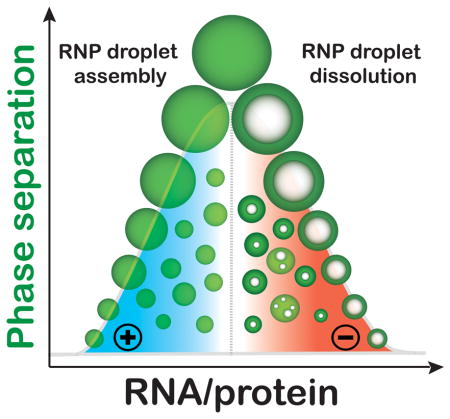
Abstract
Intracellular ribonucleoprotein (RNP) granules are membrane-less droplet organelles that are thought to regulate posttranscriptional gene expression. While liquid-liquid phase separation may drive RNP granule assembly, the mechanisms underlying their supramolecular dynamics and internal organization remain poorly understood. Here we demonstrate that RNA, a primary component of RNP granules, can modulate the phase behavior of RNPs by controlling both droplet assembly and dissolution in vitro. Monotonically increasing RNA concentration initially leads to droplet assembly via complex coacervation and subsequently triggers an RNP charge inversion, which promotes disassembly. This RNA-mediated reentrant phase transition can drive the formation of dynamic droplet substructures (vacuoles) with tunable lifetimes. We propose that active cellular processes that can create an influx of RNA into RNP granules, such as transcription, can spatiotemporally control the organization and dynamics of such liquid-like organelles.
Keywords: Electrostatic interactions, Intrinsically disordered proteins, Membrane-less organelles, Phase transitions, Vacuolated coacervates
Vacuolated Ribonucleoprotein Droplets
RNA controls the reentrant phase transition of ribonucleoproteins (RNP) to assemble and dissolve RNP droplets. During dissolution, controlled RNA flux into RNP droplets generates dynamic vacuolated substructures with tunable lifetimes.
Intracellular membrane-less organelles (MLOs), such as the nucleolus and ribonucleoprotein (RNP) granules, are dynamic liquid-like compartments commonly comprised of RNA-binding intrinsically disordered proteins (IDPs)[1]. By virtue of their fluid properties, MLOs rapidly exchange components with the surrounding environment and display dynamic patterns of assembly and dissolution, while acting as a biomolecular selectivity filter to achieve functional specificity[2]. The assembly of these MLOs is thought to be driven by liquid-liquid phase separation (LLPS) of IDPs containing low complexity sequences[2a, 3] [4]. In contrast, the disassembly of the MLOs can be achieved simply via the reversal or removal of the initial driving force[5] [6]. Here we test an alternative mechanism whereby the monotonic addition of a single component in a ternary system (polypeptide, nucleic acid, and buffer) drives RNP granule assembly, structural dynamics, and dissolution.
A number of mixed polymers are known to display a reentrant phase transition, where the coexistence of upper and lower critical solution temperatures create a window-like behavior of mutual miscibility[7]. Induced by oppositely charged counterions, similar phase behavior has been observed for bio-macromolecules such as DNA and globular proteins[8]. In response to a monotonic increase in counterion concentrations, the system undergoes two phase transitions: an initial polymer condensation, and a subsequent decondensation. Since electrostatic interactions play a significant role in RNA-protein assembly for charged IDPs[9], we hypothesize that a similar reentrant phase transition can drive the disassembly of ribonucleoprotein MLOs.
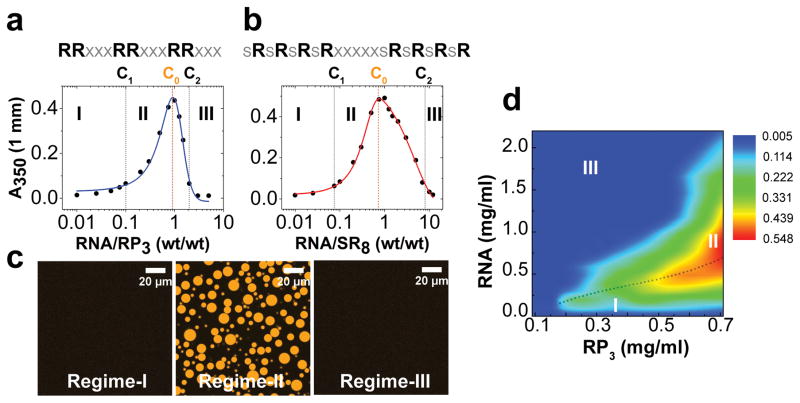
To probe whether RNP systems can undergo a reentrant phase transition, we initially study the phase behavior of two synthetic peptides (RP3[6]: Fig. 1a; SR8: Fig. 1b) containing multivalent arginine-rich linear motifs (RLMs), in the presence of single-stranded RNA (ssRNA). The design of our peptide sequences is guided by the prevalence of arginine-rich low complexity sequences in the RNA-binding proteome[9], such as the RGG/RG and SR repeat motifs[10], and their ability to potentiate LLPS via multivalent electrostatic interactions with RNA and acidic proteins[6, 11]. In the presence of a homopolymeric ssRNA (polyuridylic acid; polyU), we observe that RP3 and SR8 undergo sequential demixing and mixing phase transitions in response to increasing RNA concentration (Fig. 1; Fig. S1). These data suggest a window-like behavior for immiscibility, which is a hallmark for a reentrant phase transition[8c]. Within this window, the peptide/RNA microdroplets display liquid-like properties such as circular appearance (aspect ratio = 0.97±0.04; n =15), facile fusion, and rapid recovery of fluorescence after photobleaching (Fig. 1c; Fig. S1b, S2, S5a–f; Movie-1). We construct phase diagrams based on solution turbidity measurements (Fig. 1a, b, d), wherein three distinct regimes can be identified based on upper and lower critical concentrations, C1 and C2. At and , one homogeneous liquid phase prevails (regime-I and III, respectively), while two liquid phases coexist at (regime-II).
Figure 1. Reentrant phase transition in the mixtures of synthetic peptides containing R-rich linear motifs and RNA.
(a) and (b) Phase boundary curves for RP3/polyU and SR8/polyU mixtures, respectively, as determined by the solution turbidity. [peptide] = 200 μM in both cases. (c) Representative confocal fluorescence microscopy images of RP3/polyU mixtures that correspond to the three distinct regions of the phase diagram. (d) Projection of RP3/polyU phase diagram on the two-component concentration plane. The dotted line is the locus of the solution turbidity maxima (C0). Also highlighted are regime-I, II, and III. The color scale represents measured turbidity values at 350 nm. The corresponding turbidity plots are shown in Fig. S1a.
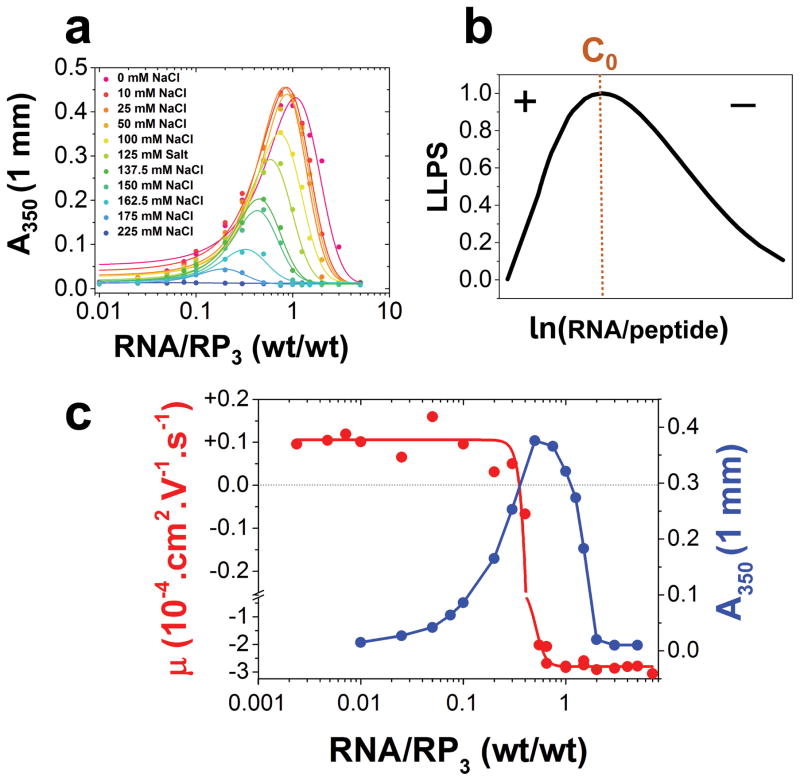
Ascertaining the molecular basis of the observed phase behavior in our peptide/RNA systems requires a closer investigation of the role of interaction electrostatics. Hypothesizing that they are the driving forces for the initial demixing transition, we determine C1 and C2 at varying monovalent salt (NaCl) concentrations. With increasing [NaCl], we observe a shrinkage of regime-II at [NaCl] > 50 mM, and complete disappearance at 225 mM NaCl (Fig. 2a; Fig. S6). The turbidity maxima, C0, also shows a marked shift towards progressively lower at [NaCl] > 50 mM (Fig. 2a, Fig. S6). These trends are expected in a system where LLPS is predominantly mediated by electrostatic attraction[12], commonly known as complex coacervation. Liquid-liquid demixing by complex coacervation has been recently implicated in intracellular MLO formation[13]. In the present case, the electrostatic interactions are mediated by the arginine side-chains (positively charged) of the peptide and the phosphate backbone of RNA (negatively charged). Increasing NaCl progressively weakens this interaction strength due to charge screening[12, 14] (Fig. S6 and the discussion therein). Simple charge screening, however, fails to explain the non-monotonic shape of our peptide/RNA phase diagrams[15] (Fig. 1a–b). To understand the physical origin of the mixing phase transition at CRNA > C0, we consider a charge inversion mechanism that predicts coacervate instability past C0 due to charge overscreening[8b]. We measure electrophoretic mobility, μ, of the peptide/RNA mixtures as a function of increasing RNA concentration utilizing dynamic light scattering (DLS). Electrophoretic mobility is expected to indicate the effective charge on the cationic peptides in the experimental buffer in regime-I, the surface charge of the peptide/RNA coacervate droplets in regime-II, and soluble peptide/RNA complexes in regime-III. In the case of a charge inversion, the net charge of the mixtures should change signs at C0 (Fig. 2b). Plots of μ vs. at a fixed [peptide] reveal that μ indeed displays a switch-like behavior at C0, shifting from positive to large negative values (Fig. 2c, Fig. S7), indicating that charge inversion accompanies a reentrant phase transition in our peptide/RNA systems. Additionally, calculations based on a simple charge inversion mechanism recapitulate the observed non-monotonic peptide/RNA phase boundary curves in our different datasets (SI Note-1) [8c]. A similar reentrant phase transition is also observed using polyadenylic acid (polyA), a short 40mer RNA (U40), and a 40mer ssDNA (T40; Fig. S3), which further confirms the role of electrostatic forces in driving the peptide/RNA phase behavior. Finally, a reverse titration experiment, where the RP3 peptide is varied monotonically against a constant RNA concentration, produces the same phase behavior (Fig. S4), which is consistent with recent spermidine-polyU phase separation observations[16].
Figure 2. Electrostatic interactions and charge inversion drive the reentrant phase behavior of peptide/RNA systems.
(a) Solution turbidity plots as a function of increasing [NaCl] (Also see Fig. S5) showing the effects of charge screening on the phase separation of the mixture. (b) Predicted RP3/polyU phase diagram using a charge inversion model (see SI Note-1). (c) Experimental detection of charge inversion at C0. Shown here is an overlay of electrophoretic mobility (red) and solution turbidity as a measure of LLPS (blue) ([RP3] = 200 μM in 10 mM Tris-HCl, pH 7.9 in all datasets).
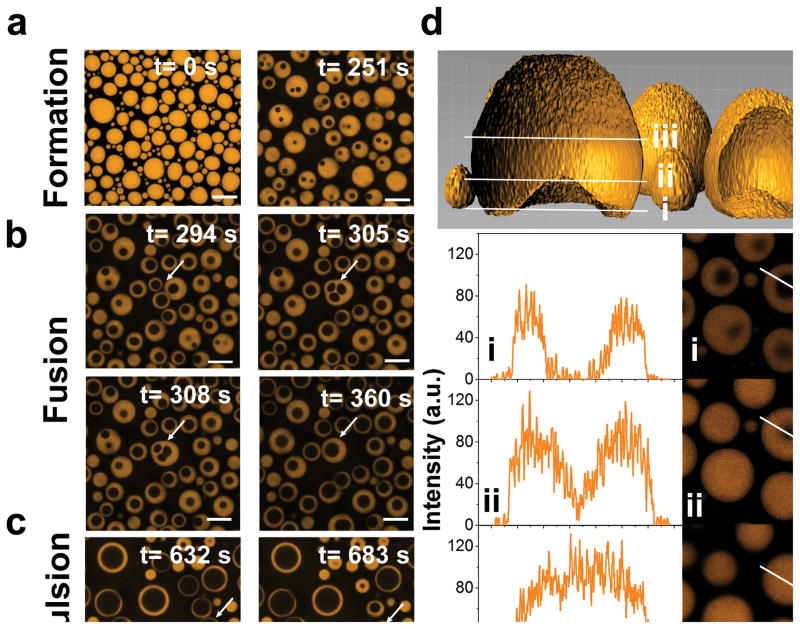
To complement the turbidity and mobility measurements, we visualize reentrant phase behavior by fluorescence and DIC microscopy. The addition of RNA into an RP3 peptide solution produces droplets at randomly nucleated sites that subsequently grow into larger droplets (Movie-1, 2). If a similar nucleation-growth mechanism is followed during the mixing phase transition, we envision that it is possible to create droplets of low-density phase within the droplets of high-density phase. To test this hypothesis, the RP3/RNA droplets (at CRNA < C0) were first allowed to grow on the microscope coverslip (Movie-1). The mixing phase transition is subsequently triggered by jumping to a final CRNA > C0 (Fig. 1a) by RNA injection into the bulk solution while monitoring the droplets on the coverslip. Two physical phenomena are registered when the system responds towards this non-equilibrium perturbation: (a) droplet shrinkage from the periphery as the excess RNA encountered the droplet surface, and (b) phase separation within the high-density droplets to form coexisting droplets of differential densities, termed as vacuoles henceforth (Movies-3, 4; Fig. 3; Figs. S8–10). The nucleation sites of these vacuoles appear to be random, as expected for a nucleation-growth mechanism, and the vacuoles show rich temporal dynamics (Movies-3, 4). Typically, the vacuoles display repetitive cycles of growth, fusion with the adjacent vacuoles, and expulsion into the bulk low-density phase upon contact with the surface of the parent RNP droplets (Movies-3, 4; Fig. 3; Fig. S8). While the overall non-equilibrium droplet dynamics lead to their complete dissolution at CRNA > C2, relatively stable droplet substructures are obtained at C2 > CRNA > C0 (SI Note-2) with a maximum vacuole lifetime of ≥ 2 hours in our experiments. Together, these data suggest that RNA influx into peptide/RNA coacervates not only triggers their disassembly but also leads to dynamic substructure formation with tunable lifetimes.
Figure 3. Dynamic droplet substructure formation during RNA-mediated mixing phase transition of RNP coacervate droplets.
(a)–(c) Confocal fluorescence microscopy images at different time-points of RP3/polyU droplets (1:0.5 wt/wt; 2 mM RP3) after injection of additional 0.75 equivalents of polyU (wt/wt) into the bulk phase. Images represent formation, fusion, and expulsion of vacuoles within RP3/polyU droplets. The corresponding video is shown in Movie-4. (d) Surface rendering of a z-stack confocal fluorescence microscopy image showing an internal vacuole within a RP3/polyU droplet. White lines represent cross-sections of the image within the vacuole (i), at the edge of the vacuole (ii), and above the vacuole (iii). (e) Fluorescence intensity plots at cross sections i, ii, and iii. The white arrows represent the distance axis (length = 14 μm). The corresponding 2D slices are shown to the right.
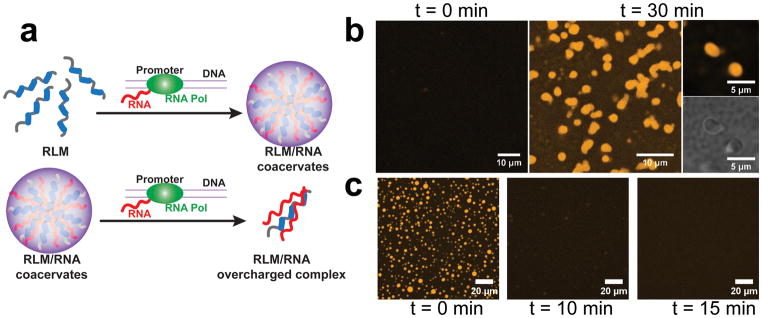
These microscopy experiments further suggest that RNA influx into a solution of the RP3 peptide can drive a complete cycle of droplet assembly and dissolution (Movie-5; Fig. S5). In a cellular context, we hypothesize that biochemical processes, such as RNA synthesis by transcription, can control RNA flux to produce the same dynamics for RLM phases. To test this, we perform transcription reactions using T7 RNA polymerase in vitro. In the presence of RP3, without any additional RNA, active RNA production by the polymerase results in the formation of RP3/RNA droplets (Fig. 4). Furthermore, when the transcription reaction is initiated in the presence of preexisting droplets of RP3/polyU, the droplets completely dissolve (Fig. 4). Control experiments show that mononucleotides, polymerase, and DNA template, at the concentrations used for the transcription reaction, have no significant effect on the phase behavior of the RP3/RNA system. Also, reaction mixtures lacking the polymerase fail to produce any effect, suggesting that the observed assembly-dissolution dynamics of the peptide/RNA droplets are solely controlled via in situ transcription by the polymerase.
Figure 4. Formation and dissolution of RP3/RNA granules by in vitro transcription.
(a) Schematic representation of reentrant phase transition in R-rich linear motif (RLM) systems by RNA synthesis in situ. RNA transcription by T7 RNA polymerase in presence of RLMs leads to the formation of phase-separated RLM/RNA droplets, while further increase in [RNA] by transcription leads to a subsequent mixing phase transition and dissolves the RLM/RNA droplets. Experimental data: (b) Confocal fluorescence and DIC microscopy images at different time-points of RP3/T7 transcription mixture. [RP3] = 1 mM. (c) Confocal fluorescence microscopy images of RP3/polyU granules (1:2 wt/wt) at different time-points after incubation with T7 transcription mixture. [RP3] = 200 μM. Control measurements in both (a) and (b) at the 60 minute time point of the mixture lacking the polymerase looked very similar to the respective images corresponding to time = 0.
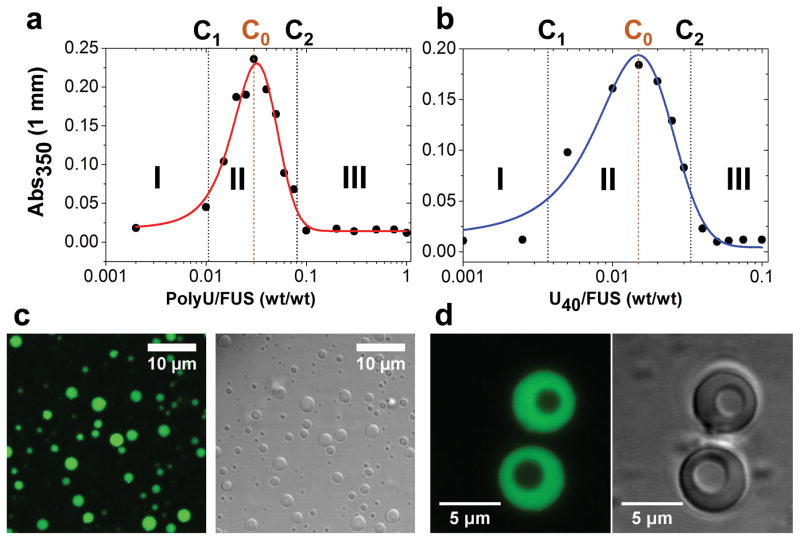
Typically, RNA-binding proteins contain more complex sequence patterns than peptides with simple amino acid compositions such as RP3 and SR8. Therefore, a complex interplay of homologous (self) and heterologous (protein-RNA) interactions are expected to shape their phase behavior[14]. To test whether RNA-binding IDPs containing multivalent RLMs can display a similar reentrant phase behavior, we study the stress granule-associated IDP, Fused in Sarcoma (FUS). Previous studies on FUS have shown that the protein undergoes RNA-mediated LLPS[17]. FUS contains two distinct low-complexity domains (LCDs). The N-terminal LCD is recognized as prion-like and mostly devoid of charges[17a] whereas the C-terminal LCD contains 23 RGG/RG repeats. Evidence suggests that FUS binds promiscuously to a large RNA pool due to these RGG/RG repeat motifs[18]. We observe that FUS undergoes LLPS via homologous interactions, which is consistent with recent reports[17]. Below the threshold concentration for homologous LLPS, FUS titrated with RNA not only triggers a demixing transition into liquid droplets but also leads to droplet dissolution, which is analogous to the phase behavior of RP3 and SR8 peptides (Fig. 5a–c; Fig. S11; Movie-6a). We propose that other RNA-binding IDPs also exhibit RNA-mediated reentrant phase behavior. LLPS data presented in a recent report on an RNA-binding protein[4] supports this proposal, while our present work offers a mechanistic basis for this observation. FUS/RNA droplets also display similar structural dynamics to that of the peptide/RNA systems. Jumping from the left (C1 < CRNA < C0) to the right (C0 < CRNA < C2) side of the phase diagram along the RNA concentration axis triggers vacuole formation with lifetimes ≥ 60 mins (Fig. 5d; Movie-6b; SI Note-2). Together, these data clearly demonstrate controlled formation of multi-compartment RNP liquid droplets that is driven by a reentrant phase transition.
Figure 5. Reentrant phase transition and dynamic droplet substructure formation in the FUS/RNA system.
(a) and (b) Phase boundary curves for FUS with polyU and U40 RNA, respectively (determined by measuring the solution turbidity). The critical concentrations (C1 and C2) and the boundary conditions (C0) are shown by vertical dotted lines, while three distinct regimes were labeled in the phase diagram. [FUS] = 10 μM in both cases. (c) DIC and confocal fluorescence microscopy images of FUS/polyU droplets (1:0.03 wt/wt) in regime-II. (d) RNA-mediated vacuole formation within FUS/polyU droplets (final ; wt/wt) using confocal fluorescence and DIC microscopy. Also, see Movie-6.
Combined, these results reveal that RNP granule assembly by complex coacervation and dissolution by RNP charge inversion can simply be controlled via a monotonic increase in RNA concentration. Reentrant phase behavior further allows for the creation of dynamic internal substructures within RNP coacervate droplets, which showcases the potential for controlled subcompartment generation within droplet-like MLOs. Nearly half a century ago, the existence of nucleolar vacuoles displaying similar dynamics was first reported [19]. Recent in vitro and in vivo studies have further reported such observations, wherein applying an external electric field[20] and overexpressing a disordered protein[21] resulted in vacuolization. Our study sheds light on a possible molecular origin of such dynamic internal substructures, suggests a fundamental cellular process that can control the reentrant phase behavior, and offers new prospects to study non-equilibrium processes in model intracellular MLOs as well as in synthetic coacervate-based protocells.
Materials and Methods
Detailed materials and methods are provided in the supplementary information section.
Supplementary Material
Acknowledgments
This work was supported by grants (RO1 GM066833 and RO1 GM115634 to A.A.D.) from the NIGMS, the NIH, the Donald E. and Delia B. Baxter fellowship (to A.N.M), and a postdoctoral fellowship (15POST22520013 to P.R.B.) from A.H.A. We thank Dr. Kaustav Bhattcharjee (senior researcher, Jadavpur University, Kolkata, India), Dr. Satamita Samanta and Dr. George Thurston (RIT) for valuable discussion on the conceptual framework of this paper, Dr. William B. Kiosses for assistance with confocal and DIC microscopy, and Nicole Chen for assistance with the DLS instrument. Author Contributions: P.R.B. conceived the idea and designed the experiments with assistance from M.M.M. and A.N.M. P.R.B., M.M.M., A.N.M. and P.L.O. performed the experiments and analyzed the data. All authors contributed in writing the manuscript.
Footnotes
Competing financial interests:
The authors declare no competing financial interests.
References
- 1.Banani SF, Lee HO, Hyman AA, Rosen MK. Nat Rev Mol Cell Biol. 2017 doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Hyman AA, Weber CA, Julicher F. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]; b) Meng F, Na I, Kurgan L, Uversky VN. International journal of molecular sciences. 2016;17 doi: 10.3390/ijms17010024. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hyman AA, Brangwynne CP. Dev Cell. 2011;21:14–16. doi: 10.1016/j.devcel.2011.06.013. [DOI] [PubMed] [Google Scholar]; d) Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]; e) Zwicker D, Decker M, Jaensch S, Hyman AA, Julicher F. Proc Natl Acad Sci U S A. 2014;111:E2636–2645. doi: 10.1073/pnas.1404855111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dundr fM. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holehouse AS, Pappu RV. Nat Mater. 2015;14:1083–1084. doi: 10.1038/nmat4459. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. Elife. 2014;3 doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aumiller WM, Jr, Keating CD. Nat Chem. 2016;8:129–137. doi: 10.1038/nchem.2414. [DOI] [PubMed] [Google Scholar]
- 7.Clark EA, Lipson JEG. Polymer. 2012;53:536–545. [Google Scholar]
- 8.a) Zhang F, Skoda MW, Jacobs RM, Zorn S, Martin RA, Martin CM, Clark GF, Weggler S, Hildebrandt A, Kohlbacher O, Schreiber F. Phys Rev Lett. 2008;101:148101. doi: 10.1103/PhysRevLett.101.148101. [DOI] [PubMed] [Google Scholar]; b) Teif VB, Bohinc K. Progress in biophysics and molecular biology. 2011;105:208–222. doi: 10.1016/j.pbiomolbio.2010.07.002. [DOI] [PubMed] [Google Scholar]; c) Nguyen TT, Rouzina I, Shklovskii BI. The Journal of chemical physics. 2000;112:2562–2568. [Google Scholar]
- 9.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 10.a) Thandapani P, O’Connor TR, Bailey TL, Richard S. Mol Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]; b) Birney E, Kumar S, Krainer AR. Nucleic Acids Research. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, Stanley CB, Nourse A, Deniz AA, Kriwacki RW. Elife. 2016;5 doi: 10.7554/eLife.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbeek JT, Voorn MJ. J Cell Physiol Suppl. 1957;49:7–22. discussion, 22–26. [PubMed] [Google Scholar]
- 13.Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, Rosen MK. Mol Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brangwynne CP, Tompa P, Pappu RV. Nat Phys. 2015;11:899–904. [Google Scholar]
- 15.Grosberg AY, Nguyen TT, Shklovskii BI. Reviews of Modern Physics. 2002;74:329–345. [Google Scholar]
- 16.Aumiller WM, Jr, Pir Cakmak F, Davis BW, Keating CD. Langmuir. 2016;32:10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- 17.a) Burke KA, Janke AM, Rhine CL, Fawzi NL. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Schwartz JC, Cech TR. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Johnson JM, Jones LE. American journal of botany. 1967;54:189–198. [PubMed] [Google Scholar]; b) Johnson JM. J Cell Biol. 1969;43:197–206. doi: 10.1083/jcb.43.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin Y, Niu L, Zhu X, Zhao M, Zhang Z, Mann S, Liang D. Nat Commun. 2016;7:10658. doi: 10.1038/ncomms10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt HB, Rohatgi R. Cell Rep. 2016;16:1228–1236. doi: 10.1016/j.celrep.2016.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.