Abstract
Posterior fossa ependymomas (EPN_PF) in children comprise two morphologically identical, but biologically distinct tumor entities. Group-A (EPN_PFA) tumors have a poor prognosis and require intensive therapy. In contrast, group-B tumors (EPN_PFB) exhibit excellent prognosis and the current consensus opinion recommends future clinical trials to test the possibility of treatment de-escalation in these patients. Therefore distinguishing these two tumor subtypes is critical. EPN_PFA and EPN_PFB can be distinguished based on DNA methylation signatures, but these assays are not routinely available. We have previously shown that a subset of poorly prognostic childhood EPN_PF exhibits global reduction in H3K27me3. Therefore, we set out to determine whether a simple immunohistochemical assay for H3K27me3 could be used to segregate EPN_PFA from EPN_PFB tumors. We assembled a cohort of 230 childhood ependymomas and H3K27me3 immunohistochemistry was assessed as positive or negative in a blinded manner. H3K27me3 staining results were compared with DNA methylation-based subgroup information available in 112 samples [EPN_PFA (n=72) and EPN_PFB tumors (n=40)]. H3K27me3 staining was globally reduced in EPN_PFA tumors and immunohistochemistry showed 99% sensitivity and 100% specificity in segregating EPN_PFA from EPN_PFB tumors. Moreover, H3K27me3 immunostaining was sufficient to delineate patients with worse prognosis in two independent, non-overlapping cohorts (n=133 and n=97). In conclusion, immunohistochemical evaluation of H3K27me3 global reduction is an economic, easily available and readily adaptable method for defining high-risk EPN_PFA from low-risk posterior fossa EPN_PFB tumors to inform prognosis and to enable the design of future clinical trials.
Keywords: Childhood ependymoma, epigenetics, H3K27me3, molecular subgrouping
Introduction
Ependymomas (EPN) are the third most common childhood brain tumor and can occur throughout the neuraxis, but most frequently arise in the posterior fossa [17]. Recent genomic, DNA methylation and gene expression studies have identified molecular subgroups within EPN occurring at different locations along the neuraxis [18, 24, 25]. Posterior fossa ependymomas (EPN_PF), previously thought of as a single entity, have been classified into EPN_PFA and EPN_PFB subgroups based on gene expression and DNA CpG island methylation profiles that correspond to crucial differences in clinical outcomes [18, 24, 27, 34]. EPN_PFA tumors frequently occur in younger children (<5 years) and carry a dismal prognosis while EPN_PFB are more common in older children (5–18 years) and adults, and carry a favorable prognosis [18, 24, 27, 34]. More recently, molecular subgrouping along with the extent of tumor resection (a previously established prognostic indicator) has been suggested to be critical in designing treatment approaches in these children. A recent study from 820 patients demonstrates that children with EPN_PFA have the best outcomes when maximal safe surgical resection is combined with localized radiotherapy, while EPN_PFB patients exhibited excellent prognosis with surgical resection[27]. Therefore, the 2017 consensus for the clinical management of EPN suggests maximal resection and radiation for EPN_PFA in contrast to the potential for de-escalation of therapy for EPN_PFB [23]. While EPN_PFA and EPN_PFB tumors most commonly occur at opposite ends of the age spectrum, children between 6–16 have similar frequencies of both tumor subgroups [27]. Therefore it is critical to stratify patients in this age group.
Delineation of EPN_PFA from EPN_PFB currently relies on methylation arrays to examine CpG island DNA methylation patterns [18, 24, 27]. However, this approach is not readily adaptable to routine clinical testing, as it requires specialized equipment, and may be difficult to validate. The absence of a high-quality, cost-effective test that can be routinely deployed in most clinical settings is a significant obstacle to widespread implementation of molecular stratification of these tumors, impacting not only clinical care resulting in overtreatment of children, but also the ability to design future clinical trials. Our goal was to develop an easy to implement immunohistochemical molecular surrogate to enable pediatric EPN_PF subgrouping. We have recently reported that a subset of childhood EPN_PF exhibit global reduction in the repressive mark histone H3 lysine 27 trimethylation (H3K27me3) that is driven by aberrant DNA methylation [1]. An immunohistochemical stain for H3K27me3 segregated posterior fossa EPN in a binary fashion, wherein a subset of tumors exhibited global loss of H3K27me3 staining similar to that observed in H3K27M mutant gliomas [1, 3, 14, 31]. Moreover, H3K27me3 negative EPN showed overlap in clinical features with EPN_PFA tumors in a small number of cases (n=14) [1]. This led us to hypothesize that global reduction in H3K27me3 is specific to EPN_PFA and can serve as an immunohistochemical surrogate to subgroup childhood posterior fossa EPN.
Materials and Methods
Study design and patients
This retrospective international study involved patient tumor sample assessment (n=230) from 22 different institutions. The only inclusion criterion was diagnosis of EPN_PF in children below 18 years of age (18 years cutoff defined by Central Brain Tumor Registry of the United States [22]) and multiple centralized neuropathological review (S.V., J.C., C. D., S. Y., M. P., Mar. S., Mat. S., Cyn. H. and A. K.) to confirm diagnosis. All studies were performed after institutional review board approval from all institutions. From these 230 tumor samples, molecular subgroup information (determined using Illumina 450K DNA methylation beadchip arrays) was available for 112 EPN (EPN_PFA=72 and EPN_PFB=40) [18, 24, 27], 14 of these cases have been previously published [1]. Tumor samples were obtained as unstained tissue sections derived from formalin fixed paraffin embedded blocks and were assembled into two cohorts. Cohort 1 (n=133): EPN samples from Children’s Hospital Los Angeles (CHLA) (n= 38), Children’s Hospital of Philadelphia (CHOP) (n= 15), The Johns Hopkins Hospital (n= 4), University of California San Francisco (UCSF) (n= 17), The Hospital for Sick Children (n= 30), German Cancer Research Center (DKFZ) (n= 25) and NYU Langone Medical Center (n= 4). Samples were in the form of full sections (n=97) or were contained in tissue microarrays (n= 36). The H3K27me3 staining and clinical data from 59 cases in cohort 1 have been previously published [1]. Cohort 2 (n=97, contained in tissue microarrays): EPN samples obtained from the University of British Columbia from the trans Canadian ependymoma study that did not overlap with the cases obtained from The Hospital for Sick Children [15]. Demographic and clinical details of patients are summarized in table 1. Patient identifiers were removed from all cases. All studies were conducted in compliance with REMARK and STARD guidelines [6, 19].
Table 1.
Demographics of childhood EPN_PF patients in cohorts 1, 2 and the combined cohort
| Characteristic | Factor | Cohort 1 (n=133) |
Cohort 2 (n=97) |
Combined |
|---|---|---|---|---|
| Age | Median Age (Interquartile Range) | 5.4 (2.14 – 10.29) | 3.1 (1.62 – 5.95) | 3.7 (1.90 – 8.89) |
| Gender | Male n (%) | 75 (56.4) | 62 (63.3) | 137 (59.3) |
| Resection | STR n (%) | 27 (20.3) | 16 (16.3) | 43 (18.6) |
| H3K27me3 status | H3K27 negative n (%) | 93 (69.9) | 81 (82.7) | 174 (75.3) |
| Grade | Grade III n (%) | 69 (51.9) | 25 (25.5) | 94 (40.7) |
| Survival | Died from disease n (%) | 48 (36.1) | 39 (39.8) | 87 (37.7) |
| Median OS (Years) | 13.8 | 16.7 | 16.7 | |
| Progression free survival | Progression n (%) | 32 (24.1) | 46 (46.9) | 78 (33.8) |
| Median PFS (Years) | NA | 3.4 | NA |
(Data not available in Cohort 1: Extent of resection n=42, grade n=5, Overall survival n=6, Progression free survival n=42. Data not available in Cohort 2: Extent of resection n=1). OS – overall survival, PFS – progression free survival; NA – not available; not defined by software because more than half of the patients did not progress throughout the study.
Immunohistochemistry and scoring
Immunohistochemistry for H3K27me3 was performed using standard immunostaining protocols as previously described [1, 31]. Inter laboratory variability was assessed in two independent institutions using two different antibodies using two independent staining platforms (Ventana and Leica Bond systems).
Institution 1
“Immunostaining was performed using the Discovery XT processor (Ventana Medical Systems). Tissue sections were blocked for 30 min in 10% normal goat serum in 2% BSA in PBS. Sections were incubated for 5 h with rabbit polyclonal anti-H3K27me3 antibody (07–449, Millipore, 0.1 µg/mL). Tissue sections were then incubated for 60 min with biotinylated goat anti-rabbit IgG (PK6101, Vector Labs) at 1:200 dilution. Blocker D, Streptavidin-HRP, and DAB detection kit (Ventana Medical Systems) were used according to the manufacturers’ instructions”.
Institution 2
Immunohistochemistry for H3K27me3 was performed using the Leica Bond RXm™ automated staining processor (Leica Biosystems, Buffalo Grove, IL). Tissue sections were cut at 5µm and dried at 70°C for 30 min. They were then dewaxed and antigen retrieval was in the Bond Rx system with Epitope Retrieval Solution 2 (pH9) for 40 min. Sections were incubated for 30 min with rabbit monoclonal anti-H3K27me3 antibody (C36B11, Cell Signaling, Danvers, MA) at a concentration of 1:150 using the standard Leica Bond protocol IHC-F. The Leica Bond Polymer Refine DAB detection kit was used according to the manufacturer’s instructions.
Positive, negative and internal controls and scoring
H3K27M mutant gliomas that exhibit global reduction in H3K27me3 and H3 Wild-type gliomas with preserved H3K27me3 staining in tumor cells were used as positive and negative controls respectively (Online Resource 1, Fig S1a–b). Non-tumor cells in the microenvironment including endothelial cells and immune cells served as internal positive controls. Five individuals scored H3K27me3 staining independently in a blinded manner based on scoring systems for H3K27M tumors [2, 31]. Sections were scored positive when more than 80% cells were positive for H3K27me3 and scored negative when they did not. Inter-reviewer variability (disagreement with consensus opinion) ranged from 0 to 3.1% with a median of 1.18%. Staining results using two independent antibodies performed in two independent laboratories were identical. To assess variability between tissue microarrays and full sections, we assessed staining results in 70 cases contained in tissue microarrays with their corresponding full sections selected from the same blocks that were used to generate the TMA. This comparison showed that staining results were identical in tissue microarrays and full sections.
Automated, blinded quantification
were performed as we have previously described [31]. “Each slide was scanned at 40× magnification using an Aperio Scanscope Scanner (Aperio Vista, CA) and viewed through the Aperio ImageScope software program. An individual blinded to the experimental design captured JPEG images from each core (circular area of 315 cm2 corresponding to the entire core) at 10× magnification on the Aperio ImageScope viewing program. Quantification of immunostaining on each JPEG was conducted using an automated analysis program with Matlab’s image processing toolbox algorithm that used color segmentation with RGB color differentiation, K-means clustering and background-foreground separation with Otsu’s thresholding. To arrive at a score the number of extracted pixels were multiplied by their average intensity for each core. The final score for a given case and marker were calculated by averaging the score of two cores vis-à-vis that marker. Quantification of the captured images was conducted using an automated analysis program with Matlab’s image processing toolbox as described previously”[31].
Statistics
Statistical analyses were performed in consultation with the University of Michigan’s department of Bioinformatics and the Division of Hematology/Oncology and Hospital for Sick Children, Toronto. Because our study was a retrospective in nature, no a priori power analysis was performed. Sensitivity, specificity, positive predictive value, negative predictive value, false positive rate, false negative rate and false discovery rate were calculated using standard definitions.
Overall survival (OS) and progression-free survival (PFS) were defined as the time from histologic diagnosis to death (OS) or the minimum of first progression or death (PFS). Kaplan-Meier curves were generated to estimate the OS and PFS; between group differences in OS or PFS were tested using log-rank tests. Univariate and multivariate Cox proportional hazard regression models were used to assess the relation between covariates and OS or PFS outcomes. All statistical analyses were performed in the R statistical environment (v3.3.2), using R packages survival (v2.40-1), rms (4.3-1) and ggplot2 (v2.2.0). Unpaired, two-tailed, Mann Whitney test with 95% confidence intervals were used to compare mean age differences and H3K27me3 quantitative values. Data are represented as the means ± standard deviation (s.d.) or median with interquartile range with 95% CI.
We evaluated the discriminatory capacity of biomarkers using Harrell’s C index and time dependent AUC evaluated at year five. To account for possible bias from using the same dataset to build and assess models, these statistics were calculated using 5-fold cross validation repeated 20 times, for 2 models: 1) H3K27me3 with covariates and 2) EPN_PFA/ EPN_PFB subgroup with covariates. Time dependent AUC was calculated following Heagerty and Zheng, 2005 [10].
Results
H3K27me3 immunohistochemistry distinguishes EPN_PFA from EPN_PFB tumors
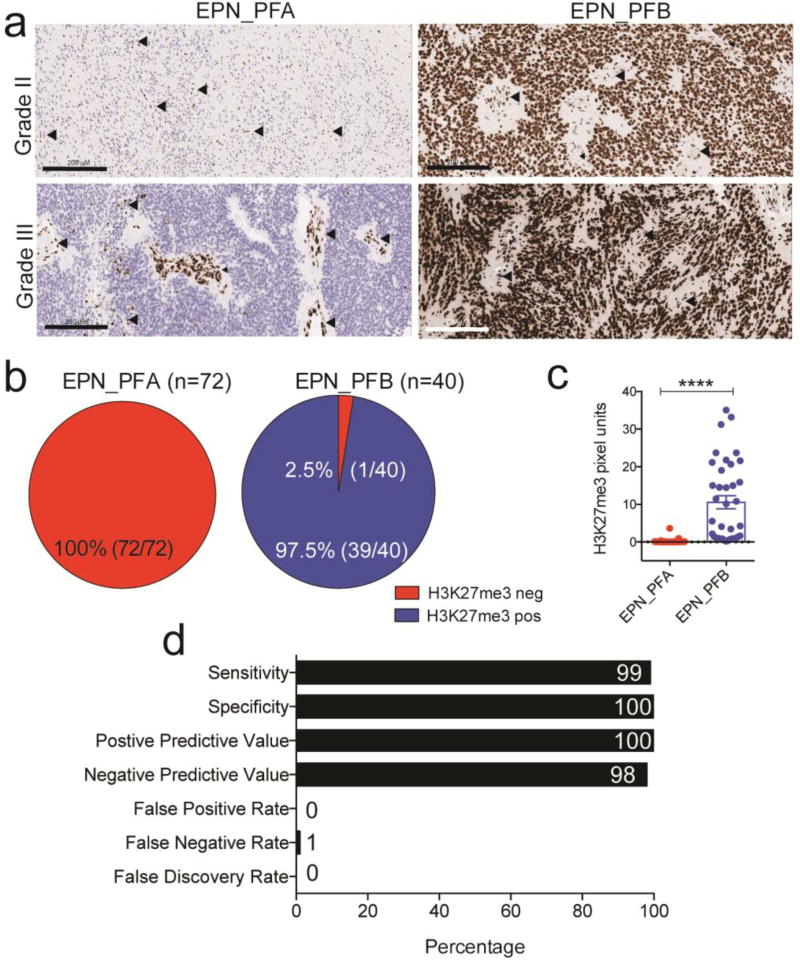
We first determined the relationship between H3K27me3 immunostaining and EPN molecular subgroups in a cohort (n=112) of previously classified childhood posterior fossa EPNs [18, 24, 27] using H3K27M mutant and H3 wild type gliomas as controls [2, 3, 14, 31]. H3K27me3 immunostaining was assessed as negative or positive by five blinded observers. These studies showed that all 72 (100%) EPN_PFA were negative for H3K27me3 staining, while 39 of 40 (97.5%) EPN_PFB were positive and 1 of 40 (2.5%) of EPN_PFB was negative for H3K27me3 (Online resource 1-Fig. S1 and Fig 1a–b). As an objective measure, an individual blinded to the study design captured and quantified images from each case using MATLAB algorithms we have previously standardized [31]. Global reduction in H3K27me3 in EPN_PFA compared to EPN_PFB was confirmed by the quantification data (Fig. 1c). One outlier was identified. It was a tumor from a 12 year-old female in cohort 1, which was H3K27me3 negative (Online resource 1-Fig S1e). This tumor was confirmed to be an EPN_PFB based on DNA methylation analyses. Overall, H3K27me3 staining showed 99% sensitivity and 100% specificity in determining the predefined molecular subgroup. (Fig 1d).
Figure. 1. H3K27me3 immunohistochemistry segregates EPN_PFA from EPN_PF B tumor.
a. Representative images of H3K27me3 immunostaining in grade II and grade III EPN_PFA and EPN_PFB. Arrowheads (black) indicate cells within the microenvironment of tumor including endothelial cells that stained positive for H3K27me3 and were used as internal positive control. Scale bars represents 200 µM
b. Percent distribution of H3K27me3 negative (Red) and H3K27me3 positive (Blue) cases in predefined EPN_PFA and EPN_PFB tumors. Numbers in parentheses indicate the number of tumors studied.
c. Quantification of H3K27me3 immunostaining in EPN_PFA and EPN_PFB. Each point represents a single tumor. Error bars represent mean +/− S.D.
d. Percent sensitivity, specificity, positive predictive value, negative predictive value, false positive rate, false negative rate and false discovery rate for detection of childhood posterior fossa EPN_PFA vs. EPN_PFB molecular subgroups using H3K27me3 immunostaining.
Global reduction in H3K27me3 immunostaining relates with poor prognosis
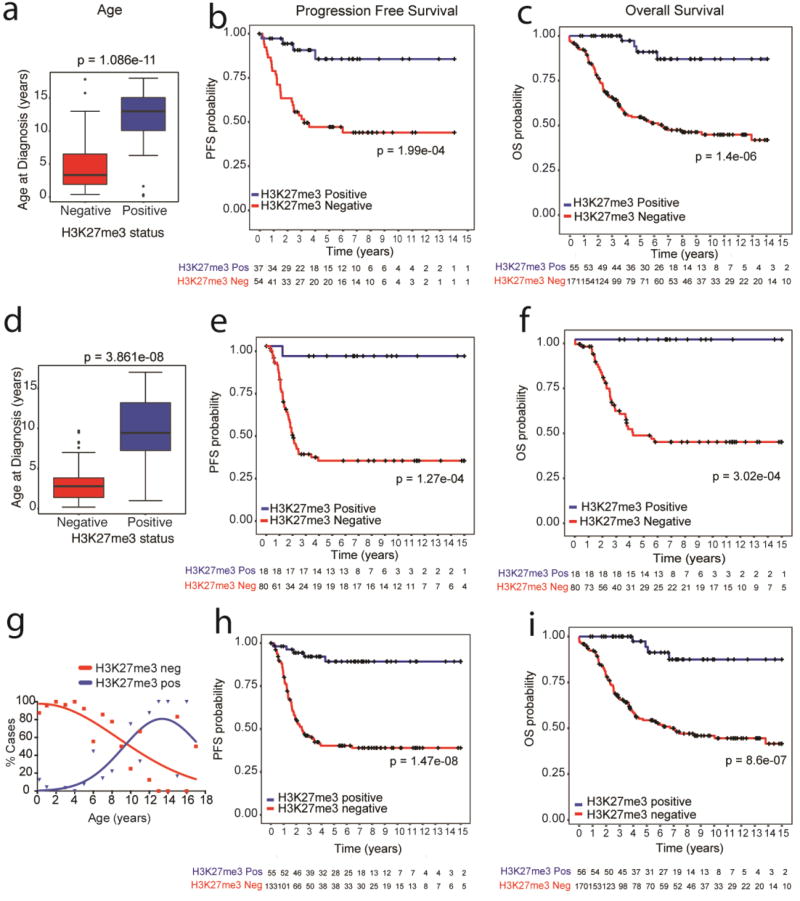
We next sought to determine if H3K27me3 immunostaining was sufficient to prognosticate EPN_PF when the molecular subgroup was not known. To address this we divided the 230 samples into two independent, retrospective, non-overlapping cohorts (cohort 1, n=133 and cohort 2, n=97, Table 1 and Online resource 1-Fig S2) Sections were stained for H3K27me3, and scored as positive or negative according to our validated method. All analyses were performed in a blinded manner (including blinding of the molecular subgroup information available in 112 of these cases). Rare EPN_PF exhibit H3K27M mutations [7]. Screening for H3K27M using a mutant specific antibody [2, 32] did not reveal any H3K27M mutant tumors in both cohorts. Univariate and multivariate analyses with other reported variables including age, sex, tumor grade, and the extent of resection were performed [27]. H3K27me3-negative tumors mainly occurred in young children below 5 years of age (median age= 3.35 years, p< 0.0001 in cohort 1 and median age= 2.8 years, p < 0.0001 in cohort 2) as reported with EPN_PFA tumors (Fig 2a, d) [24, 27, 34]. In contrast, H3K27me3-positive tumors occurred predominantly in older children and adolescents above 5 years (median age=13 years in cohort 1 and median age=9.7 years in cohort 2) as described in EPN_PFB tumors [24, 27, 34] (Fig 2a, d). The comparative age distribution of H3K27me3 negative and positive tumors demonstrated opposite trends as observed with EPN_PFA and EPN_PFB tumors [24, 27, 34] (Fig 2g). Moreover, H3K27me3 negative tumors exhibited significantly worse progression free survival as compared to H3K27me3 positive tumors in either cohort (p < 0.0001 in cohort 1 and cohort 2), and overall survival (p < 0.0001 in cohort 1 cohort 2) or in the combined cohort (PFS and OS p< 0.0001) (Fig 2, b, c, e, f, h and i). Similar survival differences have been as observed between EPN_PFA and EPN_PFB tumors [24, 27, 34].
Figure. 2. Global loss of H3K27me3 immunostaining relates with younger age and poor prognosis in two independent cohorts.
a. Box and whisker plot of age distribution of H3K27me3 negative and H3K27me3 positive tumors in cohort 1.
b–c. Progression free survival (PFS, B) or overall survival (OS, C) of EPN stratified by H3K27me3 staining in cohort 1.
d. Box and whisker plot of age distribution of H3K27me3 negative and H3K27me3 positive tumors in cohort 2.
e–f. Progression free survival (PFS, E) or overall survival (OS, F) of EPN stratified by H3K27me3 staining in cohort 2. The numbers below the X-axis indicates the number of persons at risk at each time point.
g. Percentage of H3K27me3 negative and H3K27me3 positive tumors (Y-axis) plotted against corresponding patient age (X-axis).
h–i. Progression free survival (PFS, H) or overall survival (OS, I) of EPN stratified by H3K27me3 staining in combined cohorts 1 and 2.
For a and d, data are plotted as median with interquartile range with 95% CI and each point represents a single tumor. Data was analyzed using unpaired, two-tailed Mann Whitney test. For survival curves, the numbers below the X-axis indicates the number of persons at risk at each time point. Data was analyzed using the Log-Rank test.
Multivariate Cox proportional hazards regression analyses were performed in the combined cohort (Table 2) because the H3K27me3 positive arm showed very few deaths or disease progression in each individual cohort (Fig 2), precluding multivariate analyses. H3K27me3 immunostaining status was a significant predictor of overall survival and progression free survival (OS, p< 0.05, PFS, p< 0.01) (Table 2). Extent of resection has previously been shown to be a strong predictor of survival in posterior fossa EPN and our results confirmed this finding [8, 26, 27, 29, 33]. Furthermore, we compared the discriminatory capacity of H3K27me3 negative / positive with that of EPN_PFA / EPN_PFB status as biomarkers using Harrell’s C index and time dependent AUC evaluated for 5 year survival. Both the C-index and AUC showed similar values for H3K27me3 negative / positive and EPN_PFA/ EPN_PFB subgroups for overall survival and progression free survival in both univariate and multivariate analyses with differences of less than .01 for the multivariate models (Table 3).
Table 2.
Multivariate Cox proportional hazards analysis in the combined cohort
| Overall Survival (n=184) | |||
| HR | 95% CI | p value | |
| Age | 0.911 | (0.819, 1.014) | 0.089 |
| Cohort (2 vs 1) | 1.428 | (0.839, 2.431) | 0.189 |
| Extent of resection | 2.196 | (1.268, 3.803) | 0.005 |
| Gender | 1.246 | (0.750, 2.069) | 0.396 |
| Grade | 1.389 | (0.824, 2.341) | 0.218 |
| H3K27me3 negative | 6.797 | (1.423, 32.475) | 0.016 |
| Progression Free Survival (n=169) | |||
| HR | 95% CI | p value | |
| Age | 0.879 | (0.784, 0.985) | 0.026 |
| Cohort (2 vs 1) | 1.579 | (0.926, 2.690) | 0.093 |
| Extent of resection | 2.706 | (1.568, 4.670) | <0.001 |
| Gender | 1.332 | (0.806, 2.183) | 0.266 |
| Grade | 1.891 | (1.140, 3.138) | 0.014 |
| H3K27me3 negative | 17.043 | (2.150, 135.100) | 0.007 |
| Overall Survival (n=224) without extent of resection | |||
| HR | 95% CI | p value | |
| Age | 0.969 | (0.902, 1.041) | 0.388 |
| Cohort (2 vs 1) | 1.050 | (0.672, 1.640) | 0.831 |
| Gender | 1.070 | (0.695, 1.647) | 0.760 |
| Grade | 1.561 | (0.995, 2.449) | 0.052 |
| H3K27me3 negative | 6.231 | (2.056, 18.879) | 0.001 |
| Progression Free Survival (n=188) without extent of resection | |||
| HR | 95% CI | p value | |
| Age | 0.917 | (0.839, 1.002) | 0.057 |
| Cohort (2 vs 1) | 1.451 | (0.893, 2.359) | 0.133 |
| Gender | 1.071 | (0.678, 1.690) | 0.769 |
| Grade | 1.809 | (1.119, 2.924) | 0.016 |
| H3K27me3 negative | 4.125 | (1.437, 11.841) | 0.008 |
Table 3.
C-index and AUC analysis
| Overall Survival | Progression Free Survival |
|||
|---|---|---|---|---|
| AUC | C index | AUC | C index | |
| Multivariate | ||||
| H3K27me3 status with covariates | 0.691 | 0.704 | 0.757 | 0.755 |
| EPN subgroup with covariates | 0.713 | 0.764 | 0.748 | 0.779 |
| Covariates only | 0.671 | 0.678 | 0.726 | 0.727 |
| Univariate | ||||
| H3K27me3 status | 0.623 | 0.627 | 0.667 | 0.650 |
| EPN subgroup | 0.666 | 0.678 | 0.726 | 0.714 |
Discussion
Our understanding of the molecular drivers of pediatric brain tumors has dramatically advanced over the last ten years. Recent studies have revealed molecular heterogeneity within brain tumors that inform clinical outcome, and have the potential to drive fundamentally different and personalized approaches to patient care [16, 17]. In posterior fossa EPN both molecular subgrouping and the extent of resection are critical predictors of patient outcome [23, 24, 27]. Moreover, while EPN_PFA are more common in young children, both EPN_PFA and EPN_PFBs occurred at comparable frequencies in children between 5 and 16 years of age [27]. A recent study demonstrates that majority of EPN_PFB did not recur after gross total resection. This has led to the suggestion for future clinical trials to test if de-escalation of therapy including avoiding harmful radiation therapy may be beneficial for these patients [23, 27]. In contrast, EPN_PFA tumors behave more aggressively and the current consensus recommendation is that these patients be treated by maximal safe tumor resection followed by localized adjuvant radiation therapy [23, 27]. Because posterior fossa EPN subgrouping has such a strong prognostic impact in children and adolescents, it has been recommended as part of the routine workup for the care of ependymoma patients [23]. Another important prognostic indicator in ependymomas is the gain isochromosome 1q seen in 20–30% of cases [5, 8, 9, 12, 13, 21]. In the current study information on 1q status was not available in the majority of cases. In future studies we will determine the relationship between H3K27me3 and 1q status.
EPN_PFA, but not EPN_PFB tumors show higher methylation of CpG islands with many of the corresponding genes converging on polycomb repressive complex 2 (PRC2) and H3K27me3 regulated pathways [18]. H3K27me3 is enriched at unmethylated CpG islands [20, 28, 35] and we have previously demonstrated that CpG island methylation inversely drives H3K27me3 levels so that EPN_PFA with high CpG island methylation exhibit global reduction in H3K27me3 levels compared to EPN_PFB [1]. We have extended this observation to demonstrate that H3K27me3 immunostaining stratifies childhood EPN_PF in a binary fashion into H3K27me3 negative tumors and H3K27me3 positive tumors corresponding to EPN_PFA and EPN_PFB, respectively.
The current standard for the identification of EPN_PF molecular subgroups relies on DNA methylation profiling using methylation arrays [18, 23, 24, 27]. DNA methylation arrays are not easily adoptable for widespread clinical testing, as this requires equipment, reagents and personnel beyond the resources of most clinical laboratories. Further, regulatory standards may present variably challenging hurdles for the development and validation of clinical tests based on DNA methylation profiling. In contrast, immunohistochemistry is routinely used in clinical practice worldwide and does not require additional equipment, infrastructure or personnel. As such it is a highly cost effective and widely reproducible platform for clinical testing that can be readily adapted for detection of molecular surrogates. For example, immunohistochemistry for a mutant specific IDH1 R132H is part of the routine workup for infiltrating gliomas in adults [4], as is SMARCB1 and H3K27M immunostains as molecular surrogates to diagnose rhabdoid tumors [11] and pediatric midline gliomas [2, 17, 32] in children. Moreover, H3K27me3 immunohistochemistry is already currently used in many neuropathology laboratories as a molecular surrogate to identify H3K27M mutant gliomas [2, 30, 31] and PRC2 mutant malignant peripheral nerve sheath tumors and can be easily adapted for use in childhood EPN_PFs [2, 30, 31]. Therefore, immunohistochemical staining for H3K27me3 is cost effective, highly sensitive and specific molecular surrogate to establish the molecular subgroup childhood posterior fossa ependymomas and can be used to inform prognosis, design and implement molecular-guided therapies to reduce adverse effects of radiation therapy in children and to facilitate the development of much-needed clinical trials.
Supplementary Material
Acknowledgments
We thank Dr. Paul Mischel for insightful comments. This work was supported by grants from NCI K08 CA181475 (S.V.), Mathew Larson Foundation (SV), Sidney Kimmel Foundation (SV), Doris Duke Foundation (SV); Making Headway Foundation (M.S. and M.A.K.), the Sohn Conference Foundation (M.S. and M.A.K.), the Friedberg Charitable Foundation (M.S. and M.A.K.) and Canadian Children’s Cancer & Blood Disorders- C17 grant (JH). Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA046592 by the use of the following Cancer Center Shared Resource(s): Biostatistics. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of interests: The authors declare no competing interests
Author contributions: PP, JC, VR, YS, MJS and SR analyzed data. YS and MJS are biostatisticians. PP, AJR and SV wrote the manuscript. DH and FY performed H3K27me3 staining. JC, DH, CD, SY, MrS, FJR, MaS, CH, CMH, AK, AJR and SV are neuropathologists and evaluated tumor samples. PP, JC, AB, CC and JMB captured samples in a blinded manner. Cases and clinical data were provided by CD, SY, JH, AM, MP, SJC, MrS, FJR, MS, MAK, AMS, CMH, ASC, BW, DJ, LLC, SZ, DE, MaS, KS, NJ, PDM, MK, SMP, MDT, CH, AK and AJR. VR, MDT, LC, MK, SMP and AK performed DNA methylation analyses to subgroup tumors. All authors read and edited the manuscript.
References
- 1.Bayliss J, Mukherjee P, Lu C, Jain SU, Chung C, Martinez D, Sabari B, Margol AS, Panwalkar P, Parolia A, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8:366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechet D, Gielen GG, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, et al. Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol. 2014;128:733–741. doi: 10.1007/s00401-014-1337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Capper D, Weissert S, Balss J, Habel A, Meyer J, Jager D, Ackermann U, Tessmer C, Korshunov A, Zentgraf H, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter M, Nicholson J, Ross F, Crolla J, Allibone R, Balaji V, Perry R, Walker D, Gilbertson R, Ellison DW. Genetic abnormalities detected in ependymomas by comparative genomic hybridisation. Br J Cancer. 2002;86:929–939. doi: 10.1038/sj.bjc.6600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HC, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6:e012799. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gessi M, Capper D, Sahm F, Huang K, von Deimling A, Tippelt S, Fleischhack G, Scherbaum D, Alfer J, Juhnke BO, et al. Evidence of H3 K27M mutations in posterior fossa ependymomas. Acta Neuropathol. 2016;132:635–637. doi: 10.1007/s00401-016-1608-3. [DOI] [PubMed] [Google Scholar]
- 8.Godfraind C, Kaczmarska JM, Kocak M, Dalton J, Wright KD, Sanford RA, Boop FA, Gajjar A, Merchant TE, Ellison DW. Distinct disease-risk groups in pediatric supratentorial and posterior fossa ependymomas. Acta Neuropathol. 2012;124:247–257. doi: 10.1007/s00401-012-0981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granzow M, Popp S, Weber S, Schoell B, Holtgreve-Grez H, Senf L, Hager D, Boschert J, Scheurlen W, Jauch A. Isochromosome 1q as an early genetic event in a child with intracranial ependymoma characterized by molecular cytogenetics. Cancer Genet Cytogenet. 2001;130:79–83. doi: 10.1016/s0165-4608(01)00465-4. [DOI] [PubMed] [Google Scholar]
- 10.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 11.Judkins AR, Eberhart CG, Wesseling P, Hasselblatt M. Atypical teratoid/rhabdoid tumor. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, von Deimling A, editors. WHO Classification of Tumours of the Central Nervous System Revised 4rth Edition edn. World Health Organization; City: 2016. pp. 209–212. [Google Scholar]
- 12.Kilday JP, Mitra B, Domerg C, Ward J, Andreiuolo F, Osteso-Ibanez T, Mauguen A, Varlet P, Le Deley MC, Lowe J, et al. Copy number gain of 1q25 predicts poor progression-free survival for pediatric intracranial ependymomas and enables patient risk stratification: a prospective European clinical trial cohort analysis on behalf of the Children's Cancer Leukaemia Group (CCLG), Societe Francaise d'Oncologie Pediatrique (SFOP), and International Society for Pediatric Oncology (SIOP) Clin Cancer Res. 2012;18:2001–2011. doi: 10.1158/1078-0432.CCR-11-2489. [DOI] [PubMed] [Google Scholar]
- 13.Korshunov A, Witt H, Hielscher T, Benner A, Remke M, Ryzhova M, Milde T, Bender S, Wittmann A, Schottler A, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010;28:3182–3190. doi: 10.1200/JCO.2009.27.3359. [DOI] [PubMed] [Google Scholar]
- 14.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 Activity by a Gain-of-Function H3 Mutation Found in Pediatric Glioblastoma. Science. 2013;340:857–861. doi: 10.1126/Science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li AM, Dunham C, Tabori U, Carret AS, McNeely PD, Johnston D, Lafay-Cousin L, Wilson B, Eisenstat DD, Jabado N, et al. EZH2 expression is a prognostic factor in childhood intracranial ependymoma: a Canadian Pediatric Brain Tumor Consortium study. Cancer. 2015;121:1499–1507. doi: 10.1002/cncr.29198. [DOI] [PubMed] [Google Scholar]
- 16.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, et al. International Society of Neuropathology-Haarlem Consensus Guidelines, for Nervous System Tumor Classification and Grading. Brain Pathol. 2014 doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 18.Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn S, Stutz AM, Wang X, Gallo M, Garzia L, Zayne K, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM Statistics Subcommittee of the NCIEWGoCD. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Oncol. 2005;2:416–422. [PubMed] [Google Scholar]
- 20.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendrzyk F, Korshunov A, Benner A, Toedt G, Pfister S, Radlwimmer B, Lichter P. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res. 2006;12:2070–2079. doi: 10.1158/1078-0432.CCR-05-2363. [DOI] [PubMed] [Google Scholar]
- 22.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pajtler KW, Mack SC, Ramaswamy V, Smith CA, Witt H, Smith A, Hansford JR, von Hoff K, Wright KD, Hwang E, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017;133:5–12. doi: 10.1007/s00401-016-1643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, Lee R, Tatevossian RG, Phoenix TN, Thiruvenkatam R, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollack IF, Gerszten PC, Martinez AJ, Lo KH, Shultz B, Albright AL, Janosky J, Deutsch M. Intracranial ependymomas of childhood: long-term outcome and prognostic factors. Neurosurgery. 1995;37:655–666. doi: 10.1227/00006123-199510000-00008. discussion 666-657. [DOI] [PubMed] [Google Scholar]
- 27.Ramaswamy V, Hielscher T, Mack SC, Lassaletta A, Lin T, Pajtler KW, Jones DT, Luu B, Cavalli FM, Aldape K, et al. Therapeutic Impact of Cytoreductive Surgery and Irradiation of Posterior Fossa Ependymoma in the Molecular Era: A Retrospective Multicohort Analysis. J Clin Oncol. 2016;34:2468–2477. doi: 10.1200/JCO.2015.65.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson PL, Zeltzer PM, Boyett JM, Rorke LB, Allen JC, Geyer JR, Stanley P, Li H, Albright AL, McGuire-Cullen P, et al. Survival and prognostic factors following radiation therapy and chemotherapy for ependymomas in children: a report of the Children's Cancer Group. J Neurosurg. 1998;88:695–703. doi: 10.3171/jns.1998.88.4.0695. [DOI] [PubMed] [Google Scholar]
- 30.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ, Sullivan LM, Martinez D, Perry A, Lewis PW, Thompson CB, et al. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128:743–753. doi: 10.1007/s00401-014-1338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vera-Bolanos E, Aldape K, Yuan Y, Wu J, Wani K, Necesito-Reyes MJ, Colman H, Dhall G, Lieberman FS, Metellus P, et al. Clinical course and progression-free survival of adult intracranial and spinal ependymoma patients. Neuro Oncol. 2015;17:440–447. doi: 10.1093/neuonc/nou162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witt H, Mack SC, Ryzhova M, Bender S, Sill M, Isserlin R, Benner A, Hielscher T, Milde T, Remke M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.