Abstract
Sporulation is a strategy widely utilized by a wide variety of organisms to adapt to changes in their individual environmental niches and survive in time and/or space until they encounter conditions acceptable for vegetative growth. The spores produced by bacteria have been the subjects of extensive studies, and several systems such as Bacillus subtilis have provided ample opportunities to understand the molecular basis of spore biogenesis and germination. In contrast, the spores of other microbes, such as fungi, are relatively poorly understood. Studies of sporulation in model systems such as Saccharomyces cerevisiae and Aspergillus nidulans have established a basis for investigating eukaryotic spores, but very little is known at the molecular level about how spores function. This is especially true among the spores of human fungal pathogens such as the most common cause of fatal fungal disease, Cryptococcus neoformans. Recent proteomic studies are helping to determine the molecular mechanisms by which pathogenic fungal spores are formed, persist, and germinate into actively growing agents of human disease.
Keywords: Spores, germination, fungal pathogenesis, proteomics, sexual development
Spores are essential for survival of diverse organisms
Sporulation occurs in organisms across the tree of life from bacteria and protozoa to plants and fungi and facilitates both survival in response to adverse growth conditions and dispersal to new, more hospitable environments (Driks 2002; Kessin 2010; Wyatt et al. 2013). Typically, spores are metabolically quiescent, stress resistant, and poised for propagation (germination) in response to suitable growth conditions. These features have been observed in eubacteria and eukaryotes with discoveries of viable bacterial spores embedded in amber tens of millions of years old and culturable fungal spores found in ocean sediments and arctic ice dated in the hundreds of thousands of years old. Several species of bacterial spores have even been shown to survive the vacuum of outer space (Cano and Borucki 1995; Brown and Hovmøller 2002; Damare and Raghukumar 2007; D’Elia et al. 2008). These amazing abilities showcase how spores facilitate survival of diverse organisms in myriad and varied environments.
To understand how spores are imbued with such robust survival traits, they have been studied extensively in several model systems. The best understood systems of sporulation come from bacteria in which specific developmental programs drive cellular differentiation and environmental adaptation (Driks 2002). Three classes of Gram-positive bacteria, Bacilli, Clostridia, and Negativicutes, include members that produce endospores with dramatically increased resistance to a wide range of environmental stresses, such as heat, desiccation, and ultraviolet (UV) radiation (Setlow 2007; Galperin et al. 2012). In these organisms, resistance to harsh environmental conditions results from both specialized, multi-layered surface structures and unique spore composition (Setlow 2006; Henriques and Moran 2007). In the well-studied system of Bacillus subtilis critical compositional properties include low core water content, high levels of dipicolinic acid (DPA) in chelation with divalent cations (CaDPA), abundant small acid-soluble spore proteins (SASP), and the intrinsic stability of spore proteins overall (Setlow 2006). It is hypothesized that all of these specialized features work in concert to contribute to overall spore resistance and stability in the face of harsh environmental conditions.
This robust ability of spores to persist then provides an opportunity for spores to reestablish vegetative growth in response to appropriate conditions. Given a proper stimulus, spores will break dormancy (germinate) and transition back into a growing cell (Setlow 2014). In B. subtilis, there are several major events that take place during spore germination, including the release of monovalent cations and CaDPA, degradation of peptidoglycan, and expansion of the spore core (Setlow 2013). Proteomic studies on B. subtilis spores show that spores harbor proteins required for stress resistance, anabolism, and cell signaling -- all consistent with the idea that spores resist environmental insults and are poised for rapid growth in response to germination signals (Liu et al. 2004; Mao et al. 2011). These classes of protein components of spores appear to be fairly consistent across diverse bacterial species; however, the nature and efficacy of conditions and compounds that provide the signal to germinate vary widely, as do the molecular components used to sense and signal germination (Setlow 2013; Paredes-Sabja et al. 2014; Bhattacharjee et al. 2016). Even among individual spores within the same population, there can be significant heterogeneity in the time taken to germinate when stimulated by nutrients, varying from minutes to hours or even days (Stewart and Setlow 2013). This variation is likely to influence the outcome of spore survival and epitomizes the gaps in understanding of bacterial spore biology. Understanding the properties of bacterial spores and the conditions under which they resume vegetative growth has been and will continue to be important for determining not only bacterial survival but also the resilience of microbes and other spore-forming organisms across the tree of life.
Most of what is known about spore biology is from bacterial systems, but sporulation of protists and fungi is also common. In fact, most of the spore-forming organisms on earth are fungi. Fungi have evolved complex and robust methods to produce spores and are the reigning champions of spore dispersal. Fungal spores have been shown to circumnavigate the globe via wind currents, leading to nearly ubiquitous representation of fungi among all ecosystems on the planet (Brown and Hovmøller 2002; Kessin 2010; Wyatt et al. 2013).
Fungal sporulation has been investigated in detail in the model yeast Saccharomyces cerevisiae, in which sporulation is initiated when diploid cells are starved for nitrogen in the presence of a non-fermentable carbon source (Engebrecht 2003). This developmental program couples meiosis to spore biogenesis within one single mother cell and includes several coordinated events: 1) chromosome alignment, recombination, and segregation, 2) prospore membrane formation to envelope four newly-produced haploid nuclei, and 3) cell wall assembly outside the four maturing spores (Neiman 2005, 2011; Piccirillo et al. 2016). This mechanism by which spores are formed is one of the best understood, but it represents only one of thousands of strategies that different fungi use to produce spores (Engebrecht 2003). Among Aspergillus species, for example, sporulation occurs via both sexual and asexual development, producing thousands of ascospores or conidia per fruiting body (van Leeuwen et al. 2013). Among mushrooms such as Coprinopsis cinerea, spores are produced during sexual reproduction and released through the gills of fruiting bodies (Pukkila 2011). In plant pathogens such as Ustilago maydis, diploid spores are produced in planta and disperse prior to meiosis (Vollmeister et al. 2012). As such, fungal spores can represent mitotic products or meiotic products; they can be haploid, diploid, or multinucleate; they can be contained within an ascus, produced in chains, or released individually. In all cases, however, they are specialized to survive adverse conditions in their own environmental niches and thus share several key spores features, including relative metabolic quiescence, stress resistance, and germination capability (Herman and Rine 1997; Brown and Hovmøller 2002; Wyatt et al. 2013; Krijgsheld et al. 2013).
Like bacterial spores, there are also two general categories of factors in fungal spores that contribute to their specialized functions and properties: structural and compositional. In S. cerevisiae, the spore wall consists of four layers from inside to outside: mannan, β-1,3-glucans, chitosan, and dityrosine molecules (Smits et al. 2001). The outer two layers are not present in the cell walls of vegetative yeast and are the major contributors of resistance of spores to organic solvents, heat, and digestive enzymes (Briza et al. 1990; Pammer et al. 1992; Neiman 2005). In addition to a relatively thick coat, spores possess protective small molecules including sugars (trehalose), sugar alcohols (glycerol, mannitol), betaine, and amino acids. Finally, proteins such as heat shock proteins and hydrophilins provide generally superior environmental stress resistance to various types of fungal spores (Wyatt et al. 2013).
While the production of spores can be life-saving for microbial species, the potential for survival can be realized only if spores can reinitiate vegetative growth in response to appropriate conditions (germinate). As in bacteria, fungal germination is an essential differentiation process in which spores transition from a relatively dormant particle to a vegetatively growing cell. Fungal germination generally involves a specific sequence of events, including changes in cellular absorbance and weight (water uptake), breakdown of stored carbohydrates (trehalose and mannitol), remodeling of the cell wall, and expansion and elongation of tightly packed spore contents (Rousseau et al. 1972; d’Enfert 1997; Krijgsheld et al. 2013; Novodvorska et al. 2013).
There are a wide variety of environmental signals (germinants or inhibitors) that trigger or inhibit germination, depending on the spore type. For example, uredospores of some rust fungi initiate germination when exposed to pure water (Dijksterhuis 2003). Untimely germination of Penicillium paneum conidiospores is inhibited by a self-produced volatile compound (Chitarra et al. 2004). Spores of S. cerevisiae germinate readily in response to the presence of a fermentable carbon source (Herman and Rine 1997), whereas spores of Talaromyces macrosporus require both nutrients and a rigorous external trigger of high temperature (85°C) or pressure (between 400 and 800 MPa) (Dijksterhuis et al. 2002; Dijksterhuis and Teunissen 2004). All of these signals represent the diverse situations under which fungi must grow, and they epitomize the diversity of fungal biology in general. However, it is not yet clear how universal or specialized each strategy is with respect fungal germination or how conserved the molecular mechanisms of fungal spore germination are overall.
To begin to understand molecular events that occur during germination, global gene expression profiling of germinating spores has been carried out in several fungi. In A. fumigatus and A. niger, increases in the transcript levels of genes involved in biosynthesis of proteins, RNA turnover, and respiratory metabolism occur during germination of conidia (Lamarre et al. 2008; van Leeuwen et al. 2013; Novodvorska et al. 2013, 2016; Hagiwara et al. 2016). Similarly, in S. cerevisiae, the overall trend of transcriptional changes during germination showed increases in genes involved in metabolism of glucose and other nutrients and a transient up-regulation of genes related to protein folding and transport (Joseph-Strauss et al. 2007; Geijer et al. 2012). These changes in gene expression were all consistent with a release from stress responses and a transition to cellular proliferation. These observed changes in gene expression were very similar to the changes in gene expression that take place when S. cerevisiae cells grown to stationary phase are transferred to fresh medium and re-enter vegetative growth. This metabolic similarity, along with the apparent absence of germination-specific gene groups suggests the possibility that germination is not necessarily a specific differentiation process from one cell type to another, but rather, it is primarily a regulated metabolic adaptation (Geijer et al. 2012).
This hypothesis is also supported by the fact that the handful of genes in fungi discovered to be important for spore germination, such as members of the Ras/mitogen-activated protein kinase pathway and the transcription factor Ume6, all play critical roles during other stages of the life cycle and are not specific to germination (Herman and Rine 1997; Strich et al. 2011). In fact, a large-scale genetic screen of the S. cerevisiae deletion set identified 158 strains with defects in growth from the spore state, including genes involved in meiotic chromosome behavior, carbon metabolism, vesicle transport, nutrient sensing, and cell wall integrity, but their specific roles during germination are not clear (Deutschbauer et al. 2002). A more recent screen for germination mutants identified two new genes, TRF4 and ERG6, which function in the TRAMP complex to degrade mRNA and in ergosterol biosynthesis, respectively (Kloimwieder and Winston 2011). Both genes play roles in vegetative growth. Thus, in S. cerevisiae it seems that either 1) there may be no exclusively germination-specific molecular machinery; instead, adaptation of existing signaling pathways and regulatory components may be the keys to fungal spore germination, or 2) specific components of the germination process are sufficiently important for this survival process that there is redundancy in the system that prevents their detection in screens carried out to date. In either case, understanding the specific components or combinations of existing components that govern germination will provide fundamental insights into eukaryotic responses to changing environmental conditions and the strategies for the development of new cell types in response to those conditions.
Pathogenic Fungal Spores
As discussed above, sporulation is a strategy widely utilized by fungi to survive adverse conditions and disperse to new territories. The specialized processes and properties leading to spore formation, persistence, and germination facilitate adaptation of spore-forming organisms to their individual environmental niches, and in some cases, enable fungal spores to become infectious agents of plants, animals, and humans (Nemecek et al. 2006; Mallozzi et al. 2010; Logan 2012; Wyatt et al. 2013).
Extensive studies have been carried out in a handful of model plant-pathogenic fungi, where germination of spores, either sexual or asexual, initiates their infectious life cycles with their respective plant host tissues (Dean et al. 2012). The focus on mechanisms of pathogenesis has generated a tremendous amount of information on plant-pathogen interactions and benefited the control of plant fungal diseases (Perez-Nadales et al. 2014). However, a molecular understanding of the intrinsic properties of those infectious spores is relatively sparse, despite their important roles within the life cycle. Spores from only a few plant fungal pathogens have been subjected to compositional analyses, including Uromyces appendiculatus, a rust fungus infecting wheat and beans, Blumeria graminis f. sp. hordei, an ascomycete causing barley powdery mildew, Magnaporthe oryzae, the causative agent of rice blast disease, several Puccinia rust basidiomycetes, and Colletotrichum acutatum that infects several commercially valuable fruit crops (Cooper et al. 2006; Noir et al. 2009; Gokce et al. 2012; El-Akhal et al. 2013; Quecine et al. 2016; Zhang et al. 2015; Beinhauer et al. 2016). Proteomic studies of these spores showed that proteins involved in common processes such as protein homeostasis and metabolism are readily detectable in those spores, which is consistent with stress resistance and rapid growth during germination; however, one would expect proteins involved in those processes to also be important for vegetative growth. Without a side-by-side comparison to their vegetative counterparts, it is challenging to assign particular importance to any give class of spore proteins. Future studies to compare and contrast protein content between spores and vegetative cells types may provide necessary insights into the importance of detected spore proteins in plant pathogenic spores.
Similarly, spores of animal and human fungal pathogens are also poorly understood. Fungi such as Histoplasma capsulatum, Blastomyces dermatitidis, Aspergillus fumigatus, Coccidioides immitis, Sporothrix shenkii, Penicillium marneffei, and Cryptococcus neoformans are all environmental fungi that can cause disease in animals and humans, and the most common route of infection is through the inhalation of cells from environmental sources into the lung (Nemecek et al. 2006). Spores are likely infectious particles for all of these pathogens; however, very little is known about their basic spore biology. Understanding the fundamental properties of environmental fungi and how they survive promises to inform the mechanisms by which they infect and cause disease in humans.
To determine the molecular mechanisms underlying the distinct properties of spores, proteomic studies have been carried out on human fungal pathogen spores using their vegetative counterparts for comparison. In A. fumigatus, proteins that are more abundant in conidia (asexual spores) compared to mycelia (vegetative form) have functions associated with reactive oxygen intermediates (ROI) detoxification, pigment (melanin) biosynthesis, and conidial rodlet layer formation, which all likely account for the extreme stress resistance of conidia (Teutschbein et al. 2010). However, different enrichment patterns of spore proteins have been observed in other studies. For example, proteins related to ROI detoxification and some heat shock proteins are more abundant in vegetatively growing cells than in conidiospores in the related, non-pathogenic A. nidulans (Oh et al. 2010). Another study identified A. fumigatus conidia-enriched proteins involved in sporulation, cell wall biosynthesis, and secondary metabolite biosynthesis, in addition to those related to stress responses (Suh et al. 2012). It is possible that these differences reflect different life cycles within their respective environmental niches and diverse environmental conditions under which each species produces spores.
In Trichophyton rubrum, the most common dermatophyte causing fungal skin infections, a proteomic analysis of spores alone yielded proteins involved in common processes, including protein synthesis, folding and degradation, metabolism and energy production (Leng et al. 2008). The presence of these proteins is generally interpreted as providing the specialized potential of spores for stress resistance and rapid growth during germination, but one would expect proteins involved in those processes to also be required for vegetative growth. Again, it remains to be determined which proteins are overrepresented relative to other cell types before the significance of any given spore protein can be assigned.
More recently, a comprehensive proteomic comparison between spores and yeast was carried out in Cryptococcus neoformans, which is the most common cause of fatal fungal disease in humans (Brown et al. 2012; Huang et al. 2015). Highly sensitive nanoscale liquid chromatography/mass spectrometry was used to determine the overall proteomic compositions of spores and yeast, resulting in the identification of eighteen spore-enriched proteins. The in-depth comparison between spores and yeast showed that spore-enriched proteins are associated with processes such as transcriptional regulation and chromosome organization, rather than the aforementioned categories of proteins associated with stress resistance. In contrast, yeast-enriched proteins are largely involved in RNA processing, general metabolism, and energy production. Thus, comparing the proteomes of two different cell types (yeast vs. spores) resulted in a distinct set of spore-enriched proteins whose functions could be evaluated specifically in the context of spore biology.
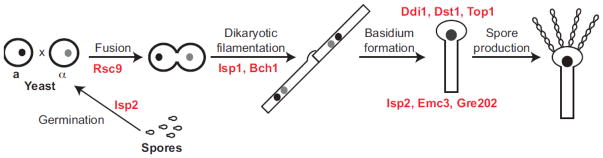
Functional studies using molecular genetics were carried out to determine the biological roles of 18 proteins that were specifically overrepresented in spores and not in yeast. Surprisingly, deletion of genes encoding these proteins did not result in discernible decreases in stress resistance of mutant spores. Instead, eight genes that encode spore-enriched proteins were found to be required for sexual development and spore biogenesis, and only one (ISP2) was involved in a spore-specific process (germination) (Figure 1). There were no clear patterns for the observed phenotypes based on the predicted molecular functions of the proteins. For example, the predicted chromatin remodeling complex component RSC9 is required for cellular fusion (mating) between haploid yeast during early sexual development. Chromatin and chromatin remodeling are important for many cellular processes (Cairns et al. 1996; Damelin et al. 2002; Marguerat et al. 2012; Hayles et al. 2013; Nakazawa et al. 2016), but the rsc9Δ strains of C. neoformans do not show overt phenotypes outside of sexual development, and the specificity of a role for Rsc9 in mating is not obvious.
Figure 1.
Proteins enriched in spores relative to yeast in the human fungal pathogen C. neoformans act primarily during sexual development. Rsc9 is involved in cell fusion (mating), Isp1 and Bch1 are required for efficient filamentation, Ddi1, Dst1, and Top1 are required for spore formation, Isp2, Emc3, and Gre202 modulate the number of spores produced. Isp2 was the only protein in the analysis with a spore-specific function; it is required for rapid germination of spores into yeast.
This is also true for the filamentation genes BCH1 and ISP1 and the spore formation genes TOP1, DST1, DDI1, EMC3, and GRE202. The spore-enrichment of these proteins with critical functions during the developmental process that leads to spore production suggests two (non-exclusive) models for formation of the non-structural spore proteome: 1) Proteins are produced in response to specific needs at various stages of sexual development and persist until spore production, eventually making their way into mature spores (passive deposition), or 2) proteins are produced when needed during development and are maintained or resynthesized during spore biogenesis for specific packaging into spores to serve spore-specific functions (active deposition).
A passive deposition model implies that the protein of interest is important in development (and only during development), and its overrepresentation in spores is a simple consequence of a stable protein in the fruiting body cytoplasm “ending up” in spores as a consequence of the packaging process. This model has been proposed in the formation of Bacillus anthracis spores in which proteins produced during spore formation and are ultimately present in spores do not appear to play additional roles in spore biology. This idea is further supported by the finding that spore proteins in B. subtilis are generally more stable than those encoded by the genome at large(Liu et al. 2004; Bergman et al. 2006).
In contrast, an active deposition model would involve the packaging of specific proteins into spores to facilitate spore-specific processes. Akin to the “maternally derived” transcripts and proteins identified in developmental models of larger eukaryotes, spore components could be “basidium-derived” and poise spores for spore-specific processes such as germination. For example, in the absence of Isp2, C. neoformans spores germinate into yeast, but they experience a two-hour delay in the initiation of vegetative growth. The Isp2 protein appears to be actively deposited into spores from the basidium, remain stable through the morphological changes that occur over 12 hours, and then function (finally) in the process of vegetative growth initiation. This active deposition of a protein into spores that functions at a much later stage of cellular differentiation is consistent with the idea that spore composition reflects function and can inform the pathways and processes critical for spore survival.
The behavior of isp2Δ strains also challenges the idea (at least in C. neoformans) that germination is simply a modified cell cycle or general exit from quiescence. Several lines of evidence support the hypothesis that germination is a specific developmental program with specialized components: First, Isp2 does not show sequence similarity to any other proteins in any other organisms, suggesting that it is not a core cell cycle component. Second, isp2Δ mutants do not have strong vegetative growth phenotypes; they and show a modest slow-growth phenotype as yeast on solid agar but grow like wild type strains in liquid culture and do not show any other vegetative growth defects. Third, they do not show any defects in re-initiating vegetative growth from stationary phase cultures (a relatively quiescent state). Instead, isp2Δ strains show phenotypes during sexual development and germination - they produce 50% more spores than wild type strains and exhibit a slow-germination phenotype on both solid agar and in liquid culture. These phenotypes indicate that the primary role of this spore-enriched protein is to function in spore biology and facilitate efficient germination. It remains to be determined precisely how Isp2 controls entry into vegetative growth, but the discovery of Isp2 also provides the impetus to consider that germination is a microbe-specific process that is distinct from more conserved eukaryotic growth mechanisms.
This idea has great appeal for medical mycology because of the limited therapeutics available for treating invasive fungal diseases. There is a tremendous clinical need for low toxicity, effective antifungal drugs, but one complication is that fungi and humans share significant similarity at the molecular level. If molecular components of germination were to be specific to fungi and distinct from humans, germination could be a useful target for new treatments that are less toxic to humans and more effective against fungi than the current arsenal of drugs. If spores that enter the lungs of a susceptible patient cannot germinate into a vegetative growth form, then they cannot cause invasive disease. Most mortality from fungi is the result of disseminated disease and fulminant fungal growth that overwhelms the host. The prevention of germination among spore-forming human fungal pathogens could provide an opportunity to stop fungal disease before it starts. Thus, the development of germination inhibitors as drugs to treat vulnerable patients prophylactically holds great promise because this strategy could prevent disease and decrease the overall burden of invasive fungi.
Conclusions
Future investigations of the molecular mechanisms underlying fungal spore germination, (such as determining the defects of isp2Δ and other mutants in C. neoformans) will provide novel insights into the biology of pathogenic spores. Because germination is a process universal among all spore-producing fungi and likely conserved due to its important roles in fungal survival, it is reasonable to speculate that there will be germination mechanisms in common across diverse fungi. Continued studies to establish a broader understanding of germination have the potential to identify targets for anti-germinants with the capacity for broad spectrum activity against the spores of fungi that cause diseases of plants, animals, and humans.
Understanding germination will also inform fungal biology more generally. Recent decades have seen the increasing spread of fungi to new environments on Earth (and even in outer space). As global temperatures have risen, fungal ranges have expanded and led to opportunities for fungal pathogens to cause diseases in new environments such as White Nose Syndrome in North American bats and Coffee Rust Disease in coffee plants in Latin America. The intertwined nature of fungal and human biology now exists off-planet with fungi living on wiring in the International Space Station and causing allergies in astronauts. All of these examples likely result from the spread of fungal spores, which came into contact with a germinant and established vegetatively growing populations. As spores continue to circle the globe on wind currents or in spacecraft and land in ever-expanding environments, understanding the molecular mechanisms by which they respond to germinants and grow will become essential in efforts to predict and/or mitigate the broad-reaching effects of fungi on Earth and beyond.
Acknowledgments
This work was supported by National Institutes of Health R01 grant AI089370 to CMH and The Hartwell Foundation Individual Biomedical Research Award to CMH.
References
- Beinhauer J, Raus M, Hanzalová A, et al. Intact spore MALDI-TOF mass spectrometry and proteomic analysis of Puccinia pathogenic fungi. Biochim Biophys Acta. 2016;1864:1093–1103. doi: 10.1016/j.bbapap.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, et al. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee D, McAllister KN, Sorg JA. Germinants and their receptors in Clostridia. J Bacteriol. 2016;198:2767–2775. doi: 10.1128/JB.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza P, Breitenbach M, Ellinger A, Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13–165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Brown JKM, Hovmøller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/S0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber. Science. 1995;268:1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- Chitarra GS, Abee T, Rombouts FM, et al. Germination of Penicillium paneum conidia is regulated by 1-octen-3-ol, a volatile self-inhibitor. Appl Environ Microbiol. 2004;70:2823–2829. doi: 10.1128/AEM.70.5.2823-2829.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Garrett WM, Campbell KB. Shotgun identification of proteins from uredospores of the bean rust Uromyces appendiculatus. Proteomics. 2006;6:2477–2484. doi: 10.1002/pmic.200500630. [DOI] [PubMed] [Google Scholar]
- Damare S, Raghukumar C. Fungi and macroaggregation in deep-sea sediments. Microb Ecol. 2007;56:168–177. doi: 10.1007/s00248-007-9334-y. [DOI] [PubMed] [Google Scholar]
- Damelin M, Simon I, Moy TI, et al. The genome-wide cocalization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell. 2002;9:563–573. doi: 10.1016/S1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JaL, Pretorius ZA, et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Elia T, Veerapaneni R, Rogers SO. Isolation of microbes from Lake Vostok accretion ice. Appl Environ Microbiol. 2008;74:4962–4965. doi: 10.1128/AEM.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Enfert Fungal spore germination: Insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Gen and Biol. 1997;21(2):163–172. doi: 10.1006/fgb.1997.0975. [DOI] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijksterhuis J. Confocal microscopy of Spitzenkörper dynamics during growth and differentiation of rust fungi. Protoplasma. 2003;222:53–59. doi: 10.1007/s00709-003-0006-6. [DOI] [PubMed] [Google Scholar]
- Dijksterhuis J, Driel KG, vanSanders MG, et al. Trehalose degradation and glucose efflux precede cell ejection during germination of heat-resistant ascospores of Talaromyces macrosporus. Arch Microbiol. 2002;178:1–7. doi: 10.1007/s00203-002-0410-x. [DOI] [PubMed] [Google Scholar]
- Dijksterhuis J, Teunissen PGM. Dormant ascospores of Talaromyces macrosporus are activated to germinate after treatment with ultra high pressure. J Appl Microbiol. 2004;96:162–169. doi: 10.1046/j.1365-2672.2003.02133.x. [DOI] [PubMed] [Google Scholar]
- Driks A. Overview: Development in bacteria: spore formation in Bacillus subtilis. Cell Mol Life Sci CMLS. 2002;59:389–391. doi: 10.1007/s00018-002-8430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Akhal MR, Colby T, Cantoral JM, et al. Proteomic analysis of conidia germination in Colletotrichum acutatum. Arch Microbiol. 2013;195:227–246. doi: 10.1007/s00203-013-0871-0. [DOI] [PubMed] [Google Scholar]
- Engebrecht J. Cell signaling in yeast sporulation. Biochem Biophys Res Commun. 2003;306:325–328. doi: 10.1016/S0006-291X(03)00983-5. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Mekhedov SL, Puigbo P, et al. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol. 2012;14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijer C, Pirkov I, Vongsangnak W, et al. Time course gene expression profiling of yeast spore germination reveals a network of transcription factors orchestrating the global response. BMC Genomics. 2012;13:554. doi: 10.1186/1471-2164-13-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce E, Franck WL, Oh Y, et al. In-depth analysis of the Magnaporthe oryzae conidial proteome. J Proteome Res. 2012;11:5827–5835. doi: 10.1021/pr300604s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Takahashi H, Kusuya Y, et al. Comparative transcriptome analysis revealing dormant conidia and germination associated genes in Aspergillus species: an essential role for AtfA in conidial dormancy. BMC Genomics. 2016;17:358. doi: 10.1186/s12864-016-2689-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, et al. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3:130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Herman PK, Rine J. Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 1997;16:6171–6181. doi: 10.1093/emboj/16.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hebert AS, Coon JJ, Hull CM. Protein composition of infectious spores reveals novel sexual development and germination factors in Cryptococcus. PLoS Genet. 2015;11:e1005490. doi: 10.1371/journal.pgen.1005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N. Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks. Genome Biol. 2007;8:R241. doi: 10.1186/gb-2007-8-11-r241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin RH. Dictyostelium: evolution, cell Biology, and the development of multicellularity, reissue edition. Cambridge University Press; Cambridge: 2010. [Google Scholar]
- Kloimwieder A, Winston F. A screen for germination mutants in Saccharomyces cerevisiae. G3. 2011;1:143–149. doi: 10.1534/g3.111.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijgsheld P, Bleichrodt R, van Veluw GJ, et al. Development in Aspergillus. Stud Mycol. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre C, Sokol S, Debeaupuis J-P, et al. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics. 2008;9:417. doi: 10.1186/1471-2164-9-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng W, Liu T, Li R, et al. Proteomic profile of dormant Trichophyton Rubrum conidia. BMC Genomics. 2008;9:303. doi: 10.1186/1471-2164-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bergman NH, Thomason B, et al. Formation and composition of the Bacillus anthracis endospore. J Bacteriol. 2004;186:164–178. doi: 10.1128/JB.186.1.164-178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan Na. Bacillus and relatives in foodborne illness. J Appl Microbiol. 2012;112:417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- Mallozzi M, Viswanathan V, Vedantam G. Spore-forming Bacilli and Clostridia in human disease. Future Microbiol. 2010;5:1109–1123. doi: 10.2217/fmb.10.60. [DOI] [PubMed] [Google Scholar]
- Mao L, Jiang S, Wang B, et al. Protein profile of Bacillus subtilis spore. Curr Microbiol. 2011;63:198–205. doi: 10.1007/s00284-011-9967-4. [DOI] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189:737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek JC, Wüthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- Noir S, Colby T, Harzen A, et al. A proteomic analysis of powdery mildew (Blumeria graminis f. sp. hordei) conidiospores. Mol Plant Pathol. 2009;10:223–236. doi: 10.1111/j.1364-3703.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novodvorska M, Hayer K, Pullan ST, et al. Transcriptional landscape of Aspergillus niger at breaking of conidial dormancy revealed by RNA-sequencing. BMC Genomics. 2013;14:246. doi: 10.1186/1471-2164-14-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novodvorska M, Stratford M, Blythe MJ, et al. Metabolic activity in dormant conidia of Aspergillus niger and developmental changes during conidial outgrowth. Fungal Genet Biol. 2016;94:23–31. doi: 10.1016/j.fgb.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YT, Ahn C-S, Kim JG, et al. Proteomic analysis of early phase of conidia germination in Aspergillus nidulans. Fungal Genet Biol. 2010;47:246–253. doi: 10.1016/j.fgb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Pammer M, Briza P, Ellinger A, et al. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast Chichester Engl. 1992;8:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Nadales E, Almeida Nogueira MF, Baldin C, et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet Biol. 2014;70:42–67. doi: 10.1016/j.fgb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S, Kapros T, Honigberg SM. Phenotypic plasticity within yeast colonies: differential partitioning of cell fates. Curr Genet. 2016;62:467–473. doi: 10.1007/s00294-015-0558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila PJ. Coprinopsis cinerea. Curr Biol. 2011;21:R616–R617. doi: 10.1016/j.cub.2011.05.042. [DOI] [PubMed] [Google Scholar]
- Quecine MC, Leite TF, Bini AP, et al. Label-free quantitative proteomic analysis of Puccinia psidii uredospores reveals differences of fungal populations infecting eucalyptus and guava. PLOS ONE. 2016;11:e0145343. doi: 10.1371/journal.pone.0145343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P, Halvorson HO, Bulla LA, Julian GS. Germination and outgrowth of single spores of Saccharomyces cerevisiae viewed by scanning electron and phase-contrast microscopy. J Bacteriol. 1972;109:1232–1238. doi: 10.1128/jb.109.3.1232-1238.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. Summer meeting 2013 – when the sleepers wake: the germination of spores of Bacillus species. J Appl Microbiol. 2013;115:1251–1268. doi: 10.1111/jam.12343. [DOI] [PubMed] [Google Scholar]
- Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Setlow P. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits GJ, van den Ende H, Klis FM. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiol Read Engl. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- Stewart K-AV, Setlow P. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J Bacteriol. 2013;195:3575–3582. doi: 10.1128/JB.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Khakhina S, Mallory MJ. Ume6p is required for germination and early colony development of yeast ascospores. FEMS Yeast Res. 2011;11:104–113. doi: 10.1111/j.1567-1364.2010.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-J, Fedorova ND, Cagas SE, et al. Development stage-specific proteomic profiling uncovers small, lineage specific proteins most abundant in the Aspergillus Fumigatus conidial proteome. Proteome Sci. 2012;10:30. doi: 10.1186/1477-5956-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutschbein J, Albrecht D, Pötsch M, et al. Proteome profiling and functional classification of intracellular proteins from conidia of the human-pathogenic mold Aspergillus fumigatus. J Proteome Res. 2010;9:3427–3442. doi: 10.1021/pr9010684. [DOI] [PubMed] [Google Scholar]
- van Leeuwen MR, Krijgsheld P, Bleichrodt R, et al. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud Mycol. 2013;74:59–70. doi: 10.3114/sim0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmeister E, Schipper K, Baumann S, et al. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol Rev. 2012;36:59–77. doi: 10.1111/j.1574-6976.2011.00296.x. [DOI] [PubMed] [Google Scholar]
- Wyatt TT, Wösten HAB, Dijksterhuis J. Fungal spores for dispersion in space and time. Adv Appl Microbiol. 2013;85:43–91. doi: 10.1016/B978-0-12-407672-3.00002-2. [DOI] [PubMed] [Google Scholar]
- Zhang R, Ma ZH, Wu BM. Multiple displacement amplification of whole genomic DNA from urediospores of Puccinia striiformis f. sp. tritici. Curr Genet. 2015;61:221–230. doi: 10.1007/s00294-014-0470-x. [DOI] [PubMed] [Google Scholar]