and fig. S3C). Similar results were also obtained with HCT116XBP1null cells (Fig. 1, D and E, and fig. S3C). It should be noted that XBP1-u overexpression did not affect the levels of XBP1-s and its downstream genes ATF4, CHOP, and BIP (fig. S3, A and D). Together, these results strongly indicate that XBP1-u, but not XBP1-s, is a negative regulator of p21.
XBP1-u suppression induces G0-G1 arrest
Knowing that p21 is a master regulator of cell cycle progression, we investigated the role of XBP1-u in cell cycle regulation. Both XBP1 silencing and knockout significantly reduced HCT116WT cell number (Fig. 2A and fig. S4A). XBP1 suppression robustly reduced cell proliferation, as indicated by the number of ethynyl deoxyuridine (EdU)–positive cells (Fig. 2B), and largely increased the percentage of G0-G1 cells (Fig. 2C and fig. S4B, for XBP1 silencing and XBP1 knockout, respectively). Concomitantly, the expression of G1 regulatory factors cyclin D and CDK4 was suppressed in XBP1-silenced cells (Fig. 2D and fig. S4C). Furthermore, the suppression of cyclin D and CDK4 expression was also observed in XBP1 knockout and thapsigargin-treated cells (fig. S4, D and E). In contrast, the levels of these factors increased substantially following overexpression of XBP1-u, but not XBP1-s, in both XBP1-silenced HCT116WT cells (Fig. 2E and fig. S4, F and G) and HCT116XBP1null cells (fig. S4H). The same pattern was also observed for other cyclins and CDK (fig. S4, I and J).
Fig. 2. XBP1-u regulates cell cycle progression and cell proliferation.
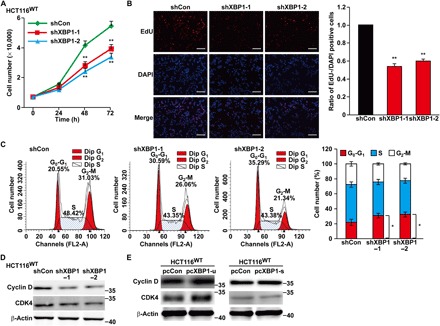
(A) Total cell number of XBP1-silenced HCT116WT cells at the indicated time points. (B) Number of proliferating XBP1-silenced HCT116WT cells, as determined by the EdU incorporation assay. Representative images (left) and the ratio of EdU-positive cells (right) are shown. Cells transfected with control vector (shCon) were used as control. 4,6′-Diamidino-2-phenylindole (DAPI) was used to stain the nuclei. Proliferating cells were calculated as the ratio of EdU-positive and DAPI-positive cells and are shown relative to the control. Scale bars, 200 μm. (C) Different cell cycle phases of XBP1-silenced HCT116WT cells. Cells were stained using propidium iodide, and the percentages were determined by flow cytometry. Representative images are shown (left), and the average percentage of cells in each cell cycle phase from three independent experiments was calculated (right). (D and E) Protein expression levels of cyclin D and CDK4 in XBP1-silenced HCT116WT cells (D) or HCT116WT cells transfected with either pcXBP1-u or pcXBP1-s (E), as analyzed by Western blotting. β-Actin was used as loading control. Quantitative data are expressed as the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 (ANOVA).
p53 plays a crucial role in XBP1-u–regulated transcription of p21
It is well known that the tumor suppressor p53 blocks cell cycle progression by positively regulating p21 transcriptional activity (13). Thus, we investigated the role of p53 in the XBP1-u–mediated regulation of p21. Both XBP1 silencing and knockout promoted the accumulation of p53 protein but not its mRNA, a pattern that was reversed by XBP1-u overexpression (Fig. 3, A and B, and fig. S5A). Next, we constructed a mutant p21 reporter vector without the p53 binding site (p21mut-luc). We found that overall luciferase activities in HCT116WT cells cotransfected with p21mut-luc were significantly lower than those in HCT116WT cells cotransfected with a wild-type p21 reporter, irrespective of XBP1 status (fig. S5B). A similar tendency was also observed in HCT116p53null cells cotransfected with a wild-type p21 reporter (fig. S5B). Moreover, XBP1 silencing failed to promote p21 mRNA expression in p53-null cells (Fig. 3C) and only slightly affected p21 protein accumulation (fig. S5C), as well as cyclins and CDKs expression (fig. S5, D and E). Concomitantly, the effect of XBP1 silencing on the characteristics of HCT116p53null cells, that is, total cell number (fig. S6, A and B), induction of G0-G1 arrest (fig. S6C), and colony-forming potential (fig. S6, D and E), was significantly weaker than the effect on the characteristics of HCT116WT cells.
Fig. 3. XBP1-u promotes p53 protein degradation.
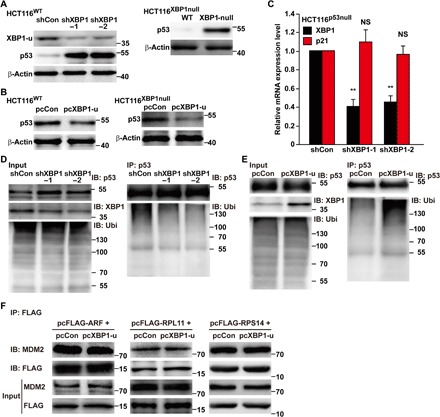
(A and B) Protein expression level of p53 in XBP1-silenced HCT116WT cells and XBP1–knocked out HCT116XBP1null cells (A) or in HCT116WT cells and HCT116XBP1null cells transfected with pcXBP1-u (B), as determined by Western blotting. (C) p21 mRNA expression level in XBP1-silenced HCT116p53null cells, as analyzed by qPCR. Data are shown relative to those of cells transfected with shCon and expressed as the mean ± SEM of three independent experiments. (D and E) p53 ubiquitination levels in HCT116WT cells transfected with pcp53, pcUbi, and shXBP1 (D) or pcXBP1-u (E) were analyzed by anti-ubiquitin immunoblotting (IB) of cell lysates immunoprecipitated with anti-p53 antibody. Ubi, ubiquitin. (F) Physical interactions between MDM2 and ARF, RPL11, or RPS14 in HCT116WT cells transfected with corresponding FLAG-conjugated overexpression vectors, MDM2 and pcXBP1-u, as determined by immunoprecipitation (IP). Cell lysates were immunoprecipitated with anti-FLAG antibody; binding was detected by immunoblotting against anti-MDM2 antibody. β-Actin was used for qPCR normalization and as Western blotting loading control. **P < 0.01 (ANOVA).
Given that p53 had been reported to be degraded by the proteasomal pathway, we investigated the role of XBP1-u in the ubiquitination of p53. XBP1 silencing significantly reduced p53 ubiquitination, whereas overexpression of XBP1-u robustly increased it (Fig. 3, D and E). These results suggest that XBP1-u regulates p21 mainly at a transcriptional level and in a p53-dependent manner via increased p53 ubiquitination. Previous studies reported that factors such as p14(ARF) and ribosomal proteins, for example, RPL11 and RPS14, could form complexes with MDM2, a negative regulator of p53 protein stability (14), and suppress its stimulation of p53 ubiquitination (15–17). Therefore, we investigated whether XBP1-u affected the binding of these factors to MDM2. As shown in Fig. 3F, overexpression of XBP1-u had no significant effect on the binding of p14(ARF), RPL11, or RPS14 to MDM2, indicating that XBP1-u did not exert its function in degrading p53 through any of these pathways.
XBP1-u regulates the p53/p21 axis by promoting MDM2 protein accumulation
Next, we investigated whether XBP1-u exerted its inhibitory effect on the p53/p21 axis through a positive regulation of MDM2. We found that silencing of XBP1, as well as knocking out XBP1 and decreasing XBP1-u level by inducing its splicing, resulted in a significant decrease in MDM2 protein level, whereas overexpressing XBP1-u, but not XBP1-s, induced its accumulation (Fig. 4A and fig. S7, A to D). Furthermore, MDM2 overexpression repressed the p21 and p53 up-regulation observed in XBP1-silenced cells (fig. S7E). Next, we constructed HCT116XBP1null/pcMDM2 as well as control (HCT116WT/pcDNA3 and HCT116XBP1null/pcDNA3) stable cell lines (Fig. 4B and fig. S7F). Analysis of total cell numbers of these cell lines revealed that MDM2 overexpression in XBP1-null cells successfully restored high cell numbers (fig. S7G). Together, these results suggest that XBP1-u most likely regulates the p53/p21 axis in an MDM2-dependent manner. Furthermore, XBP1-u/MDM2/p53 regulatory pathways could also be found in hepatocarcinoma and breast cancer cell lines (fig. S8, A and B), indicating that this mechanism is common to various human carcinomas.
Fig. 4. XBP1-u stabilizes MDM2 protein by inhibiting its self-ubiquitination.
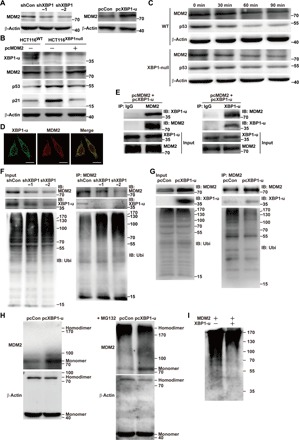
(A and B) MDM2 protein expression level in XBP1-silenced or pcXBP1-u–transfected HCT116WT cells (A) or MDM2, p53, and p21 in HCT116XBP1null/pcMDM2, HCT116XBP1null/pcDNA3, and HCT116WT/pcDNA3 stable cell lines (B), as determined by Western blotting. (C) Degradation rates of MDM2 and p53 in HCT116WT and HCT116XBP1null cells. The levels of MDM2 and p53 protein at the indicated time points after the addition of cycloheximide, which inhibits new protein synthesis, were analyzed using Western blotting. (D) Colocalization of MDM2 and XBP1-u, as determined by immunofluorescence staining. Scale bars, 20 μm. (E) Physical interaction between XBP1-u and MDM2, as determined by anti-XBP1 immunoblotting of cell lysate immunoprecipitated with anti-MDM2 antibody and vice versa. IgG, immunoglobulin G. (F and G) MDM2 ubiquitination level in HCT116WT cells transfected with shXBP1 (F) or pcXBP1-u (G), as determined by anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-MDM2 antibody. (H) Homodimerization of MDM2 in HCT116WT cells transfected with pcXBP1-u, as analyzed by DSS-induced chemical cross-linking assay with or without MG132. (I) Ubiquitination level of MDM2 in the presence or absence of XBP1-u, as analyzed by an in vitro ubiquitination assay. β-Actin was used as loading control.
However, MDM2 mRNA expression in XBP1-silenced HCT116WT cells and XBP1–knocked out (HCT116XBP1null) cells, as well as in HCT116WT cells overexpressing XBP1-u, did not show any significant changes (fig. S9, A and B). These results were intriguing, as a previous study (18) revealed that MDM2 transcription could be regulated by two promoters: the p53-irresponsive p1 promoter and the p53-responsive p2 promoter; thus, MDM2 transcription is also regulated by p53 through a feedback mechanism (19–21). Similar to the effect of XBP1 silencing on mRNA in HCT116WT cells, our results showed that XBP1 silencing did not affect the activities of both p1 and p2 reporters (fig. S9C). Next, we assessed the effect of XBP1 silencing on the MDM2 mRNA expression level in HCT116p53null cells and found that in the absence of p53, XBP1 silencing significantly suppressed MDM2 mRNA expression level (fig. S9D). The same tendency was observed for the activity of the p2 reporter (fig. S9E) in XBP1-silenced HCT116p53null cells. Thus, these results indicate that XBP1 silencing might also down-regulate MDM2 at its transcriptional level. Furthermore, these results suggest that the overall effect of XBP1 silencing on MDM2 transcriptional activity in HCT116WT cells was the accumulative effect of both XBP1 suppression and p53-induced feedback mechanism. In other words, the increase of p53 restored the decrease of MDM2 transcriptional activity caused by XBP1 suppression, resulting in the overall unchanged MDM2 mRNA expression level in XBP1-silenced HCT116WT cells.
XBP1-u enhances MDM2 protein stability by inhibiting its homodimerization and self-ubiquitination
The fact that XBP1-u could positively regulate MDM2 protein accumulation even in cells with wild-type p53, in which no significant change in overall MDM2 mRNA expression could be observed, suggested that XBP1-u might regulate MDM2 at the protein level. Ubiquitin-mediated proteasomal degradation is the canonical pathway for regulating MDM2 homeostasis and is thus critical for fine regulation of the cell cycle and proliferation (22). Time-dependent protein degradation rate assays showed that, compared to HCT116WT, the degradation rate of MDM2 in HCT116XBP1null cells was substantially faster (Fig. 4C), in contrast with the pattern for p53 degradation. Endogenous XBP1-u and MDM2 colocalized to the cytoplasm, and they could be reciprocally immunoprecipitated (Fig. 4, D and E). Moreover, silencing XBP1 significantly enhanced MDM2 ubiquitination, whereas overexpressing XBP1-u reduced it (Fig. 4, F and G).
Given that MDM2 could form homodimers and face degradation via self-ubiquitination (23, 24), we examined the effect of XBP1-u on MDM2 homodimerization and self-ubiquitination. To this end, we performed an in vivo chemical cross-linking assay using disuccinimidyl suberate (DSS), a chemical cross-linking reagent that promotes protein homodimerization (25, 26). Compared to the control, XBP1-u overexpression significantly increased MDM2 monomer level (Fig. 4H, left). The MDM2 homodimer could not be detected in control or in XBP1-u–overexpressing cells (Fig. 4H, left); however, its accumulation was significantly stronger in control cells upon treatment with the proteasome inhibitor MG132 (Fig. 4H, right). These results strongly suggest that XBP1-u inhibited the formation of the MDM2 homodimer, which, as reported previously, is unstable and undergoes proteasomal degradation (26, 27). To further confirm that XBP1-u prevented MDM2 self-ubiquitination, we examined the MDM2 self-ubiquitination potential in the presence of XBP1-u. An in vitro assay showed that the ubiquitination level of MDM2 decreased significantly in the presence of XBP1-u (Fig. 4I). Together, these results revealed that XBP1-u could form a complex with MDM2, enhance MDM2 stability by preventing its homodimerization and self-ubiquitination, and promote p53 protein degradation.
Next, we overexpressed FLAG-conjugated full-length XBP1-u, FLAG-conjugated XBP1-u N-terminal region, or FLAG-conjugated XBP1-u C-terminal region (FLAG-XBP1-u, FLAG-XBP1-u-N, and FLAG-XBP1-u-C, respectively). Immunoprecipitation with anti-FLAG antibody revealed that only the C terminus (and not the N terminus) of XBP1-u could bind MDM2 (Fig. 5A). A glutathione S-transferase (GST) pull-down assay using GST-conjugated MDM2 (GST-MDM2) and His-conjugated XBP1-u fragments (His-XBP1-u, His-XBP1-u-N, and His-XBP1-u-C for the full-length XBP1-u, the N terminus of XBP1-u, and the C terminus of XBP1-u, respectively) further confirmed the above results: Only His-XBP1-u and His-XBP1-u-C could be pulled down by GST-MDM2 (Fig. 5B). We also showed that overexpression of XBP1-u-C suppressed the formation of the MDM2 homodimer (Fig. 5C) and inhibits MDM2 self-ubiquitination (Fig. 5D). These effects could not be observed with XBP1-u-N (fig. S10, A and B). Thus, it is most likely that the C terminus of XBP1-u could bind to MDM2 and suppress its homodimerization, resulting in the decrease of MDM2 self-ubiquitination. We showed that overexpression of FLAG-XBP1-u-C was functionally sufficient to block intracellular MDM2 ubiquitination (Fig. 5E). Concomitantly, XBP1-u-C overexpression induced MDM2 protein accumulation and reduced p53 and p21 protein levels in HCT116WT and HCT116XBP1null cells (Fig. 5F). Together, these results reveal that XBP1-u could bind MDM2, suppress its ubiquitination, and thus regulate the p53/p21 pathway. Moreover, all these functions could be attributed to the C terminus of XBP1-u.
Fig. 5. The C terminus of XBP1-u suppresses MDM2 ubiquitination.
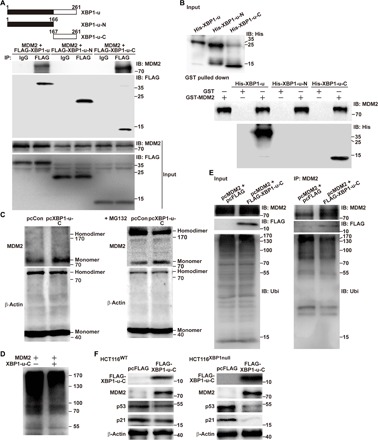
(A) In vivo binding capacity of XBP1-u fragments to MDM2, as determined by immunoprecipitation. HCT116WT cells were transfected with pcMDM2 and pcFLAG-XBP1-u, pcFLAG-XBP1-u-N, or pcFLAG-XBP1-u-C. Cell lysates were immunoprecipitated against IgG or anti-FLAG antibody. The presence of MDM2 was detected by immunoblotting. (B) Binding capacity of XBP1-u fragments to MDM2, as determined by an in vitro GST pull-down assay. His-tagged XBP1-u peptides were mixed with GST-MDM2 peptides and pulled down using a GST column. The presence of XBP1-u fragments was detected using anti-His immunoblotting. (C) Homodimerization of MDM2 in HCT116WT cells transfected with pcXBP1-u-C, as analyzed by DSS-induced chemical cross-linking assay with or without MG132. (D) Ubiquitination level of MDM2 in the presence or absence of XBP1-u-C, as analyzed by an in vitro ubiquitination assay. (E) MDM2 ubiquitination level in HCT116WT cells transfected with pcFLAG-XBP1-u-C, pcMDM2, and pcUbi, as analyzed by anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-MDM2 antibody. (F) Protein expression levels of MDM2, p53, and p21 in HCT116WT and HCT116XBP1null cells transfected with FLAG-XBP1-u-C, as determined by Western blotting. β-Actin was used as loading control.
The XBP1-u/MDM2 pathway is critical for tumorigenesis
To elucidate the pathological function of the XBP1-u/MDM2 pathway in vivo, especially during tumorigenesis, we transplanted HCT116WT/pcDNA3, HCT116XBP1null/pcDNA3, and HCT116XBP1null/pcMDM2 stable cell lines subcutaneously into BALB/c-nu/nu mice. XBP1 deficiency significantly reduced the tumorigenesis potential of HCT116 cells; however, MDM2 overexpression restored it (Fig. 6, A and B). Furthermore, Western blotting results revealed that the expression of MDM2 was negatively regulated in XBP1 knockout cells. Conversely, the expression of p53 and p21 was enhanced but was then suppressed by MDM2 overexpression (Fig. 6C). Immunohistochemistry of tissue sections from xenografted tumors further confirmed the above results (Fig. 6D).
Fig. 6. XBP1-u is involved in MDM2-mediated tumorigenesis.
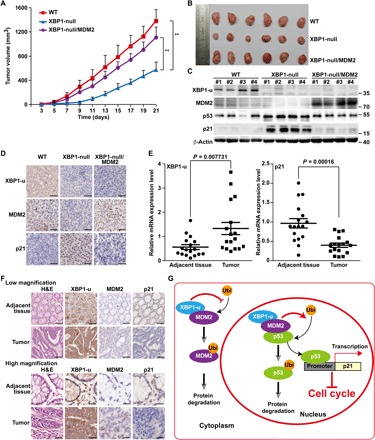
(A and B) Tumorigenesis potentials of HCT116WT/pcDNA3, HCT116XBP1null/pcDNA3, and HCT116XBP1null/pcMDM2 stable cell lines, as determined in vivo by subcutaneous injection into BALB/c-nu/nu mice (n = 6). The volume of the generated tumor was measured at the indicated time points (A), and morphological images are shown (B). (C) Protein expression levels of XBP1-u, MDM2, p53, and p21 in the generated tumors, as determined by Western blotting. β-Actin was used as loading control. (D) Immunohistochemistry staining showing the localization of MDM2, XBP1-u, and p21 in tissue sections of xenografted tumors in BALB/c-nu/nu mice injected with the indicated cell lines. Scale bars, 20 μm. (E) mRNA expression levels of XBP1-u and p21 in clinical human colon carcinoma and adjacent tissues samples (n = 17), as determined by qPCR. Data were normalized with β-actin. P values were determined by ANOVA. (F) Immunohistochemistry staining showing localization of MDM2, XBP1-u, and p21 in clinical human colon carcinoma and adjacent tissues samples. Low-magnification (top) and high-magnification (bottom) images are shown. Scale bars, 100 μm (top) and 20 μm (bottom). H&E, hematoxylin and eosin. (G) Schematic diagram showing the mechanism of XBP1-u regulation on the MDM2/p53/p21 axis.
Finally, we found that XBP1-u, MDM2, and p21 were aberrantly expressed in clinical human colon carcinoma samples (Fig. 6, E and F). XBP1-u and MDM2 demonstrated a positive correlation because both were up-regulated in tumor lesions compared to tumor adjacent tissues. At the same time, the mRNA and protein levels of XBP1-u were up-regulated in tumor tissues, whereas those of p21 were down-regulated.
We found that XBP1-u localization to the nucleus was enhanced in tumor lesions, in which expression of both XBP1 and MDM2 was up-regulated (Fig. 6F). Similarly, we found that whereas XBP1-u localized predominantly to the cytoplasm under basal condition or when it was overexpressed alone, it colocalized with MDM2 to the nucleus when both of them were overexpressed (fig. S11A). Furthermore, using sequential immunoprecipitation, we observed that XBP1-u, MDM2, and p53 interacted physically, indicating that they could form at least a tertiary complex (fig. S11, B and C). Given that MDM2 interacts with and degrades p53 protein in the nucleus, these results indicate that XBP1-u might shuttle to the nucleus upon binding to MDM2 and then be involved in p53 degradation. These mechanisms resemble the conventional and highly conserved MDM2/MDMX/p53 regulatory pathway (28, 29). Human homolog of double minute X [MDMX (also known as MDM4)], a homolog of MDM2 that could form a heteromeric complex with MDM2, inhibits MDM2 homodimerization and, as a result, suppresses MDM2 self-ubiquitination in the cytoplasm and promotes p53 degradation (23, 24). However, MDMX itself localizes only to the cytoplasm and enters the nucleus solely upon binding to MDM2 (29, 30). Despite similarities on inhibition of MDM2 homodimerization and self-ubiquitination, we found that XBP1-u could not be immunoprecipitated with MDMX (fig. S12). Thus, it is most likely that this novel and specific regulatory pathway is irrelevant to the conventional, well-known MDM2/MDMX/p53 pathway. Together, these findings reveal that XBP1-u is a novel oncogene responsible for regulating the MDM2/p53 axis through an unconventional pathway (Fig. 6G).
DISCUSSION
Tight control of cell cycle progression is critical for maintaining normal biological functions, and the p53/p21 axis plays a critical role in this regulation. Loss of control of this mechanism leads to several abnormalities, including tumors and embryonic lethality. Besides mutations in p53, which could be observed in approximately half of all human malignancies, many tumors with wild-type p53 exhibit abnormal expression of p53 regulators. MDM2 is a crucial negative regulator of p53 that promotes its degradation by the ubiquitin proteasomal degradation pathway (14). Our findings describe a novel regulatory mechanism of the MDM2/p53 pathway: XBP1-u binds to MDM2, inhibits its self-ubiquitination, stabilizes it, and thus enhances p53 degradation. This subsequently leads to the down-regulation of p21 and accelerated cell cycle progression as well as cell proliferation (Fig. 6G).
XBP1 was initially identified for its spliced active form, XBP1-s (12). In response to ER stress, the mRNA of XBP1-u is spliced to generate XBP1-s, which regulates ER homeostasis by binding to a subset of genes and components of the ER-associated degradation pathway (31). XBP1-s has been reported to be involved in various biological and physiological contexts, such as adaptive and innate immunity, circadian rhythm, and oxidative stress (32–34). However, Nekrutenko and He (35) demonstrated that XBP1 evolved in a fashion that was consistent only if both reading frames were functional. Unlike XBP1-s, the specific role of XBP1-u has remained poorly understood. This may be due to the high sequence similarity between XBP1-u and XBP1-s and to the low expression level of XBP1-u under ER stress condition, making it difficult to perform a gene function analysis specific to XBP1-u. Although recent studies have shown that XBP1-u is involved in the protection of endothelial cells from oxidative stress by promoting Akt1 (AKT serine/threonine kinase 1) phosphorylation and in the suppression of autophagy by promoting the degradation of forkhead box 1 (36, 37), knowledge of any specific, ER stress–independent functions of XBP1-u remains minimal. Our findings show that XBP1-u intersects with the MDM2/p53 pathway. The latter is a fundamental regulatory pathway critical for tumorigenesis and other biological and pathological functions, such as embryogenesis, DNA repair, and senescence (38, 39). Hence, our results elucidate a novel function of XBP1-u in pivotal biological functions.
MDM2 is an oncogene that belongs to the RING domain–containing E3 ligase family (14, 40, 41). MDM2 could shuttle between the nucleus, where it enhances p53 ubiquitination, and the cytoplasm, where its protein regulation occurs. Unlike XBP1-s, which localizes to the nucleus (42), we show here that XBP1-u is detected predominantly in the cytoplasm. Furthermore, the MDM2-stabilizing function of XBP1-u could be attributed to its C terminus, which is different from that of XBP1-s, and has been reported to have a strong nuclear exclusion signal (42). These facts strongly support our conclusion that the MDM2-stabilizing function is specific to XBP1-u. Intriguingly, when overexpressed, MDM2 could promote XBP1-u localization to the nucleus, where MDM2 functions as a negative regulator of p53. Although the detailed mechanism needs to be investigated further, these results indicate that XBP1-u might not only stabilize MDM2 protein but also be involved in MDM2-induced p53 degradation. Taking into account the fact that XBP1-u is sufficient to inhibit MDM2 self-ubiquitination in vitro and that it neither immunoprecipitates with MDMX nor affects the binding of MDM2 to factors that inhibit its ubiquitination activity such as ARF and ribosomal proteins RPL11 and RPS14, the XBP1-u/MDM2/p53 axis represents a novel pathway. Hence, although further investigation is required to elucidate whether XBP1-u competes with MDMX in stabilizing MDM2, our findings unraveled a critical, unconventional pathway for the regulation of MDM2 and p53.
Besides regulating p53, recent reports have shown that MDM2 could also promote cell proliferation and tumorigenesis in a p53-independent manner (20), raising the possibility that XBP1-u/MDM2 might also contribute to tumorigenesis in a p53-independent manner. Our results show that XBP1 silencing could still suppress cell cycle progression, cell proliferation, and colony-forming potential in p53-null cells, although the effects are significantly lower than in wild-type cells. This could be mediated, at least in part, through the induction of p21 protein accumulation. This result conforms to the notion that MDM2 could directly regulate p21 protein stability (43). Therefore, it is most likely that although p53 is critical for the involvement of XBP1-u/MDM2 in tumorigenesis, the XBP1-u/MDM2 pathway could also participate in p53-independent tumorigenic processes. This fact further highlights the importance of XBP1-u/MDM2 in regulating both cell cycle and tumorigenesis.
p53 could regulate MDM2 transcription through a feedback mechanism, which would give rise to an oscillated expression pattern of both proteins (19–21). However, our results indicate that in the presence of p53, XBP1-u did not significantly affect MDM2 mRNA expression. This intriguing result is most plausibly due to the accumulative effects of XBP1-u positive regulation on MDM2 transcription and p53-induced feedback mechanism. To our knowledge, unlike its spliced form, XBP1-s, which had been reported to act as a transcriptional factor for its downstream genes (44), there is no report regarding the transcriptional function of XBP1-u at present. The transcriptional functions of XBP1-s should be attributed to its C terminus (44), which is different from that of XBP1-u. Nevertheless, although the detailed mechanism of XBP1-u regulation on MDM2 transcription needs further investigation, the fact that MDM2 protein accumulation is suppressed by XBP1 silencing even in the presence of p53 reveals that the contribution of XBP1-u is even more significant at the protein stability level. On the other hand, the fact that XBP1-u could positively regulate MDM2 on both transcriptional and translational levels strongly suggests the clinical significance of its oncogenic function. Given that p53 down-regulation could also be observed even in many tumors with wild-type p53 (45), our results indicate that XBP1-u could mask the feedback regulation of p53 on MDM2 and thus might be involved in the aberrant high expression of MDM2 in many cancers with wild-type p53 (21).
In summary, by performing shRNA library screening, as well as in vitro and in vivo experiments, we uncover a previously unknown function of XBP1-u in regulating the MDM2/p53 axis and elucidate the new, critical role of XBP1-u in cell cycle and tumorigenesis. The fact that the MDM2/p53 pathway is also involved in other processes, including embryogenesis and senescence, indicates that the importance of XBP1-u has been underestimated and might indeed have a far-reaching impact. Finally, these findings give a new perspective regarding the regulation of the basic, conventional MDM2/p53 axis.
MATERIALS AND METHODS
shRNA expression vector and shRNA expression vector library
shRNA target sites were predicted using the algorithm for predicting specific target sites, and the shRNA expression vector library was constructed as described previously (46). The library contains 3354 shRNA expression vectors against 2065 genes (1 or 2 shRNA expression vectors with different target sequences for each gene). The specific target sites for shXBP1-1, shXBP1-2, shXBP1-3, and shXBP1-4 were GTA AGA AAT ATT ACT ATA A, AGT AAG AAA TAT TAC TAT A, GCT GGA AGC CAT TAA TGA A, and GAA CCA GGA GTT AAG ACA G, respectively; the target sites for p53 (NM_000546.5) were GCA AGA AGG GAG ACA AGA T and GAG AAG AGT TGG AAC AGA A; and the target sites for MDM2 (NM_002392.5) were GAC TAA ACG ATT ATA TGA T and AGG CAA ATG TGC AAT ACC A. The shRNA expression vectors were constructed as described previously (47). Briefly, the oligonucleotides with a hairpin structure, overhanging sequences, and a terminator were synthesized, annealed, and inserted into the Bsp MI sites of a pcPUR+U6i cassette vector containing the U6 promoter as described previously.
For XBP1-u (NM_005080.3) and XBP1-s (NM_001079539.1) overexpression vectors (pcXBP1-u and pcXBP1-s, respectively), the oligonucleotides of the coding sequences were synthesized and cloned into pUC57 (Invitrogen) before they were subcloned into the Bam HI and Not I sites of pcDNA3.1(+) (Invitrogen). For p53 and MDM2 overexpression vectors (pcp53 and pcMDM2, respectively), the corresponding regions were amplified using the Takara PrimeSTAR Max DNA Polymerase (Takara Bio) from human complementary DNA (cDNA) obtained by reverse-transcribing the total RNA extracted from HCT116WT cells using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio) and cloned into the Bam HI and Not I sites of pcDNA3.1(+). XBP1-u C-terminal (1 to 498 regions) and N-terminal (499 to 786 regions) overexpression vectors (pcXBP1-u-C and pcXBP1-u-N, respectively) were cloned in a similar way, except that pcXBP1-u and pcXBP1-s were used as templates.
For pcFLAG, the FLAG sequence was inserted into the Nhe I and Hind III sites of pcDNA3.1(+). For FLAG-tagged MDMX (NM_002393.4), FLAG-tagged ARF (NM_000077.4), FLAG-tagged RPL11 (NM_000975.3), and FLAG-tagged RPS14 (NM_001025071.1) overexpression vectors (FLAG-MDMX, FLAG-ARF, FLAG-RPL11, and FLAG-RPS14, respectively), the coding sequences were amplified from human cDNA as described above, whereas for FLAG-tagged XBP1-u, XBP1-u C terminus, and XBP1-u N terminus (FLAG-XBP1-u, FLAG-XBP1-u-C, and FLAG-XBP1-u-N, respectively), the XBP1-u, XBP1-u-C, and XBP1-u-N fragments were amplified from pcXBP1-u, pcXBP1-u-C, and pcXBP1-u-N, respectively. These fragments were then inserted into the Bam HI and Not I sites of pcFLAG.
For the p21 wild-type promoter reporter vector containing −65 to +2586 p21 promoter region (p21-luc) and MDM2 promoter reporter vectors (MDM2-p1-luc and MDM2-p2-luc, containing −2410 to −1 and +317 to +841 of MDM2 promoter regions, respectively), the corresponding promoter regions were amplified using the Takara PrimeSTAR Max DNA Polymerase (Takara Bio) from human genome DNA extracted from HCT116WT cells using the TIANamp Genomic DNA Kit (Tiangen Biotech) and cloned into the Bgl II and Hind III sites of pGL4.13 vector (Promega). p21-luc vector lacking the p53 binding site (p21mut-luc) was constructed on the basis of the site-specific mutagenesis method (48).
Cell cultures and cell lines
The wild-type and p53-null HCT116 colon carcinoma cell lines (HCT116WT and HCT116p53null, respectively) were provided by B. Vogelstein (Johns Hopkins University School of Medicine). The HepG2 human liver carcinoma cell line and the MCF-7 human breast carcinoma cell line were obtained from the Cell Bank of the Chinese Academy of Sciences, Shanghai. Cells were maintained in medium [McCoy’s 5A medium (Gibco) for HCT116 cells or Dulbecco’s modified Eagle’s medium (Gibco) for HepG2 and MCF-7 cells] supplemented with 10% fetal bovine serum (Biological Industries). All cell lines were routinely tested and found negative for mycoplasma contamination using Mycoplasma Detection Kit-QuickTest (Biotool).
The XBP1-null HCT116 (HCT116XBP1null) cell line was established using CRISPR/Cas9 technology. Briefly, human XBP-1 CRISPR/Cas9 KO Plasmid and human XBP-1 HDR Plasmid (Santa Cruz Biotechnology) were transfected into the HCT116WT cells using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, puromycin selection (0.8 μg/ml) was performed to eliminate untransfected cells for 7 days. Cell line was then established from a single clone. The elimination of XBP1 was confirmed by analyzing the sequence of the corresponding genome DNA: The nucleotides located at the +570 to +616 region (47 base pairs) of XBP1-u were deleted.
To construct the XBP1-null stable cell line overexpressing MDM2, MDM2 overexpression vector pcMDM2 was transfected into HCT116XBP1null cells. To construct control stable cells lines, pcDNA3.1(+) vector was transfected into HCT116XBP1null or HCT116WT cells. Transfection was performed using Lipofectamine 2000 according to the manufacturer’s instruction. The transfected cells were then selected using G418 (final concentration, 400 μg/ml), and stable cell lines were established from single clones.
For gene silencing experiments, cells were transfected with 2 μg of indicated shRNA expression vector using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were subjected to puromycin selection (final concentration, 1.2 μg/ml) to eliminate untransfected cells. mRNA and protein samples were collected 36 hours after puromycin selection.
For overexpression experiments, cells were transfected with 2 μg of indicated overexpression vector using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, mRNA and protein samples were collected and subjected for further analysis.
For rescue experiments, cells were transfected with 1 μg of indicated shRNA expression vector and 1 μg of overexpression vector using Lipofectamine 2000 (Invitrogen). Cells were subjected to puromycin selection (final concentration, 1.2 μg/ml) to eliminate untransfected cells. mRNA and protein samples were collected 36 hours after puromycin selection.
For thapsigargin treatment, 1 × 106 cells were seeded and cultured for 24 hours. After being treated with thapsigargin (final concentration, 1 nM) for 6 hours, mRNA and protein were collected.
Animal experiment
For the in vivo tumor study, BALB/c-nu/nu mice (male; body weight, 18 to 22 g; 6 weeks old) were purchased from the Third Military Medical University (Chongqing, China; permit number SYXK-PLA-20120031). Animal studies were carried out in the Third Military Medical University and approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University. All animal experiments conformed to the approved guidelines of the Animal Care and Use Committee of the Third Military Medical University. All efforts to minimize suffering were made.
To generate an experimental subcutaneous tumor model, BALB/c-nu/nu mice were divided into three groups (n = 6), and each group was injected subcutaneously with 5 × 106 HCT116WT/pcDNA, HCT116XBP1null/pcDNA, or HCT116XBP1null/pcMDM2 stable cells. Tumor size (V) was evaluated by a caliper every 2 days with reference to the following equation: V = a × b2/2, where a and b are the major and minor axes of the tumor, respectively (47). Mice were randomly divided into three groups (n = 6). The investigator was blinded to the group allocation and during the assessment.
Clinical human colon carcinoma specimens
Human colon carcinoma specimens were obtained from colon carcinoma patients undergoing surgery at Chongqing Cancer Institute (Chongqing, China) and stored in the Biological Specimen Bank of Chongqing Cancer Institute. Patients did not receive chemotherapy, radiotherapy, or other adjuvant therapies before the surgery. The specimens were snap-frozen in liquid nitrogen. Patients’ written informed consents were obtained, and the experiments were approved by the Institutional Research Ethics Committee of Chongqing Cancer Institute.
Dual-Luciferase Reporter System and library screening
Cells were seeded onto 24-well plates for luciferase reporter assay or 96-well plates for library screening. Twenty-four hours later, cells were cotransfected with the indicated shRNA expression vectors, reporter vector, and the Renilla luciferase expression vector (pRL-SV40, Promega) as internal control using Lipofectamine 2000. Twenty-four hours after cotransfection, the luciferase activities were then measured with the Dual-Luciferase Reporter Assay System (Promega).
RNA extraction and quantitative RT-PCR analysis
Total RNAs were extracted using TRIzol (Invitrogen) according to the manufacturer’s instruction. Each total RNA sample (1 μg) was reverse-transcribed into cDNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio), and quantitative reverse transcription PCR (qPCR) was performed to assess the mRNA expression levels with SYBR Premix Ex Taq (Takara Bio). The sequences of the primers used for quantitative RT-PCR are shown in table S2. β-Actin was used to normalize sample amplification. The results are shown relative to the expression level in the corresponding controls, which are assumed as 1.
Western blotting
For cell culture experiments, cells were collected and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer with protease inhibitor and phosphatase inhibitor cocktail (complete cocktail, Roche Applied Science). Equal amounts of the sample proteins were electrophoresed on SDS polyacrylamide gel and transferred to a polyvinylidene fluoride membrane (Millipore) with a pore size of 0.45 or 0.22 μm (for protein, ≤15 kDa). The antibodies used are listed in table S3, and immunoblotting with anti–β-actin antibody was conducted to ensure equal protein loading. The signals were measured using SuperSignal West Femto Maximum Sensitivity Substrate detection system (Thermo Fisher Scientific). Images of uncropped blots are shown in fig. S13 (A to Q). Quantification was performed using Quantity One software.
For protein degradation assay, 1 × 106 cells were seeded in a 3.5-cm dish. Cycloheximide was added at the final concentration of 200 μg/ml. Protein samples were collected at the indicated time points and were subjected to Western blotting as described above.
Quantification of total cell number
HCT116WT cells or HCT116p53null cells were first transfected with the indicated vectors using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, puromycin selection (final concentration, 1.2 μg/ml) was performed to eliminate untransfected cells. The transfected cells were seeded into 96-well plates. The cell numbers were measured by Cell Counting Kit-8 (Dojindo) at the indicated time points.
EdU incorporation assay and colony formation assay
Cells were transfected with the indicated shRNA expression vectors and selected using puromycin as indicated above. For EdU incorporation assay, after puromycin selection, EdU incorporation and staining were performed using Cell-Light EdU Apollo488 In Vitro Imaging Kit (RiboBio) according to the manufacturer’s instruction. Nuclei were stained with DAPI. Images were taken with DMI6000B (Leica). Quantification of EdU-positive and DAPI-positive cells was performed using Microsystems LAS AF-TCS MP5 (Leica), and the results are shown as the ratio of EdU-positive cells to DAPI-positive cells. For colony formation assay, 300 cells were cultured in a six-well plate for 8 days. Cells were then fixed with 30% ethanol and stained with methylene blue. The colonies were then counted. The investigator was blinded during the assessment.
Cell cycle analysis
HCT116WT cells or HCT116p53null cells were transfected with the indicated shRNA expression vectors and selected using puromycin selection as described above. Cells were subjected for starvation for 24 hours before they were incubated further for 24 hours under normal conditions and then harvested and stained with propidium iodide (KeyGen Biotech). The percentages of the cells in each cell cycle phase were determined by flow cytometry.
Immunoprecipitation and in vivo ubiquitination assay
Cells were seeded in a 10-cm dish (5 × 106 cells per dish) and transfected with 16 μg of shRNA expression vectors or overexpression vectors as described above. Total protein samples were collected and lysed with RIPA lysis buffer with protease inhibitor and phosphatase inhibitor cocktail (complete cocktail, Roche Applied Science) and cleared by centrifugation at 12,000 rpm. The supernatants were incubated at 4°C for 2 hours with protein A+G beads (Beyotime Biotechnology) in the presence of indicated antibodies or IgG as control. Then, the immunoprecipitated proteins were subjected to immunoblotting as described in Western blotting.
For ubiquitination assay, cells were transfected with the ubiquitin overexpression vector (pcUbi) and the indicated shRNA expression vector or overexpression vector, as well as the pcp53 or pcMDM2 vector (6 μg each). Twenty-four hours later, cells were treated with MG132 (final concentration, 20 μM) for 8 hours, and then the lysates were collected as described above. Immunoprecipitation was performed using the indicated antibodies, and then the immunoprecipitated proteins were subjected to immunoblotting using anti-ubiquitin antibody and other antibodies as indicated.
GST pull-down assay
His-tagged N-terminal and C-terminal fragments, as well as the full-length XBP1-u proteins, were purchased from Proteintech. The GST-MDM2 protein was purchased from Boston Biochem and mixed with GST beads (GE Healthcare Life Sciences) in the binding buffer [phosphate-buffered saline (PBS) + 2 mM EDTA + 0.05% Tween 20 + 3% milk powder] at 4°C for 1 hour. The mixtures were then washed four times with wash buffer (PBS + 2 mM EDTA + 0.05% Tween 20 + 150 mM NaCl). His-tagged XBP1 fragments were added into the mixture and mixed in the abovementioned binding buffer at 4°C for 2 hours. The mixtures were then washed four times with wash buffer as mentioned above, and the protein complexes were eluted using 1% SDS. The pulled-down proteins were subjected to Western blotting as described above.
In vivo chemical cross-linking assay
Cells were transfected with the indicated vectors. After culturing for 24 hours, cells were collected and washed with cold PBS. The cell suspensions were treated with DSS for 30 min at 25°C, and the reaction was stopped using tris-buffered saline (pH 7.5). Cells were then lysed with RIPA lysis buffer with protease inhibitor and phosphatase inhibitor cocktail (complete cocktail, Roche Applied Science) before they were subjected to Western blotting as described above. For assay with MG132, cells were treated with MG132 (final concentration, 20 μM) for 8 hours before they were collected and treated with DSS.
In vitro ubiquitination assay
Fifty nanograms of E1 (Boston Biochem), 100 ng of UBE2D2 (Boston Biochem), 5 μg of ubiquitin (Boston Biochem), and 4 μg of each protein as indicated were mixed in a buffer containing 50 mM tris (pH 7.6), 5 mM MgCl2, 2 mM adenosine triphosphate, and 2 mM dithiothreitol. The mixtures were then incubated at 30°C for 90 min, and the reactions were then stopped using 5× SDS–polyacrylamide gel electrophoresis loading buffer. The samples were resolved on 10% SDS polyacrylamide gel and subjected to Western blotting against anti-MDM2 antibody as described above.
Immunohistochemical analysis
Paraffin-embedded sections were obtained from fresh colon carcinoma and adjacent tissues or xenografted tumors (4 μm thick) using a cryostat and subjected to immunohistochemistry. Briefly, the tissue sections were incubated with primary antibodies for 1 hour. The specimens were then incubated with the corresponding second antibodies conjugated with horseradish peroxidase. Visualization was performed using a DAB (diaminobenzidine) Kit (DAKO) under a microscope. Nuclei were then counterstained with hematoxylin, followed by dehydration and coverslip mounting. The antibodies used are listed in table S2. Images were taken using Pannoramic Midi (3DHistech).
Immunofluorescence staining
Cells were seeded in 3.5-cm culture dishes (2 × 105 cells per dish), fixed for 30 min at room temperature with 4% paraformaldehyde, permeabilized for 5 min with PBS containing 0.1% Triton X-100, blocked with 1% bovine serum albumin for 1 hour, and incubated with anti–XBP1-u and anti-MDM2 antibodies for 2 hours. Cells were then incubated with fluorescent second antibodies for 1 hour. Nuclei were stained with DAPI (Beyotime Biotechnology) for 15 min. Images were taken with laser scanning confocal microscopy (Leica Microsystems TCS SP5).
Sequential immunoprecipitation
Sequential immunoprecipitation was performed as described previously (49). Briefly, HCT116WT cells seeded in a 10-cm dish (5 × 106 cells per dish) were transfected with pcFLAG-XBP1u, pcMDM2, and pcp53 (6 μg each) as described above. Total protein samples were treated as described in the “Immunoprecipitation and in vivo ubiquitination assay” section, except that after immunoprecipitation with IgG or anti-FLAG antibody, the immunoprecipitants were again immunoprecipitated with IgG or anti-MDM2 antibody. Then, the immunoprecipitants were subjected to immunoblotting as described above.
Statistical analysis
Quantification data were analyzed by one-way ANOVA conducted using SPSS Statistics v17.0. Statistical significance was defined as P < 0.05, and P < 0.01 was considered highly significant.
Supplementary Material
Acknowledgments
We thank B. Vogelstein (Johns Hopkins University School of Medicine) for providing HCT116WT and HCT116p53null cell lines. Funding: This work was supported by grants from the National Natural Science Foundation of China (31301119 and 81372202), the Natural Science Foundation of Chongqing (cstc2014jcyjA10058), the Fundamental Research Funds for the Central Universities (106112016CDJZR235516), the Graduate Scientific Research and Innovation Foundation of Chongqing (CYB17039), the National Institute of Advanced Industrial Science and Technology (AAZ30368), and the New Energy and Industrial Technology Development Organization (P00013). Author contributions: V.K. and S.W. conceived the project, designed the experiments, analyzed and interpreted the experimental results, and wrote the manuscript. M.M. designed the shRNA target sequences, constructed the shRNA expression vector library, supervised the library screening, and analyzed the data. C.H. performed most of the experiments and analyzed the experimental data. H.J. carried out Western blotting and qPCR analysis. S.M. performed library screening. H.Z. collected human clinical samples. Y.X. and X.Y. carried out part of the animal and clinical sample experiments. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/10/e1701383/DC1
fig. S1. XBP1 is a novel regulator of p21.
fig. S2. Expression levels of unspliced XBP1 (XBP1-u) and spliced XBP1 (XBP1-s).
fig. S3. Overexpression of XBP1-u suppresses p21 expression without inducing UPR.
fig. S4. XBP1-u promotes cell cycle progression and cell proliferation.
fig. S5. XBP1-u regulates p21 translation p53-dependently.
fig. S6. XBP1-u regulates cell cycle and colony-forming potential mainly through the p53 axis.
fig. S7. XBP1-u regulates p21 through the MDM2/p53 axis.
fig. S8. XBP1-u regulates the MDM2/p53/p21 axis in hepatocarcinoma and breast cancer cells.
fig. S9. p53 feedback mechanism restores XBP1-u regulation on the MDM2 mRNA expression level.
fig. S10. The N terminus of XBP1-u does not inhibit MDM2 homodimerization and self-ubiquitination.
fig. S11. Simultaneous up-regulation of XBP1-u and MDM2 induces the formation of XBP1-u/MDM2/p53 complex.
fig. S12. XBP1-u does not interact with MDMX.
fig. S13. Uncropped Western blots with the indicated areas of selection in Figs. 1 to 6 and figs. S1, S2, S4, S5, S7, S8, S10, S11, and S12.
table S1. List of the genes in the top 10% of the screening results.
table S2. Primers used for gene quantification by qPCR.
table S3. Antibodies used for Western blotting, immunohistochemistry, immunofluorescence, and immunoprecipitation.
REFERENCES AND NOTES
- 1.el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B., WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J., The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Abbas T., Dutta A., p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9, 400–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Cunto F., Topley G., Calautti E., Hsiao J., Ong L., Seth P. K., Dotto G. P., Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science 280, 1069–1072 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Kim H.-S., Heo J.-I., Park S.-H., Shin J.-Y., Kang H.-J., Kim M.-J., Kim S. C., Kim J., Park J.-B., Lee J.-Y., Transcriptional activation of p21WAF1/CIP1 is mediated by increased DNA binding activity and increased interaction between p53 and Sp1 via phosphorylation during replicative senescence of human embryonic fibroblasts. Mol. Biol. Rep. 41, 2397–2408 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Lozano G., Zambetti G. P., What have animal models taught us about the p53 pathway? J. Pathol. 205, 206–220 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Dhyani A., Machado-Neto J. A., Favaro P., Olalla Saad S. T., ANKHD1 represses p21 (WAF1/CIP1) promoter and promotes multiple myeloma cell growth. Eur. J. Cancer 51, 252–259 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Zhang P., Wong C., Liu D., Finegold M., Harper J. W., Elledge S. J., p21CIP1 and p57KIP2 control muscle differentiation at the myogenin step. Genes Dev. 13, 213–224 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiohara M., el-Deiry W. S., Wada M., Nakamaki T., Takeuchi S., Yang R., Chen D. L., Vogelstein B., Koeffler H. P., Absence of WAF1 mutations in a variety of human malignancies. Blood 84, 3781–3784 (1994). [PubMed] [Google Scholar]
- 10.Liou H. C., Boothby M. R., Finn P. W., Davidon R., Nabavi N., Zeleznik-Le N. J., Ting J. P., Glimcher L. H., A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247, 1581–1584 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Yanagitani K., Kimata Y., Kadokura H., Kohno K., Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331, 586–589 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K., XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Feng H., Gu H., Lewis D. W., Yuan Y., Zhang L., Yu H., Zhang P., Cheng H., Miao W., Yuan W., Cheng S.-Y., Gollin S. M., Cheng T., The p53–PUMA axis suppresses iPSC generation. Nat. Commun. 4, 2174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M., Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Eischen C. M., Weber J. D., Roussel M. F., Sherr C. J., Cleveland J. L., Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13, 2658–2669 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fåhraeus R., Olivares-Illana V., MDM2’s social network. Oncogene 33, 4365–4376 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Zhou X., Hao Q., Liao J., Zhang Q., Lu H., Ribosomal protein S14 unties the MDM2–p53 loop upon ribosomal stress. Oncogene 32, 388–396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zauberman A., Flusberg D., Haupt Y., Barak Y., Oren M., A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 23, 2584–2592 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barak Y., Juven T., Haffner R., Oren M., mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karni-Schmidt O., Lokshin M., Prives C., The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wade M., Li Y.-C., Wahl G. M., MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C., Wang X., Mdm2 Splice isoforms regulate the p53/Mdm2/Mdm4 regulatory circuit via RING domain-mediated ubiquitination of p53 and Mdm4. Cell Cycle 16, 660–664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai H., Lopez-Pajares V., Kim M. M., Wiederschain D., Yuan Z.-M., RING domain–mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 67, 6026–6030 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Tanimura S., Ohtsuka S., Mitsui K., Shirouzu K., Yoshimura A., Ohtsubo M., MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 447, 5–9 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Chavez J. D., Liu N. L., Bruce J. E., Quantification of protein–protein interactions with chemical cross-linking and mass spectrometry. J. Proteome Res. 10, 1528–1537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T., Zhang H., Xiong J., Yi S., Gu L., Zhou M., Inhibition of MDM2 homodimerization by XIAP IRES stabilizes MDM2, influencing cancer cell survival. Mol. Cancer 14, 65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itahana K., Mao H., Jin A., Itahana Y., Clegg H. V., Lindström M. S., Bhat K. P., Godfrey V. L., Evan G. I., Zhang Y., Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12, 355–366 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Coffill C. R., Lee A. P., Siau J. W., Chee S. M., Joseph T. L., Tan Y. S., Madhumalar A., Tay B.-H., Brenner S., Verma C. S., Ghadessy F. J., Venkatesh B., Lane D. P., The p53–Mdm2 interaction and the E3 ligase activity of Mdm2/Mdm4 are conserved from lampreys to humans. Genes Dev. 30, 281–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolezelova P., Cetkovska K., Vousden K. H., Uldrijan S., Mutational analysis of Mdm2 C-terminal tail suggests an evolutionarily conserved role of its length in Mdm2 activity toward p53 and indicates structural differences between Mdm2 homodimers and Mdm2/MdmX heterodimers. Cell Cycle 11, 953–962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z.-M., Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J. Biol. Chem. 277, 19251–19254 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., Dynlacht B. D., XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27, 53–66 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Cretenet G., Le Clech M., Gachon F., Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metab. 11, 47–57 (2010). [DOI] [PubMed] [Google Scholar]
- 33.He Y., Sun S., Sha H., Liu Z., Yang L., Xue Z., Chen H., Qi L., Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 15, 13–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwakoshi N. N., Lee A.-H., Vallabhajosyula P., Otipoby K. L., Rajewsky K., Glimcher L. H., Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4, 321–329 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Nekrutenko A., He J., Functionality of unspliced XBP1 is required to explain evolution of overlapping reading frames. Trends Genet. 22, 645–648 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Martin D., Li Y., Yang J., Wang G., Margariti A., Jiang Z., Yu H., Zampetaki A., Hu Y., Xu Q., Zeng L., Unspliced X-box-binding protein 1 (XBP1) protects endothelial cells from oxidative stress through interaction with histone deacetylase 3. J. Biol. Chem. 289, 30625–30634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y., Li X., Cai M.-Y., Ma K., Yang J., Zhou J., Fu W., Wei F.-Z., Wang L., Xie D., Zhu W.-G., XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 23, 491–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris S. L., Levine A. J., The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Montes de Oca Luna R., Wagner D. S., Lozano G., Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P., Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Kwon D.-H., Eom G. H., Ko J. H., Shin S., Joung H., Choe N., Nam Y. S., Min H.-K., Kook T., Yoon S., Kang W., Kim Y. S., Kim H. S., Choi H., Koh J.-T., Kim N., Ahn Y., Cho H.-J., Lee I.-K., Park D. H., Suk K., Seo S. B., Wissing E. R., Mendrysa S. M., Nam K.-I., Kook H., MDM2 E3 ligase-mediated ubiquitination and degradation of HDAC1 in vascular calcification. Nat. Commun. 7, 10492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida H., Oku M., Suzuki M., Mori K., pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H., Zhang Z., Li M., Zhang R., MDM2 promotes proteasomal degradation of p21Waf1 via a conformation change. J. Biol. Chem. 285, 18407–18414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shajahan A. N., Riggins R. B., Clarke R., The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 22, 241–246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hainaut P., Wiman K. G., 30 years and a long way into p53 research. Lancet Oncol. 10, 913–919 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Kasim V., Wu S., Taira K., Miyagishi M., Determination of the role of DDX3 a factor involved in mammalian RNAi pathway using an shRNA-expression library. PLOS ONE 8, e59445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu S., Kasim V., Kano M. R., Tanaka S., Ohba S., Miura Y., Miyata K., Liu X., Matsuhashi A., Chung U.-i., Yang L., Kataoka K., Nishiyama N., Miyagishi M., Transcription factor YY1 contributes to tumor growth by stabilizing hypoxia factor HIF-1α in a p53-independent manner. Cancer Res. 73, 1787–1799 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Kunkel T. A., Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto N., Yasukawa M., Nguyen C., Kasim V., Maida Y., Possemato R., Shibata T., Ligon K. L., Fukami K., Hahn W. C., Masutomi K., Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc. Natl. Acad. Sci. U.S.A. 108, 20388–20393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/10/e1701383/DC1
fig. S1. XBP1 is a novel regulator of p21.
fig. S2. Expression levels of unspliced XBP1 (XBP1-u) and spliced XBP1 (XBP1-s).
fig. S3. Overexpression of XBP1-u suppresses p21 expression without inducing UPR.
fig. S4. XBP1-u promotes cell cycle progression and cell proliferation.
fig. S5. XBP1-u regulates p21 translation p53-dependently.
fig. S6. XBP1-u regulates cell cycle and colony-forming potential mainly through the p53 axis.
fig. S7. XBP1-u regulates p21 through the MDM2/p53 axis.
fig. S8. XBP1-u regulates the MDM2/p53/p21 axis in hepatocarcinoma and breast cancer cells.
fig. S9. p53 feedback mechanism restores XBP1-u regulation on the MDM2 mRNA expression level.
fig. S10. The N terminus of XBP1-u does not inhibit MDM2 homodimerization and self-ubiquitination.
fig. S11. Simultaneous up-regulation of XBP1-u and MDM2 induces the formation of XBP1-u/MDM2/p53 complex.
fig. S12. XBP1-u does not interact with MDMX.
fig. S13. Uncropped Western blots with the indicated areas of selection in Figs. 1 to 6 and figs. S1, S2, S4, S5, S7, S8, S10, S11, and S12.
table S1. List of the genes in the top 10% of the screening results.
table S2. Primers used for gene quantification by qPCR.
table S3. Antibodies used for Western blotting, immunohistochemistry, immunofluorescence, and immunoprecipitation.


